Abstract
In the nonobese diabetes mouse, the murine type 1 diabetes susceptibility locus Idd20 interacts genetically with the diabetes resistance locus Idd19. Both Idds are located on distal mouse Chromosome 6, and previous studies on NOD.C3H congenic strains have shown that C3H alleles at Idd20 can suppress the disease-promoting effects of C3H alleles at Idd19 in both spontaneous and cyclophosphamide-induced diabetes. In this article we present the construction of novel congenic strains which, while maintaining the C3H alleles at Idd19, have allowed the candidate interval of Idd20 to be reduced from 4 to 1.8 cM. The analysis of these strains shows that Idd20 controls the progression of insulitis. Idd20 also increases the suppressive but not the pathogenic activity of splenocytes in diabetes transfer experiments. Our results suggest that the two Chromosome 6 susceptibility loci, Idd6 and Idd20, interact with the resistance locus Idd19 by regulating the activity of suppressor cells in the peripheral immune system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The nonobese diabetes (NOD) mouse (Hattori et al. 1986; Makino et al. 1980) is a well-characterized animal model of type 1 or insulin-dependent diabetes (T1D, IDDM), an autoimmune disease characterized by the progressive destruction of insulin-producing ß cells of the islets of Langerhans by infiltrating lymphocytes (Tisch and McDevitt 1996). Over 30 murine insulin dependent diabetes loci (Idd) have been genetically identified (http://www.informatics.jax.org). Congenic strains that differ from the NOD receiver strain by only a selected genetic region derived from a non-diabetes-prone parental donor strain (McAleer et al. 1995; Prochazka et al. 1989) are critically important tools for candidate gene identification when their characterization combines phenotypic studies with expression profiling, haplotype, and mutational analysis (Eckenrode et al. 2004; Lyons 2002; Rogner and Avner 2003; Wicker et al. 1995).
Several Idd loci have been identified on mouse Chromosome 6 (de Gouyon et al. 1993; Ghosh et al. 1993; Melanitou et al. 1998), and these have been further defined by the analysis of a series of congenic strains carrying C3H/HeJ genomic material for distal Chromosome 6 introgressed onto the NOD/Lt genetic background (Rogner et al. 2001). NOD/Lt alleles at the Idd6 locus distal to 70 cM of mouse Chromosome 6 confer susceptibility to T1D (Carnaud et al. 2001; Ghosh et al. 1993; Rogner et al. 2001), while NOD alleles at the Idd19 locus, centered around 55 cM, confer resistance (Melanitou et al. 1998; Rogner et al. 2001). A third diabetes-associated locus on mouse Chromosome 6, Idd20, which maps between 35 and 41 cM, interacts with Idd19 in an epistatic manner and can suppress Idd19 effects in both spontaneous and cyclophosphamide-induced diabetes. Idd20 has also been shown to genetically control the acceleration of diabetes incidence induced by deficiency of the programmed cell death 1 (PD-1, Pdcd1) gene, which encodes an immunoinhibitory receptor belonging to the CD28/cytotoxic T-lymphocyte-associated antigen-4 family (Wang et al. 2005).
Recently, we have undertaken a detailed phenotypic analysis of the Idd6 locus containing congenic strain NOD.C3H 6.VIII (Rogner et al. 2001), which shows resistance to the spontaneous development of diabetes. We have shown that splenocytes of Idd6 congenic mice confer enhanced disease protection in diabetes transfer assays (Rogner et al. 2006). In this article we show that C3H alleles at the Idd19 locus abolish this Idd6-mediated mechanism of protection which can be largely restored by the presence of C3H alleles at the Idd20 locus. This study, building on the analysis of the genetic interaction of Idd20 with Idd19, has allowed the reduction of the Idd20 candidate interval from 4 to 1.8 cM. The characterization of disease-relevant tissues for Idd20 action coupled with this reduction of the genetic interval should facilitate the identification of candidate genes for Idd20.
Material and methods
Mice
F1 males of the congenic strain NOD.C3H 6.I, carrying C3H alleles at and distal of D6Mit69 (Rogner et al. 2001), were backcrossed to NOD/Lt mice. Recombinant mice were identified at backcross generation BC12, rendered homozygous for the C3H intervals by intercrossing, and maintained in our animal house by brother-sister mating. The animal studies were approved by the relevant institutional review boards.
