Abstract
The ‘Nanguo’ pear (Pyrus ussuriensis) fruit is typically climacteric, and ethylene is the main factor controlling the ripening of climacteric fruit. Whether the actin cytoskeleton is involved in ethylene-mediated fruit ripening remains unclear. In this study, we characterized an actin-related protein, PuARP4. The expression of PuARP4 was evaluated in young leaves, stems, flowers, and roots as well as in fruits. Expression of PuARP4 decreased during fruit development and ripening, and it was inhibited by Ethephon treatment but induced by 1-MCP treatment. To explore the network of PuARP4 function in ‘Nanguo’ pear fruit ripening, we screened a cDNA library from ‘Nanguo’ pear fruits using PuARP4 as bait. PuPME1 (pectin methylesterase 1) was identified as a potential interactor of PuARP4; PuPME1 has been found to degrade the pectin of cell walls. This direct interaction was further confirmed by a yeast two-hybrid system and pull-down analyses. Analysis of the expression of PuPME1 showed that it could be regulated by ethylene. Our results indicated that PuARP4 was involved in ethylene-mediated fruit ripening and might cooperate with PuPME1 to regulate the ripening process. Our results provide a new link between fruit ripening and the cytoskeleton and will provide a new platform for research on ethylene-mediated fruit ripening. The possible mechanisms underlying this process are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cytoskeleton is part of the cellular structure and plays important roles in maintaining cell shape, in the transport of various materials, and in cell division. Its role in cell signal transduction has attracted increasing attention.
The actin cytoskeleton is highly unstable, being able to depolymerize or polymerize in response to various environmental factors both in vivo and in vitro. And actin polymerization plays an important role in apical growth in hyphae and pollen tubes (Zepeda and others 2014). These cytoskeletal proteins can respond to signals such as hormones and pathogens to regulate cell physiology. The actin filaments of Commelina communis L. guard cells have been localized to the cell cortex, radiating from the stomatal pore, but with abscisic acid treatment, the actin began to disintegrate within a few minutes and was completely disintegrated in 1 h (Soon and Youngsook 1997). In vetch root hairs, the number of actin filaments increased dramatically within 3–15 min of treatment with the host-specific Nod factor (de Ruijter and others 1999).
Actin-related protein (ARP) is a type of actin that has 20–60% similarity to traditional actin proteins (McKinney and others 2002). There are at least eight ARPs in Arabidopsis, most of which have orthologues in other organisms (Kandasamy and others 2003). ARPs play important roles in flower morphological development and in cell motility. Arabidopsis plants in which AtARP4 was silenced showed early flowering and increased infertility (Kandasamy and others 2005). Although the functions of most ARPs are not clear, several have been identified as components of various chromatin-modifying complexes. The arp2/3 complex in Arabidopsis functions in cell motility and transmembrane transport processes (Goley and Welch 2006) and has also been implicated in the control of root elongation (Dyachok and others 2008). The arp3 of the arp2/3 complex was shown to function in root gravitropism by affecting amyloplast sedimentation in Arabidopsis (Zou and others 2016). Plants lacking arp2 exhibited decreased mitochondria movement and hypersensitivity to salt, indicating that the arp2/3 complex regulates mitochondrial-dependent Ca2+ in response to salt stress (Zhao and others 2013). In Medicago truncatula root nodules, arp3 protein and actin were shown to be spatially associated with maturing symbiosomes (Gavrin and others 2015). Arps are known to participate in numerous processes, but whether they are involved in fruit ripening and softening remains unclear.
