Abstract
Susceptibility to autoimmune diabetes is a complex genetic trait. Linkage analyses exploiting the NOD mouse, which spontaneously develops autoimmune diabetes, have proved to be a useful tool for the characterization of some of these traits. In a linkage analysis using 3A9 TCR transgenic mice on both B10.BR and NOD.H2k backgrounds, we previously determined that both the Idd2 and Idd13 loci were linked to the proportion of immunoregulatory CD4-CD8- double negative (DN) T cells. In addition to Idd2 and Idd13, five other loci showed weak linkage to the proportion of DN T cells. Of interest, in an interim analysis, a locus on chromosome 12 is linked to DN T cell proportion in both the spleen and the lymph nodes. To determine the impact of this locus on DN T cells, we generated two congenic sublines, which we named Chr12P and Chr12D for proximal and distal, respectively. While 3A9 TCR:insHEL NOD.H2k-Chr12D mice were protected from diabetes, 3A9 TCR:insHEL NOD.H2k-Chr12P showed an increase in diabetes incidence. Yet, the proportion of DN T cells was similar to the parental 3A9 TCR NOD.H2k strain for both of these congenic sublines. A genome-wide two dimensional LOD score analysis reveals genetic epistasis between chromosome 12 and the Idd13 locus. Altogether, this study identified further complex genetic interactions in defining the proportion of DN T cells, along with evidence of genetic epistasis within a locus on chromosome 12 influencing autoimmune susceptibility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Immune responses targeted to self-tissues are responsible for a broad spectrum of autoimmune diseases (Theofilopoulos et al. 2017). Complex genetic traits contribute to susceptibility to most autoimmune diseases, including autoimmune diabetes (Polychronakos and Li 2011). The NOD mouse model, which spontaneously develops autoimmune diabetes, has facilitated the identification of over 30 Insulin-dependent diabetes (Idd) loci linked to autoimmune susceptibility (Chen et al. 2018; Driver et al. 2011; Maier and Wicker 2005; Ridgway et al. 2008). Extensive characterization of some Idd loci demonstrates that a given locus can encompass multiple susceptibility alleles (Driver et al. 2011).
The generation of congenic mice has proven to be an effective tool to determine the contribution of Idd loci on autoimmune diabetes susceptibility. Very early on it was demonstrated that while NOD congenic mice bearing resistance alleles at Idd1, which encompasses the MHC locus, are fully protected from diabetes onset (Podolin et al. 1993), other loci confer only partial protection (Wicker et al. 1995). Beyond quantifying the impact of genetic loci on diabetes susceptibility, congenic mice have facilitated the identification of gene-gene interactions as well as genetic epistasis (Chen et al. 2018; Ridgway et al. 2008). For instance, NOD mice–bearing resistance alleles at both Idd3 and Idd5 loci show increased diabetes resistance relative to NOD mice congenic for either locus (Hill et al. 2000). Additionally, careful dissections of genetic loci by generating congenic sublines have revealed gene-gene interactions within a locus. For example, the Idd5 locus has been divided into five subloci, each distinctly contributing to diabetes susceptibility (Hunter et al. 2007). Moreover, congenic mice have allowed to determine the impact of resistance alleles on immune phenotypes, such as the production and function of regulatory T cells (Chen et al. 2008; Kachapati et al. 2012; Sgouroudis et al. 2008; Tellier et al. 2006; Yamanouchi et al. 2007).
The NOD mouse has thus been effectively used to identify loci linked to autoimmune diabetes susceptibility and to understand how these loci influence immune tolerance (Chen et al. 2018; Hamilton-Williams et al. 2016). Conversely, linkage analysis of defects in immune tolerance can also identify loci linked to autoimmune susceptibility. For example, the defects in central tolerance in NOD mice represent a complex trait, linking to many loci including Idd13 (Liston et al. 2004). Similarly, T cell-destructive insulitis is linked to Idd5 and Idd13 (Fox et al. 2000), whereas the proportion of NKT cells, thought to contribute to autoimmune susceptibility in NOD mice, linked to loci that overlap with various Idd loci (Esteban et al. 2003; Tsaih et al. 2013).