Assessment of glucosuria and adoptive transfer of diabetes
Spontaneous cumulative diabetes incidence was monitored weekly for a period of 30 weeks by glucosuria assessment (Diabur test, Roche). In transfer experiments, 107 splenocytes from diabetic or aged NOD donor mice were injected intravenously into immunodeficient NOD/Scid mice. In cotransfer assays the 107 diabetogenic NOD splenocytes were injected together with 2 × 107 donor splenocytes obtained from 8-week-old mice. Recipients were monitored for diabetes for ten weeks after transfer. Time-to-event distributions were calculated by Kaplan-Meier estimation and compared by log-rank tests over the period of observation.
To study induction of diabetes by cyclophosphamide (CY), animals were first injected with 200 mg/kg CY at eight weeks of age and the injection was repeated two weeks later. Glucosuria was tested two weeks (T1) and four weeks (T2) after the initial injection of CY. Statistical analysis at T1 and T2 was performed using the Fisher’s exact test.
Histopathology of the pancreas
Pancreata were excised, fixed in Bouin’s solution, and processed for paraffin embedding. Four sections (5 μm) taken at 100-μm intervals were stained using hematein- eosin-safranin. At least 20 islets per specimen were analyzed. Histology grades for single pre-diabetic animals were defined as follows: 1 = normal islets, 2 = peri-insulitis with less than 20% of the islets destroyed, 3 = insulitis with 20%–50% of the islets destroyed, 4 = extensive insulitis with more than 50% of the islets destroyed. Statistical analysis was performed using the Fisher’s exact test.
Insulin autoantibody (IAA) determination
The 96-well filtration plate micro-IAA assay (Yu et al. 2000) was performed as previously described (Thebault-Baumont et al. 2003). Statistical analysis was performed using Mann-Whitney and analysis of variance (ANOVA) tests.
Immunofluorescence staining
Islet-infiltrating leukocytes were isolated as previously described (Faveeuw et al. 1995). Cells were pelleted in 96-well plates and stained for 30 min on ice in 20 μl of phosphate-buffered saline (PBS) supplemented with 2% fetal calf serum and 5 mM sodium azide, using biotin-, PE-, FITC-, PerCP-, or APC-labeled reagents (Pharmingen, BD Bioscience, France) at optimal concentration. Where appropriate, a secondary staining step using fluorochrome-conjugated streptavidin was performed. Cells were washed twice and resuspended in PBS containing 1% formaldehyde. Flow cytometry analysis was performed using a FACS calibur and CellQuest software (BD Biosciences, Grenoble, France). The sample size for data collection was 10,000 cells. Pooled data were compared as mean ± SD and by unpaired Student t tests.
Results
Reduction of the genetic interval for Idd20
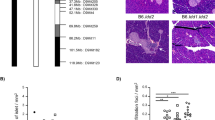
Novel congenic strains for Chromosome 6 were derived by backcrossing males of the NOD.C3H strain 6.I, originally generated at backcross generation BC7, to NOD/Lt female mice. 6.I carries C3H alleles at and distal to D6Mit69, located at 35.15 cM. Six recombinants were obtained by intercrossing a heterozygous male at backcross generation BC12. At this level of backcross, donor-strain genome material from the recipient strain outside of the differential segment is expected to be fully replaced. The C3H intervals of the recombinants were fixed by intercrossing. Congenic strain 6.1 was shown to carry C3H alleles between D6Mit256 and D6Mit15, strain 6.2 between D6Mit100 and D6Mit15, and strain 6.3 between D6Mit100 to D6Mit55. C3H alleles of strain 6.5 covered the D6Mit322-D6Mit15 interval, while strain 6.6 carried C3H alleles for the small region between D6Mit322 and D6Mit31. The strain 6.4 was generated as an internal control strain and does not carry C3H alleles for Chromosome 6 (Fig. 1). In case there are small segments of donor genome introgressed during backcrossing that were missed by genome scans and so may influence diabetes susceptibility, these segments would, most likely, be shared by 6.4 and the other congenic strains. Strain 6.4 therefore is a better control than simply using the progenitor strain, NOD/Lt.