Fruit softening occurs predominantly by disassembly of the cell wall and dissolution of the middle lamella, which attenuates cell–cell adhesion (Goulao and Oliveira 2008; Mercado and others 2011). A previous study showed that actin could regulate the accumulation of cell wall components (Meagher and others 1999). Pectins are the major components of the primary cell wall and middle lamella, determining fruit texture and quality. Pectin depolymerization was reported as the main reason for the loss of fruit firmness (Guo and others 2015). Pectin methylesterase (PME) is one type of cell wall hydrolase and the PME catalyzes the hydrolytic de-esterification of pectins, finally leading to tissue softening during fruit ripening (Guo and others 2015). ‘La France’ pear (Pyrus communis L.) possesses four PME genes, PcPME1-4, of which PcPME4 has been suggested to be involved in fruit softening during storage (Sekine and others 2006). Expression of PME in ‘Golden Delicious’ apple (Malus domestica Borkh.) fruits increased rapidly after harvest and was significantly regulated by ethylene (Wei and others 2010). MdPME2 has been suggested to be related to fruit softening (Segonne and others 2014). In addition to playing a role in the fruit ripening process, PME genes affect root development and phloem fibre development. When RcPME1 was inhibited, the normal separation of pea (Pisum sativum cv. Little Marvel) root border cells from the root tip into the external environment was prevented (Wen and others 1999). Eleven and 15 PMEs were found to be expressed in early and late fibres of Linum usitatissimum, respectively (Pinzón-Latorre and Deyholos 2013).
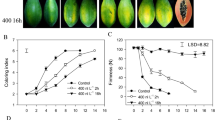
The ‘Nanguo’ pear (Pyrus ussuriensis) fruit is typically climacteric, and ethylene is the main factor controlling the ripening of climacteric fruit. During ripening, the fruits become yellow and produce large amounts of ethylene, accompanied by rapid softening, but the soluble solids contents and the titratable acidities of the fruits show no obvious changes (Supplemental Fig. 1 and Fig. 1). Previously, we compared the transcriptomes of pre- and post-climacteric ‘Nanguo’ pear fruits, and we found that the expression of an ARP4 gene, PuARP4, was down-regulated during fruit ripening. In this study, we analysed the expression pattern of PuARP4 during fruit development and ripening, as well as in Ethephon- and 1-MCP-treated fruits. Moreover, using PuARP4 as bait, we screened a yeast two-hybrid (Y2H) cDNA library constructed with ‘Nanguo’ pear fruit cDNA. A PME gene named PuPME1 was identified as a potential interactor with PuARP4.
Materials and Methods
Plant Materials and Treatments
Young leaves, stems, flowers, and fruits were collected from mature ‘Nanguo’ pear (P. ussuriensis) trees and grafted onto ‘Shanli’ (P. ussuriensis Maxim.) rootstock. The plants were grown at the experimental farm of Shenyang Agricultural University (Shenyang, China). Roots were collected from the ‘Shanli’ pear. Fruits were sampled every 30 days from 30 days after full bloom (DAFB) until commercial harvest. The fruits harvested at 134 DAFB (commercial maturity, 16 September 2014) were stored at room temperature (RT, 24 °C) for 15 days and sampled every 5 days. In addition, two other groups of ‘Nanguo’ fruits collected at 134 DAFB were subjected to 1-MCP (1μL/L) or Ethephon (1000 ppm) treatment according to Tan and others (2013). After treatment, the fruits were held at RT for 15 days and samples were collected every 5 days for RNA extraction. At each sampling point, five fruits were sampled for measurement of ethylene production and flesh firmness, after which the fruits were sliced, pooled, frozen in liquid N2, and stored at −80 °C for later use.
Measurement of Ethylene Production Rates, Flesh Firmness, Soluble Solids and Titratable Acidity
For measurement of ethylene production rates, intact fruits were enclosed in an airtight container (0.86 L 24 °C) equipped with septa, and 1 ml of headspace gas was sampled using a syringe. The ethylene concentration was measured with a gas chromatograph (Agilent 7890 A, USA), equipped with a flame ionization detector according to the methods of Tan and others (2013). Five fruits per sample were measured.