We study an immunoregulatory T cell subset, lacking both CD4 and CD8 expression (Hillhouse et al. 2013; Hillhouse and Lesage 2013). These double negative (DN) T cells are cytotoxic and, while not fully understood, they appear to carry out their immunoregulatory function by eliminating autoreactive immune cells (Hillhouse et al. 2013; Hillhouse and Lesage 2013; Juvet and Zhang 2012; Martina et al. 2015). Of note, immunoregulatory DN T cells are more abundant in diabetes-resistant mice, suggesting that their abundance contributes to autoimmune resistance (Dugas et al. 2010; Hillhouse et al. 2010). To facilitate the study of DN T cells, we take advantage of the 3A9 TCR transgenic model. This TCR recognizes a peptide from hen egg lysozyme (HEL) in the context of I-Ak (Allen et al. 1987). Using 3A9 TCR B10.BR and NOD.H2k mice, we previously performed a linkage analysis to the proportion of 3A9 TCR DN T cells in the lymph nodes (Collin et al. 2014). We observed significant linkage to the Idd2 locus. By generating NOD.B10-Idd2 congenic mice, we validated the impact of B10 resistance alleles at the Idd2 locus in defining DN T cell proportion as well as diabetes susceptibility (Collin et al. 2014). We also identified various suggestive loci linked to DN T cell proportion, which included Idd13 (Collin et al. 2014). Using congenic mice, we validated that the Idd13 locus influenced DN T cell proportion (Dugas et al. 2014) and subsequently demonstrated that NOD mice–bearing resistance alleles at both Idd2 and Idd13 loci had an increased proportion of DN T cells relative to single congenic mice (Collin et al. 2018). Still, the combination of Idd2 and Idd13 resistance alleles was not sufficient to fully restore DN T cell proportion and number in all lymphoid organs (Collin et al. 2018), prompting us to investigate the impact of other suggestive loci linked to DN T cell proportion.
Among the suggestive loci linked to DN T cell proportion, we turned our attention to the locus on mouse chromosome 12. Indeed, an interim linkage analysis, presented in this study, suggested that a locus on mouse chromosome 12 was linked to DN T cell proportion in the spleen, skin-draining lymph nodes, and pancreatic lymph nodes. Hence, we began to generate NOD.B10-Chr12 congenic mice prior to the end of the linkage analysis. However, as more F2 mice were included in the linkage analysis, the importance of the chromosome 12 linkage to DN T cell proportion decreased. Having generated the NOD.B10-Chr12 congenic mice, we decided to assess their susceptibility to diabetes. We found that 3A9 TCR:insHEL NOD.B10-Chr12 congenic mice are partially resistant to diabetes (Collin et al. 2015). The chromosome 12 locus did not coincide with known Idd loci but closely overlapped with Nbwa1, a locus linked to autoimmune susceptibility and autoantibody production (Rigby et al. 2004). Therefore, we exploited the NOD.B10-Chr12 congenic mice to validate the impact of this locus on autoantibody production (Collin et al. 2015).
Here, using NOD.B10-Chr12 congenic mice, we further investigate the impact of the chromosome 12 locus with suggestive linkage to the proportion of DN T cells. We find that autoimmune diabetes resistance alleles on chromosome 12 are not sufficient to increase the proportion of DN T cells in lymphoid organs. However, the congenic sublines reveal genetic epistasis in modulating autoimmune diabetes susceptibility, where B10 alleles within the chromosome 12 locus either increase or decrease disease susceptibility.