Genetic map of distal Chromosome 6 summarizing the genetic intervals carried by the novel NOD.C3H congenic strains relative to the localization of disease-relevant genes and QTLs. NOD alleles of the congenic strains are indicated as light bars, C3H alleles as dark bars. Numbers indicate the D6Mit markers that are closest to the end of the interval. The calculation of genetic and physical distances is based on the NCBI M. musculus Genome Build 34.1. Previously defined candidate regions are indicated as light-shaded fields, newly refined regions as dark-shaded fields. Hrh1 = histamine receptor H 1; Tnfrsf1a = tumour necrosis factor receptor superfamily, member 1a; Cd4 = CD4 antigen; Klrb1c = killer cell lectin-like receptor subfamily B member 1C; Cd69 = CD69 antigen; Ht6.9 = T-cell clone BDC-6.9 antigen; Nidd3n = non-insulin-dependent diabetes mellitus 3 in NSY mice; Cia3 = collagen-induced arthritis QTL 3; Bhr5 = bronchial hyperresponsiveness 5; Insq8 = insulin QTL 8; Lbw4 = lupus NZB × NZW 4.
We first selected for congenic strains that showed strongly reduced spontaneous diabetes incidence over an eight-month period. Two of the above NOD.C3H congenic strains lacked significant protection over this period (strain 6.3, 24 females, 14 males, p = 0.25; strain 6.6, 19 females, 13 males, p = 0.35). Therefore, these strains were not retained for further detailed analysis of spontaneous diabetes and related subphenotypes. Strains 6.1 showed almost complete protection in females and males (Fig. 2). with an incidence of close to 0% at 8 months of age (p < 0.0001). Strain 6.1 carries C3H alleles at an interval covering the previously defined Idd6 candidate region distal of D6Mit113 and including the marker D6Mit256 and the natural killer complex (NKC). This observation is consistent with previous findings that the D6Mit254 (60.55 cM) to D6Mit14 (71.1 cM) interval is protective against type I diabetes in NOD.C56BL/6 congenic strains (Bergman et al. 2003; Carnaud et al. 2001; Dallas-Pedretti et al. 1995).
Cumulative diabetes incidence in females and males of BC12 NOD.C3H congenic strains. P values are as follows: females: 6.4–6.5, p < 0.0001; 6.2–6.4, p = 0.008; 6.2–6.5, p = 0.010; 6.1–6.4, p < 0.0001; 6.1–6.5, p = 0.089; 6.1–6.2, p = 0.007; males: 6.4–6.5, p = 0.004; 6.2–6.4, p = 0.018; 6.2–6.5, p = 0.290; 6.1–6.4, p = 0.017; 6.1–6.5, p = 0.471; 6.1–6.2, p = 0.205. n indicates the number of animals.
Strain 6.5 was also strongly protected compared with the control strain 6.4 (p < 0.0001 for females). Strain 6.2, which differs from 6.5 only by NOD alleles between D6Mit322 and D6Mit100, was significantly less protected against diabetes when compared with strain 6.5 (p = 0.01 for females). The originally defined interval for Idd20 mapped to between D6Mit69/D6Mit322 (35.15/35.2 cM, both excluded) and D6Mit326 (41 cM, excluded). The comparison of strain 6.5 with strain 6.2 allows the originally defined candidate region for Idd20 to be reduced to an interval of 1.8 cM [NCBI M. musculus Genome (Build 34.1)] and both the D6Mit322 and D6Mit100 markers to be excluded from the candidate region.
Strain 6.6 covers the Idd20 interval, but as we have previously described (Rogner et al. 2001), C3H alleles at Idd20 alone are not protective against spontaneous diabetes. Idd20 acts as a suppressor of the Idd19 locus and Idd20-mediated protection therefore can be detected only by its genetic interaction with diabetes resistance alleles at the Idd19 locus. However, we were able to confirm the previous finding of resistance against CY-induced diabetes for the 6.6 strain (15 mice, 0% at T1, 20% at T2, p = 0.014) compared with the 6.3 strain (17 mice, 17.6% at T1, 76.5% at T2) and 6.4 strain (n = 19, 36.8% at T1, 63.3% at T2). This result confirms that the previously defined Idd20-mediated resistance against CY-induced diabetes colocalizes with the resistance against spontaneous diabetes to the D6Mit322-D6Mit100 interval.