Flesh firmness was measured with a portable pressure tester (FT-327, Facchini, Italy) fitted with an 11-mm-diameter probe. Four skin discs (approximately 2.5 cm in diameter) were removed from opposite sides of each fruit. In a single smooth motion, the probe was pressed into the tissue of the cut surface to a depth of 8–9 mm. Five fruits per sample were measured.
Contents of soluble solids were measured using a refractometer (Atago, PAL-1, Japan). The fruits were ground and filtered, and 0.3 ml of juice was then assayed. Five fruits per sample were measured.
The titratable acidity was measured using acid-base titration. Five fruits per sample were measured.
Quantitative RT-PCR
Total RNA was extracted using a modified CTAB method (Gasic and others 2004). One μg of total RNA was used to synthesize first-strand cDNA using a PrimeScript First-Strand cDNA Synthesis Kit (Takara, Japan).
Quantitative RT-PCR (qRT-PCR) was conducted as described by Tan and others (2013). Specific primers for each gene were designed using Primer 5 and are listed in Supplementary Table S1. The pear actin gene was used as an internal control. Reactions were run in triplicate.
Sequence Analysis
BLAST searches and structural analyses were conducted using the National Center for Biotechnology Information database (NCBI, http://www.ncbi.nlm.nih.gov/) and the Genome Database for Rosaceae (GDR, https://www.rosaceae.org/node/1).
Construction and Screening of the Y2H Library
The Y2H library was constructed using ‘Nanguo’ pear fruits collected in 2012. Total RNA was isolated from four fruit samples (75 DAFB, 130 DAFB, 145 DAFB, and 10 DAH) as described above. The library was constructed with the Make Your Own Mate & Plate Library System (Clontech, CA, USA, Cat. No. 630490). First-strand cDNA was synthesized from 2 µg of mRNA using oligo-dT primers (CDSIII Primer). The cDNA was amplified by long-distance PCR (LD-PCR) with 5′ and 3′ PCR primers for 20 cycles to generate double-stranded cDNA. The double-stranded cDNA was then purified using a CHROMA SPIN TE-400 column. After purification, 3.8 µg of double-stranded cDNA was ligated into SmaI-linearized pGADT7-Rec cloning vector (Clontech) and subsequently transformed into yeast strain Y187 to generate the cDNA library for a Y2H assay using the Yeastmaker Yeast Transformation System 2 (Clontech) according to the manufacturer’s instructions.
The ORF of PuARP4 was cloned into the pGBKT7 vector (Clontech) and confirmed by sequencing with the T7 primer. The resulting recombinant plasmid was then transformed into the yeast strain Y2H Gold using the Yeastmaker Yeast Transformation System 2 (Clontech).
The pGBKT7-PuARP4 was used as bait to screen the Y2H library with the GAL4-based Matchmaker Gold Yeast Two-Hybrid System (Clontech) according to the manufacturer’s instructions. Briefly, 4 ml of overnight cultures of the bait strain (SD/-Trp liquid medium), 1 ml of library aliquot (1.2 × 107 cells), and 45 ml 2× YPDA medium were combined in a 2 l flask and incubated at 30 °C for 20 h with shaking at 50 rpm and then checked for zygote formation. If zygotes appeared, the cells were collected and resuspended in 10 ml of 0.5× YPDA medium. The mated cell cultures were spread on DDO/X/A plates (containing double-dropout SD medium that lacked tryptophan and leucine but was supplemented with X-α-Gal) and kept at 30 °C. Positive colonies that appeared within 5 days were streaked onto QDO/X/A plates (containing double-dropout SD medium that lacked tryptophan, leucine, histidine, ade but was supplemented with X-α-Gal) for high-stringency screening. Colonies that appeared within 5 days on these QDO/X/A plates were predicted to be positive hybrids, and the plasmids were rescued using the Easy Yeast Plasmid Isolation Kit (Clontech). The plasmids were then transformed into the Escherichia coli strain TOP10 and plated on LB plates with 100 mg/ml ampicillin to select the prey plasmids. To identify the prey plasmids, the inserts were sequenced using the T7 primer.