Materials and methods
Mice
3A9 TCR and insHEL transgenes on B10.BR and NOD.H2k backgrounds have been previously described (Lesage et al. 2002). 3A9 TCR F2 mice result from the second-generation intercross between B10.BR and 3A9 TCR NOD.H2k mice. The NOD.H2k-Chr12 congenic line was obtained by backcrossing B10.BR mice to the NOD.H2k for eight generations as previously reported (Collin et al. 2015). As two congenic sublines were derived from the NOD.H2k-Chr12 congenic line, we hereafter refer to this line as NOD.H2k-Chr12L, where L stand for long. At the fifth generation backcross, the two congenic sublines arose; NOD.H2k-Chr12D and NOD.H2k-Chr12P, where D and P stand for distal and proximal, respectively. Specifically, the NOD.H2k-Chr12P lost B10 alleles at the D12Mit184 marker at the fifth backcross. The loci were delimited by testing various polymorphic markers by PCR. An Illumina low-density linkage, serviced through the Center of Applied Genomics at the Hospital for Sick Children (Toronto, Canada), showed absence of B10 alleles outside of Chr12 intervals. All of the mouse strains were maintained in the animal facility within the Maisonneuve-Rosemont Hospital Research Center. The Maisonneuve-Rosemont Hospital ethics committee, overseen by the Canadian Council for Animal Protection, approved the experimental procedures.
Flow cytometry
Spleens and lymph nodes were processed as described (Collin et al. 2014). Skin-draining lymph nodes include a pool of axial, brachial, and inguinal lymph nodes. DN T cells were identified as CD4- CD8- 3A9 TCR+ cells. Each flow cytometry sample was assessed individually for quality. Some samples were excluded either based on poor quality staining or low cell viability. In the 176 3A9 TCR transgenic F2 mice for the linkage analysis, all spleen samples passed the quality control and were included. Five samples from skin-draining lymph nodes and 21 samples from pancreatic lymph nodes did not pass the quality control, such that 171 and 155 samples were included for skin-draining and pancreatic lymph nodes, respectively. Pancreatic lymph nodes are technically more difficult to harvest, and cell viability is influenced by the degree of pancreatic tissue tear during extraction, explaining the higher number of pancreatic lymph nodes that did not pass the quality control.
Linkage analysis
Genomic tail DNA of 3A9 TCR transgenic F2 mice were subjected to the Illumina mouse low-density linkage panel, testing 377 SNPs, serviced through The Center for Applied Genomics at the Hospital for Sick Children in Toronto. Samples were processed in two batches: the first batch contained 96 samples, and the second batch included 80 samples. Marker location (in Mb) was determined using the NCBI build m37. The LOD scores were obtained as described (Collin et al. 2014). The significant and suggestive thresholds are based on permutation testing (n = 10 000, significant and suggestive loci exceeded the 95th percentile, P ≤ 0.05 and the 37th percentile P ≤ 0.63, respectively). The data for chromosome 5 was excluded from our analysis, because the 3A9 TCR transgene is located on chromosome 5. Code has been deposited on Github repository (https://github.com/slesageLab/DNT_F2_Collin).
Diabetes incidence
In spontaneous mouse models of autoimmune diabetes, such as in the 3A9 TCR:insHEL mice, the degree of insulitis and the progression to overt diabetes is stochastic (Anderson and Bluestone 2005; Chen et al. 2018; Lesage et al. 2002; Mullen 2017). To quantify the diabetes incidence, 3A9 TCR:insHEL female mice were monitored daily for overt signs of diabetes and every 2 weeks for urine glucose levels. None of the 3A9 TCR single transgenic mice developed diabetes. After two successive urine glucose positive tests, overt diabetes was confirmed by blood glucose levels > 17 mmol/L. The first urine glucose positive test defines the time of diabetes onset. The mice were sacrificed within 1 week of detection of high blood glucose or when they reached > 34 weeks of age. Pancreases from mice in the diabetes incidence as well as non-diabetic 8- to 14-week-old mice were collected, conserved in formalin for at least 48 h at room temperature, and sent for paraffin inclusion. H&E and quantification of anti-HEL autoantibody levels were performed as described (Collin et al. 2015; Collin et al. 2014).
Statistics
Comparisons of two experimental groups were tested for significance using a non-parametric Mann–Whitney U test with a minimal threshold of 0.05. Significance for the F2 genotype distribution differences was tested with a one-way ANOVA test and Bonferroni post hoc test. Statistical significance of the differences between the diabetes incidence curves was determined with the Mantel-Cox log rank test with a minimal threshold of 0.05. Statistical analyses and the F2 distribution were obtained using the SPSS 19.0 software (IBM, Montreal, Québec, Canada).