Idd20 controls insulitis progression
To understand in more detail the mechanisms of diabetes protection mediated by Idd20, we compared several diabetes-related subphenotypes in strains 6.5 and 6.2. We had previously shown that Idd20 mediates resistance against CY-induced insulitis. To evaluate if Idd20 was involved in insulitis progression in spontaneously developing diabetes, we undertook a histopathologic analysis of the pancreas of 12-week-old females. This study revealed that the 6.5 strain was significantly protected against peri-insulitis and insulitis (p < 0.001) when compared with the 6.4 NOD control strain. Interestingly, we noticed that strain 6.1 also showed reduced progression from mild to the severe stages of insulitis (p < 0.001). Because no such observation has been made for the Idd6 locus at distal Chromosome 6 (Rogner et al. 2006), we concluded that C3H alleles at the D6Mit256-D6Mit57 interval may contribute to this protection. This genetic interval contains both the NKC (Carnaud et al. 2001) and the gene encoding for the BDC6.9 antigen (Dallas-Pedretti et al. 1995). While the C3H-derived interval of strain 6.1 was completely covered by the C3H-derived intervals of strains 6.5 and 6.2, the protective effect was absent in the 6.2 strain carrying C3H alleles at the Idd19 locus and was present in strain 6.5. Such a result would be expected if the insulitis protection mediated by Idd20 depended on the interaction of Idd20 with Idd19 (p < 0.001 for comparison of 6.2 and 6.5, Fig. 3). The analysis of strain 6.6 revealed that C3H alleles at Idd20 alone in strain 6.6 were insufficiently protective, suggesting that Idd20 acts as suppressor of diabetes resistance alleles at Idd19 in spontaneous diabetes and spontaneous developing insulitis.
Pancreas histopathology and anti-insulin antibody (IAA) levels of 12-week-old female mice. p values for histology scores compared with those of strain 6.4 (n = 19) are as follows: 6.1 (n = 21), p < 0.001; 6.2 (n = 18), p = 0.311; 6.3 (n = 20), p = 0.844; 6.5 (n = 18), p < 0.001; 6.6 (n = 19), p = 0.113. Differences in IAA levels were not significant (p > 0.4 for all groups compared to 6.4 in Mann-Whitney test).
The difference in the extent of insulitis between the two strains prompted us to test for more subtle differences in the leukocyte subsets invading the islets. We analyzed the islet infiltrate of six 12-week-old female mice by FACS. The frequencies of infiltrating lymphocytes and nonlymphoid cells were not found to be significantly different in strains 6.2, 6.4, and 6.5 (p > 0.05, Table 1), suggesting that the aggressiveness of the autoimmune response to ß cells does not depend on the ratios of islet-infiltrating cells.
Anti-insulin antibody levels in the serum were similarly present in females of strains 6.4, 6.2, and 6.5 at 12 weeks of age (Fig. 3). This result indicates that the resistance to insulitis mediated by Idd20 does not involve major alterations in the immune response to insulin.
Idd20 does not control the pathogenicity of splenocytes
We wanted to test the hypothesis that the pathogenic activity of splenocytes could be reduced in strain 6.5 compared with strain 6.2. Splenocytes from aged NOD mice are expected to induce diabetes rapidly in immunodeficient recipients because of the progressive decrease in the activity of regulatory T cells and the enhanced pathogenicity of CD25 T cells with age (Gregori et al. 2003; Rohane et al. 1995). Adoptive transfer of splenocytes to NOD/Scid mice was performed using prediabetic 15-week-old male and female donors. It should be noted that aged prediabetic mice allow a better estimation of the diabetogenic T-cell pool size than diabetic mice which most likely harbor equivalent numbers of effector T cells independent of their genotype. Splenocytes from diabetic NOD mice were used as positive control and allowed a rapid induction of diabetes some three weeks or more after transfer. Diabetes developed with similar kinetics when donor cells of prediabetic 6.4, 6.5, and 6.2 mice were used (Fig. 4), with diabetes being observed from five weeks onward. Our results suggest that splenocytes from aged 6.5 mice induce diabetes as efficiently as cells from aged 6.4 NOD control and 6.2 mice, with all showing a delay of at least one week compared with that obtained using splenocytes from diabetic NOD mice. This delay is likely linked to the use of nonpurified splenocytes in the transfer experiment, which contain both effector and persistent regulatory T cells. We concluded that the presence of C3H alleles at the Idd20 locus does not markedly reduce the pathogenic activity of the splenocyte population.