Yeast Two-Hybrid Assays
The CDS of PuPME1 was cloned into a pGADT7 vector (Clontech, CA, USA). This vector then was co-transformed into Y2H Gold with the pGBKT7-PuARP4 vector. After growing on SD/-Leu/-Trp medium at 30 °C for 3–4 days, the clones were grown on SD/-Ade-His-Leu-Trp at 30 °C for 3–4 days and then stained with X-α-gal (Clontech) to visualise their interaction.
Pull-Down and Western Blot Assays
Pull-down and Western blot assays were performed as described by Yuan and others (2014) using His and GST antibodies. The CDS of PuARP4 was cloned into a pGEX-4T-1 vector (CW Biotech) for expression of GST-tagged fusion protein, and the CDS of PuPME1 was cloned into the pEASY-E1 vector (TransGen Biotech) to express a His-tagged fusion protein. All vectors were transformed into E. coil strain BL21 or BL21(DE3) (Transgen). The proteins were purified as described by Yuan and others (2014).
For the pull-down assay, purified His-PuPME1 protein was adsorbed onto Ni–NTA resin, and an equivalent amount of purified GST-PuARP4 protein was added to the column. After incubation at 4 °C for 1 h, the column was washed twice with soluble binding buffer to remove unbound proteins. Bound proteins were then eluted with elution buffer. The eluted proteins were boiled for 5 min, separated by 12% SDS-PAGE, and blotted onto a nitrocellulose membrane. A Western blot was performed using an anti-GST antibody. GST was used as a negative control.
Results
Cloning and Expression Analysis of PuARP4
‘Nanguo’ pear (P. ussuriensis) fruit undergo typical climacteric changes during ripening, with firmness dropping rapidly and large amounts of ethylene being produced (Fig. 1). In our previous study, we obtained a fragment of the ARP4 gene from ‘Nanguo’ pear; this gene showed down-regulation during fruit ripening. We subsequently cloned its full-length cDNA and genomic DNA. Both the cDNA and genomic DNA sequence were found to be identical to that obtained from the NCBI (National Center for Biotechnology Information) database, and the gene was named PuARP4. The cDNA of PuARP4 was 1335 bp in length and encoded 445 amino acids.
The expression profile of PuARP4 was then examined in the roots, stems, leaves, flowers, and fruits of ‘Nanguo’ pear. PuARP4 was expressed not only in fruits but also in all the other tissues that were examined, with the highest expression level found in roots (Fig. 2a). In fruit, the expression of PuARP4 decreased gradually during development and ripening (Fig. 2b). To ascertain whether ethylene influenced the expression of PuARP4, we treated ‘Nanguo’ pear fruits with Ethephon and 1-MCP (an ethylene antagonist). The results showed that expression of PuARP4 was induced by 1-MCP and inhibited by Ethephon (Fig. 2b).
The expression profile of PuARP4. a Expression patterns of PuARP4 in different tissues. Young leaves, stems, flowers, and fruits were collected from a ‘Nanguo’ pear tree, and roots were sampled from the ‘Shanli’ pear. b Expression of PuARP4 during fruit development and ripening. Fruits were collected at 30, 60, 90, and 120 DAFB. H indicates harvest day (0). Fruits collected at harvest day were treated with Ethephon and 1-MCP, stored at room temperature and then sampled every 5 days. Values are mean ± SD of three biological replicates
Screening of a Y2H Library
We constructed a cDNA library from ‘Nanguo’ pear fruit for Y2H screening. To explore the network of PuARP4 in ‘Nanguo’ pear fruit ripening, we used PuARP4 as a bait to screen the cDNA library. Fifty potential interactors were obtained, one of which was PME1 (pectin methylesterase 1). We then cloned the full-length cDNA of this PME1 gene and named it PuPME1. The PuPME1 cDNA was 1854 bp in length and encoded 617 amino acids.