Results
Diabetes-susceptible mice have a lower proportion of DN T cells, and injecting DN T cells reduces diabetes incidence in diabetogenic TCR transgenic mouse models (Dugas et al. 2010; Ford et al. 2007). This suggests that their abundance contributes to autoimmune resistance (Collin et al. 2014; Dugas et al. 2010; Dugas et al. 2014; Hillhouse et al. 2010). To identify the genetic loci that determine the proportion of DN T cells in lymphoid organs, we had previously generated 176 3A9 TCR transgenic F2 mice from a B10.BR to NOD.H2k outcross (Collin et al. 2014). We had focused our linkage analysis of 3A9 DN T cell proportion to the skin-draining lymph nodes of F2 mice (Collin et al. 2014). In addition to skin-draining lymph nodes, we had performed flow cytometry on the spleen and pancreatic lymph nodes. We have returned to this dataset and analyzed the proportion of 3A9 DN T cells in the spleen and in the pancreatic lymph nodes. Similar to our previous observations in the skin-draining lymph nodes (Collin et al. 2014), we find a normal distribution of the proportion of 3A9 DN T cells in both the spleen and pancreatic lymph nodes (Fig. 1a, b), supporting the view that 3A9 DN T cell proportion is regulated by a multigenic trait.
Interim LOD score analysis suggests that a locus on chromosome 12 is linked to 3A9 DN T cell proportion in the spleen and lymph nodes. The distribution of the 3A9 TCR F2 cohort relative to the proportion of 3A9 DN T cells in the spleen of 176 F2 mice (a) and the pancreatic lymph nodes of 155 F2 mice (b). Midway genome-wide LOD score plot (R/Qtl) for the proportion of 3A9 DN T cells in the spleen of 96 F2 mice (significant threshold is 3.64, suggestive threshold is 2.17) (c), in the skin-draining lymph nodes of 95 F2 mice (suggestive threshold is 2.17) (d), and in the pancreatic lymph nodes of 94 F2 mice (significant threshold is 3.58, suggestive threshold is 2.16) (e). The data presented in panel (d) is an interim analysis of previously published data (Collin et al. 2014). The full and dotted lines indicate the significant (P = 0.05) and the suggestive threshold, respectively
To perform the linkage analysis, we subjected the DNA samples of the 3A9 TCR transgenic F2 mice to the Illumina Golden Gate low-density platform, which provides a genome-wide SNP genotyping. As the platform can run 96 samples at a time, the 176 samples were run in two distinct batches. The first batch was processed as soon as we had collected DNA from the first 96 mice. A midway analysis of this first batch showed a significant linkage of the proximal region of chromosome 12 to the proportion of 3A9 DN T cells in the spleen (Fig. 1c). This same locus was suggestively linked to the proportion of 3A9 DN T cells in the skin draining and pancreatic lymph nodes (Fig. 1d, e). In addition to chromosome 12, we found that other loci were suggestively linked to 3A9 DN T cell proportion, in line with a multigenic trait. Because we observed that a locus on chromosome 12 was linked to the proportion of 3A9 DN T cells in the spleen, skin-draining lymph nodes, and pancreatic lymph nodes, we initiated the generation of a NOD mouse congenic for this locus on chromosome 12 to determine whether B10 alleles at this locus would be sufficient to impact the proportion of 3A9 DN T cells on the NOD background.
Meanwhile, we phenotyped and genotyped another 80 3A9 TCR transgenic F2 mice. In the combined LOD score for all 176 mice, the chromosome 12 locus was no longer linked to 3A9 DN T cell proportion in the spleen, but it remained suggestively linked to the proportion of 3A9 DN T cells in both the skin-draining and pancreatic lymph nodes (Fig. 2) (Collin et al. 2014). Additionally, the Idd2 locus on mouse chromosome 9 was strongly linked to 3A9 DN T cell proportion in both skin-draining and pancreatic lymph nodes, with suggestive linkage in the spleen (Fig. 2) (Collin et al. 2014). Hence, we generated a 3A9 TCR NOD.H2k-Idd2 congenic mouse and showed that B10 alleles at the Idd2 locus increased 3A9 DN T cell proportion (Collin et al. 2014). Other suggestive loci linked to 3A9 DN T cell proportion include loci on chromosomes 1 (skin-draining LN), chromosome 2 (Idd13, lymph nodes), chromosome 4 (lymph nodes), chromosome 6 (spleen and lymph nodes), and chromosome 18 (pancreatic lymph nodes).