Diabetes transfer and cotransfer results. No significant differences were found among 6.4, 6.5, and 6.2 mice in diabetes transfer. Significant differences were found in cotransfer experiment for 6.5 mice, p = 0.041 against 6.4 and p = 0.029 against 6.2. The p value for comparison of 6.5 against strain 6.VIII is p = 0.082; for 6.4 against 6.VIII, p = 0.001; and for 6.4 against the diabetic animals (Db), p = 0.007. n indicates the number of recipients.
Idd20 enhances the suppressive activity of splenocytes
We next tested the hypothesis that Idd20 contributes to the control of the suppressive activity of the peripheral immune system. Splenocytes from 8-week-old mice have been described to contain few diabetogenic cells and to inhibit efficiently diabetes in cotransfer with diabetogenic cells (Boitard et al. 1989; Lepault and Gagnerault 2000). A total of 2 × 107 splenocytes isolated from mice of each strain were injected together with 107 splenocytes from diabetic NOD mice into recipient mice. Diabetes incidence was observed weekly for 10 weeks after adoptive transfer. Compared with the transfer of diabetogenic cells alone, which induced diabetes in 90% of the recipients at 10 weeks after transfer, even coinjection of splenocytes from 6.4 control mice was shown to confer some protection against diabetes (p < 0.01).
While coinjection of splenocytes from young 6.2 mice was not significantly more protective than that of 6.4, spleen cells from strain 6.5 provided statistically significant (p < 0.05) higher protection. We conclude that while 6.4, 6.2, and 6.5 splenocytes all contain suppressor cells able to control the development of diabetes, the 6.5 spleen cells exhibit significantly higher protective activity. Interestingly, this suppressive activity was nearly as important as that we have recently described for the Idd6-containing strain 6.VIII [p = 0.082 for comparison of strains 6.VIII and 6.5, Fig. 4. (Rogner et al., 2006)].
To investigate if the increased suppressive activity of splenocytes in strain 6.5 compared with that in strains 6.2 and 6.4 was associated with numerical changes in the immune cell population, we performed FACS analysis on thymocytes and splenocytes of six 8-week-old female mice. No major changes in the CD4+ T cell, CD8+ T cell, or the regulatory CD4+CD25+ T-cell population, in either the central or the peripheral immune system, were observed (p > 0.05, Table 1). Similar findings were made for B cells with an average numeration percentage of 35% and for CD4+CD62L+ regulatory T cells with an average numeration percentage of 14.6% in the spleen (data not shown). This suggests that the increased suppressive activity of 6.5 splenocytes may involve increased activity rather than major quantitative alterations in the presence of particular cellular subsets.
Discussion
The Idd20 locus on mouse Chromosome 6 appears to genetically control accelerated type 1 diabetes disposition. C3H alleles at Idd20 suppress progression of diabetes and insulitis mediated by C3H alleles at the Idd19 locus. Idd20 also influences cyclophosphamide (CY)-induced diabetes and insulitis (Fig. 5). Cyclophosphamide has been described as accelerating autoimmune diabetes in the NOD mouse at different levels, including through the critical targeting of a regulatory T-cell subset, the exacerbation of pro-Th1 IFN-γ production, and the promotion of inflammation in pancreatic islets (Ablamunits et al. 1999a, 1999b). The Idd20 locus may interfere with one or several stages of this process.