Expression Profiles of PuPME1
We examined the expression profiles of PuPME1 in different tissues. PuPME1 was found to be expressed in all the tissues that were examined, with the highest levels found in fruits (Fig. 3a).
The expression patterns of PuPME1. a Expression patterns of PuPME1 in different tissues. Young leaves, stems, flowers, and fruits were collected from a ‘Nanguo’ pear tree, and roots were sampled from the ‘Shanli’ pear. b Expression of PuPME1 during fruit development and ripening. Fruits were collected at 30, 60, 90, and 120 DAFB. H indicates harvest day (0). Fruits collected at harvest day were treated with Ethephon and 1-MCP, stored at room temperature and then sampled every 5 days. Values are mean ± SD of five biological replicates
Expression profiles of PuPME1 in different stages of ‘Nanguo’ pear fruit development and ripening were also examined. PuPME1 expression increased gradually as fruit development progressed (Fig. 3b).
The Effect of Ethylene and 1-MCP Treatments on PuPME1
To understand whether the expression of PuPME1 was regulated by ethylene, we measured the expression levels of PuPME1 in 1-MCP and Ethephon-treated ‘Nanguo’ pear fruits. Ethephon induced the expression of PuPME1, while 1-MCP significantly inhibited PuPME1 expression compared to that in untreated fruits (Fig. 3b).
Confirmation of the Interaction Between PuARP4 and PuPME1
The Y2H screening showed that PuPME1 was a potential interactor with PuARP4. We therefore confirmed their interaction by co-transformation in yeast cells. The CDS of PuPME1 without signal peptides was cloned into a pGADT7 vector, which was then co-transformed with pGBKT7-PuARP4 into the yeast strain Y2H Gold. The results showed that PuARP4 was able to interact with PuPME1 in yeast cells (Fig. 4a). Next, PuARP4 and PuPME1 proteins were purified and a pull-down assay was conducted. This assay showed that the proteins interacted with each other (Fig. 4b).
Interaction between PuARP4 and PuPME1 by yeast two-hybrid (Y2H) and pull-down assay. a Y2H assay of PuARP4 with PuPME1. The AD and BD fusions were co-transformed into the yeast strain Y2H Gold. p53 and SV40 were used as positive controls, and pGBKT7 and pGADT7 were used as negative controls. b Pull-down analysis of PuARP4 and PuPME1. Purified His-PuPME1 was used as bait. Bound proteins were detected with an anti-GST antibody. GST was used as a negative control
Discussion
Fruit softening is the most important sign that fruit is beginning to break down, and the softening rate directly determines the fruit storage quality. ‘Nanguo’ pear is a typical climacteric fruit that produces high levels of ethylene during ripening, accompanied by a rapid decrease in firmness (Huang and others 2014).
The actin cytoskeleton plays an important role in many cell physiological processes and can respond to stimuli such as hormones and pathogens. Ethylene is the main factor controlling the ripening of ‘Nanguo’ pear fruit. Based on our previous research, we found a gene, PuARP4, the expression level of which was down-regulated during fruit ripening. The ARP protein family is one kind of actin proteins with 20–60% similarity to traditional actin proteins (McKinney and others 2002). ARPs have been shown to function in flower morphological development and in cell motility. In this study, the expression of PuARP4 could be inhibited by ethylene treatment (Fig. 2b), indicating that ethylene transmitted a signal to the actin cytoskeleton, which responded to this signal by reducing the levels of actin cytoskeletal proteins, ultimately leading to fruit softening and ripening. However, the mechanism by which the ethylene signal influences the actin cytoskeleton requires further research.
It was previously reported that actin could regulate the accumulation of cell wall components (Meagher and others 1999). Using PuARP4 as bait, we screened the ‘Nanguo’ pear fruit cDNA library and obtained a PME gene, PuPME1. PME is a cell wall hydrolase and is responsible for the degradation of pectin in the cell wall. The expression of PuPME1 in fruits was higher than that in the other tissues that were examined. During fruit ripening, the expression of PuPME1 increased gradually, which would lead to increased degradation of cell wall pectin and contribute to fruit softening. Furthermore, the expression of PuPME1 was significantly induced by Ethephon and inhibited by 1-MCP treatment, indicating the involvement of PuPME1 in ethylene-mediated fruit ripening.
Several studies have demonstrated the involvement of PME genes in fruit ripening, such as PcPME4 in ‘La France’ pear (Sekine and others 2006). PcPME4 was expressed at high levels at day 0 in fruits subjected to long-term storage, indicating its involvement in fruit softening during storage (Sekine and others 2006). Based on our results, we speculated that there may exist two opposite processes during ethylene-mediated fruit ripening process, less accumulation of cell wall components due to lower expression of PuARP4 and increased cell wall pectin degradation by PuPME1 due to higher expression of PuPME1. And PuPME1 could interact with PuARP4, indicating that PuARP4 might cooperate with PuPME1 during the fruit ripening process. However, how the interaction between PuARP4 and PuPME1 to regulate fruit ripening together still needs further research. These results provide a new link between the actin cytoskeleton and fruit ripening.
In conclusion, PuARP4 is involved in ethylene-mediated fruit ripening and can interact with PuPME1, which is involved in cell wall pectin degradation.
References
de Ruijter NCA, Bisseling T, Emons AMC (1999) Rhizobium nod factors induce an increase in sub-apical fine bundles of actin filaments in Vicia sativa root hairs within minutes. Mol Plant Microbe Interact 12:829–832
Dyachok J, Shao MR, Vaughn K, Bowling A, Facette M, Djakovic S, Clark L, Smith L (2008) Plasma membrane-associated SCAR complex subunits promote cortical F-actin accumulation and normal growth characteristics in Arabidopsis roots. Mol Plant 1:990–1006. doi:10.1093/mp/ssn059
Gasic K, Hernandez A, Korban SS (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep 22:437–438. doi:10.1007/BF02772687
Gavrin A, Jansen V, Ivanov S, Bisseling T, Fedorova E (2015) ARP2/3-mediated actin nucleation associated with symbiosome membrane is essential for the development of symbiosomes in infected cells of medicago truncatula root nodules. Mol Plant Microbe Interact 28:605–614
Goley ED, Welch MD (2006) The Arp2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol 7:713–726. doi:10.1038/nrm2026
Goulao LF, Oliveira CM (2008) Cell wall modifications during fruit ripening: when a fruit is not the fruit. Trends Food Sci Technol 19:4–25. doi:10.1016/j.tifs.2007.07.002
Guo S, Sun H, Zhang H, Liu J, Ren Y, Gong G, Jiao C, Zheng Y, Yang W, Fei Z, Xu Y (2015) Comparative transcriptome analysis of cultivated and wild watermelon during fruit development. PLoS ONE 10(6):e0130267
Huang G, Li T, Li X, Tan D, Jiang Z, Wei Y, Li J, Wang A (2014) Comparative transcriptome analysis of climacteric fruit of Chinese Pear (Pyrus ussuriensis) reveals new insights into fruit ripening. PLoS ONE 9:e107562. doi:10.1371/journal.pone.0107562
Kandasamy MK, McKinney EC, Meagher RB (2003) Cell cycle-dependent association of Arabidopsis actin-related proteins AtARP4 and AtARP7 with the nucleus. Plant J 33:939–948. doi:10.1046/j.1365-313X.2003.01691.x