Summary of the genetic interactions of the Idd6, Idd19, and Idd20 loci on distal mouse Chromosome 6 in NOD.C3H congenic strains and description of the associated subphenotypes according to this work and the results published in Rogner et al. (2001, 2006). CY = cyclophosphamide.
Recently, an independent genetic study has revealed that Idd20 is one of five genetic loci that control diabetes incidence in PD-1-deficient NOD mice. PD-1 (programmed cell death 1, Pdcd1) is an immunoinhibitory receptor belonging to the CD28/cytotoxic T-lymphocyte-associated antigen-4 family, which acts as an immunoregulatory receptor. PD-1 is expressed on the surface of activated T cells, B cells, and monocytes and prevents autoimmunity by inhibiting activation of self-reactive lymphocytes upon interaction with its ligands PD-L1 and PD-L2. Mice that are deficient in PD-1 spontaneously develop lupus-like autoimmune disease and autoimmune dilated cardiomyopathy on C57BL/6 and BALB/c genetic backgrounds. In NOD mice, PD-1 deficiency specifically accelerates the onset and frequency of type 1 diabetes, with strong T-helper 1 polarization of T cells infiltrating into pancreatic islets. These results have suggested that PD-1 deficiency accelerates autoimmune predisposition of the background strain, leading to the induction of different forms of autoimmune diseases, each depending on the genetic background of the strain. The genetic interaction of PD-1 deficiency with Idd20 may suggest that Idd20 or a gene within the locus is involved in determination of organ specificity against β cells of the pancreatic islets (Wang et al. 2005).
Our present analysis suggests that Idd20 controls the suppressive activity of splenocytes but does not reduce the splenocyte pathogenicity. This C3H allele-mediated suppressive effect may directly result in inhibition of the progression of insulitis and diabetes development. Further genetic and phenotypic analysis may be necessary to show if Idd20 contains several genes that influence different subphenotypes or if a unique gene with pleiotropic effects is involved in conferring a diversity of subphenotypes. At present, no gene known to be involved in type 1 diabetes or in the PD-1 pathway has been shown to localize to the Idd20 interval. Systematic efforts to analyze the gene content of the refined candidate region will therefore be crucial to identify these genes and their causal polymorphisms.
References
Ablamunits V, Elias D, Cohen IR (1999a) The pathogenicity of islet-infiltrating lymphocytes in the non-obese diabetic (NOD) mouse. Clin Exp Immunol 115, 260–267
Ablamunits V, Quintana F, Reshef T, Elias D, Cohen IR, et al. (1999b) Acceleration of autoimmune diabetes by cyclophosphamide is associated with an enhanced IFN-gamma secretion pathway. J Autoimmun 13, 383–392
Bergman ML, Duarte N, Campino S, Lundholm M, Motta V, et al. (2003) Diabetes protection and restoration of thymocyte apoptosis in NOD Idd6 congenic strains. Diabetes 52, 1677–1682
Boitard C, Yasunami R, Dardenne M, Bach JF (1989) T cell-mediated inhibition of the transfer of autoimmune diabetes in NOD mice. J Exp Med 169, 1669–1680
Carnaud C, Gombert J, Donnars O, Garchon H, Herbelin A (2001) Protection against diabetes and improved NK/NKT cell performance in NOD.NK1.1 mice congenic at the NK complex. J Immunol 166, 2404–2411
Dallas-Pedretti A, McDuffie M, Haskins K (1995) A diabetes-associated T-cell autoantigen maps to a telomeric locus on mouse chromosome 6. Proc Natl Acad Sci USA 92, 1386–1390
de Gouyon B, Melanitou E, Richard MF, Requarth M, Hahn IH, et al. (1993) Genetic analysis of diabetes and insulitis in an interspecific cross of the nonobese diabetic mouse with Mus spretus. Proc Natl Acad Sci USA 90, 1877–1881