Kandasamy MK, Deal RB, McKinney EC, Meagher RB (2005) Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J 41:845–858. doi:10.1111/j.1365-313X.2005.02345.x
McKinney EC, Kandasamy MK, Meagher RB (2002) Arabidopsis contains ancient classes of differentially expressed actin-related protein genes. Plant Physiol 128:997–1007. doi:10.1104/pp.010906
Meagher RB, McKinney EC, Vitale AV (1999) The evolution of new structures: clues from plant cytoskeletal genes. Trends Genet 7:278–284
Mercado JA, Pliego-Alfaro F, Quesada MA (2011) Fruit shelf life and potential for its genetic improvement. In: Jenks MA, Bebeli PJ (eds) Breeding for fruit quality. Wiley, Oxford, pp 81–104
Pinzón-Latorre D, Deyholos MK (2013) Characterization and transcript profiling of the pectin methylesterase (PME) and pectin methylesterase inhibitor (PMEI) gene families in flax (Linum usitatissimum). BMC Genomics 14:742. doi:10.1186/1471-2164-14-742
Segonne SM, Bruneau M, Celton J-M, Le Gall S, Francin-Allami M, Juchaux M, Laurens F, Orsel M, Renou JP (2014) Multiscale investigation of mealiness in apple: an atypical role for a pectin methylesterase during fruit maturation. BMC Plant Biol 14:375. doi:10.1186/s12870-014-0375-3
Sekine D, Munemura I, Gao M, Mitsuhashi W, Toyomasu T, Murayama H (2006) Cloning of cDNAs encoding cell-wall hydrolases from pear (Pyrus communis) fruit and their involvement in fruit softening and development of melting texture. Physiol Plant 126:163–174. doi:10.1111/j.1399-3054.2006.00583.x
Soon OKE, Youngsook L (1997) Actin filaments of guard cells are reorganized in response to light and abscisic acid. Plant Physiol 5:1491–1498
Tan D, Li T, Wang A (2013) Apple 1-aminocyclopropane-1-carboxylic acid synthase genes, MdACS1 and MdASC3a, are expressed in different systems of ethylene biosynthesis. Plant Mol Biol Rep 31:204–209. doi:10.1007/s11105-012-0490-y
Wei J, Ma F, Shi S, Qi X, Zhu X, Yuan J (2010) Changes and postharvest regulation of activity and gene expression of enzymes related to cell wall degradation in ripening apple fruit. Postharvest Biol Technol 56:147–154. doi:10.1016/j.postharvbio.2009.12.003
Wen F, Zhu Y, Hawes MC (1999) Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11:1129–1140
Yuan H, Meng D, Gu Z, Li W, Wang A, Yang Q, Zhu Y, Li T (2014) A novel gene, MdSSK1, as a component of the SCF complex rather than MdSBP1 can mediate the ubiquitination of S-RNase in apple. J Exp Bot 65:3121–3131. doi:10.1093/jxb/eru164
Zepeda I, Sánchez-López R, Joseph G, Bañuelos L, Hernández-Barrera A, Sánchez F, Quinto C, Cárdenas L (2014) Visualization of highly dynamic F-Actin plus ends in growing Phaseolus vulgaris root hair cells and their responses to rhizobium etli Nod factors. Plant Cell Physiol 55(3): 580–592
Zhao Y, Pan Z, Zhang Y, Qu X, Zhang Y, Yang Y, Jiang X, Huang S, Yuan M, Schumaker Karen S, Guo Y (2013) The actin-related protein2/3 complex regulates mitochondrial-associated calcium signaling during salt stress in Arabidopsis. Plant Cell 25:4544–4559
Zou J, Zheng Z, Xue S, Li H, Wang Y, Le J (2016) The role of Arabidopsis actin-related protein 3 in amyloplast sedimentation and polar auxin transport in root gravitropism. J Exp Bot. doi:10.1093/jxb/erw294
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.

Rights and permissions
About this article
Cite this article
Yuan, H., Zhang, L., Jiang, Z. et al. Characterization of Ripening-Related PuARP4 in Pear (Pyrus ussuriensis). J Plant Growth Regul 36, 766–772 (2017). https://doi.org/10.1007/s00344-017-9680-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-017-9680-z