Eckenrode SE, Ruan Q, Yang P, Zheng W, McIndoe RA, et al. (2004) Gene expression profiles define a key checkpoint for type 1 diabetes in NOD mice. Diabetes 53, 366–375
Faveeuw C, Gagnerault MC, Lepault F (1995) Isolation of leukocytes infiltrating the islets of Langerhans of diabetes-prone mice for flow cytometric analysis. J Immunol Methods 187, 163–169
Ghosh S, Palmer SM, Rodrigues NR, Cordell HJ, Hearne CM, et al. (1993) Polygenic control of autoimmune diabetes in nonobese diabetic mice. Nat Genet 4, 404–409
Gregori S, Giarratana N, Smiroldo S, Adorini L (2003) Dynamics of pathogenic and suppressor T cells in autoimmune diabetes development. J Immunol 171, 4040–4047
Hattori M, Buse JB, Jackson RA, Glimcher L, Dorf ME, et al. (1986) The NOD mouse: recessive diabetogenic gene in the major histocompatibility complex. Science 231, 733–735
Lepault F, Gagnerault MC (2000) Characterization of peripheral regulatory CD4+ T cells that prevent diabetes onset in nonobese diabetic mice. J Immunol 164, 240–247
Lyons P (2002) Gene-expression profiling and the genetic dissection of complex disease. Curr Opin Immunol 14, 627
Makino S, Kunimoto K, Muraoka Y, Mizushima Y, Katagiri K, et al. (1980) Breeding of a non-obese, diabetic strain of mice. Jikken Dobutsu 29, 1–13
McAleer MA, Reifsnyder P, Palmer SM, Prochazka M, Love JM, et al. (1995) Crosses of NOD mice with the related NON strain. A polygenic model for IDDM. Diabetes 44, 1186–1195
Melanitou E, Joly F, Lathrop M, Boitard C, Avner P (1998) Evidence for the presence of insulin-dependent diabetes-associated alleles on the distal part of mouse chromosome 6. Genome Res 8, 608–620
Prochazka M, Serreze DV, Worthen SM, Leiter EH (1989) Genetic control of diabetogenesis in NOD/Lt mice. Development and analysis of congenic stocks. Diabetes 38, 1446–1455
Rogner UC, Avner P (2003) Congenic mice: cutting tools for complex immune disorders. Nat Rev Immunol 3, 243–252
Rogner UC, Boitard C, Morin J, Melanitou E, Avner P (2001) Three loci on mouse chromosome 6 influence onset and final incidence of type I diabetes in NOD.C3H congenic strains. Genomics 74, 163–171
Rogner UC, Lepault F, Gagnerault MC, Vallois D, Morin J, et al. (2006) The diabetes type 1 locus Idd6 modulates activity of CD4+CD25+ regulatory T-cells. Diabetes 55, 186–192
Rohane PW, Shimada A, Kim DT, Edwards CT, Charlton B, et al. (1995) Islet-infiltrating lymphocytes from prediabetic NOD mice rapidly transfer diabetes to NOD-scid/scid mice. Diabetes 44, 550–554
Thebault-Baumont K, Dubois-Laforgue D, Krief P, Briand JP, Halbout P, et al. (2003) Acceleration of type 1 diabetes mellitus in proinsulin 2-deficient NOD mice. J Clin Invest 111, 851–857
Tisch R, McDevitt H (1996) Insulin-dependent diabetes mellitus. Cell 85, 291–297
Wang J, Yoshida T, Nakaki F, Hiai H, Okazaki T, et al. (2005) Establishment of NOD-Pdcd1-/- mice as an efficient animal model of type I diabetes. Proc Natl Acad Sci USA 102, 11823–11828
Wicker LS, Todd JA, Peterson LB (1995) Genetic control of autoimmune diabetes in the NOD mouse. Annu Rev Immunol 13, 179–200
Yu L, Robles DT, Abiru N, Kaur P, Rewers M, et al. (2000) Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proc Natl Acad Sci USA 97, 1701–1706
Acknowledgments
The authors thank Corinne Veron for excellent technical assistance and Bruno Lucas for help with the FACS analysis. This work was supported by grants from the Juvenile Diabetes Research Foundation International (1-2000-600), and recurrent funding from the CNRS, INSERM, and the Pasteur Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Morin, J., Boitard, C., Vallois, D. et al. Mapping of the murine type 1 diabetes locus Idd20 by genetic interaction. Mamm Genome 17, 1105–1112 (2006). https://doi.org/10.1007/s00335-006-0076-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-006-0076-9