3A9 DN T cell proportion is a multigenic trait. Genome-wide LOD score plot (R/Qtl) for the proportion of 3A9 DN T cells in the spleen of 176 F2 mice (significant threshold is 3.52, suggestive threshold is 2.10) (a), in the skin-draining lymph nodes of 171 F2 mice (significant threshold is 3.52, suggestive threshold is 2.12) (b), or in the pancreatic lymph nodes of 155 F2 mice (significant threshold is 3.56, suggestive threshold is 2.13) (c) is shown. The full and dotted lines indicate the significant (P = 0.05) and the suggestive threshold, respectively. The data in panel (b) was previously published (Collin et al. 2014)
To determine the potential contribution of these loci in defining the proportion of 3A9 DN T cells, we segregated the 3A9 TCR transgenic F2 mice according to their genotype for each of the suggestive linkages (Fig. 3). We selected the SNPs showing the highest LOD score for all of the suggestive loci. For some of these SNPs, the average proportion of 3A9 DN T cells was higher in 3A9 TCR transgenic F2 homozygous for NOD alleles when compared to B10 alleles. This was the case for the SNPs rs3658044 on chromosome 1, rs3023025 on chromosome 4, and rs6320743 on chromosome 18 (Fig. 3). In contrast, segregation of 3A9 TCR transgenic F2 mice based on B10 and NOD alleles for SNPs on chromosomes 2, 6, and 12 concurred with the phenotype observed in the parental strains, namely, that 3A9 TCR transgenic F2 mice bearing NOD alleles at these loci had fewer 3A9 DN T cells than those carrying B10 alleles for all lymphoid organs (Fig. 3). Altogether, these results demonstrate that the proportion of 3A9 DN T cells is determined by a complex trait.
Complex gene interactions regulate the proportion of 3A9 DN T cells in lymphoid organs. 3A9 DN T cell proportion in F2 mice segregated according to their genotype (a) in the spleen (n = 176), (b) skin draining lymph nodes (n = 171), and (c) pancreatic lymph nodes (n = 155) for the indicated SNP. B/B, F2 mice homozygous for B10.BR alleles (black circles); B/N, F2 mice heterozygous for B10.BR and NOD.H2k alleles (grey diamonds); N/N, F2 mice homozygous for NOD.H2k alleles (empty circles) for each indicated SNP. The SNP chromosomal position is indicated in the figure based on NCBI build m37. Each dot represents data for an individual mouse. P value, *< 0.05, **< 0.01, ***< 0.001 according to one way ANOVA with Bonferroni post hoc tests
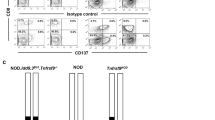
NOD.H2k-Chr12 mice were generated by backcrossing for eight generations (Collin et al. 2015). The resulting backcross yielded three strains. The congenic strain carrying the longest interval was named NOD.H2k-Chr12L, where L stands for long. The two other strains were respectively named NOD.H2k-Chr12P and NOD.H2k-Chr12D, where P and D stand for proximal and distal, respectively. Various genetic markers were used to define the congenic intervals by PCR genotyping (Fig. 4).
Delimitation of the congenic intervals. The B10.BR congenic intervals in the 3A9 TCR NOD. H2k-Chr12L congenic line and Chr12P and Chr12D congenic sublines, based on microsatellite marker testing by PCR. The markers are listed on the figure, and marker position is based on NCBI build m37. The chromosome 12 is fully depicted for all three mouse strains. White depicts regions of NOD origin, black of B10 origin, and grey of undefined origin
The Chr12D locus almost perfectly coincides with the Nbwa1 locus linked to autoantibody production (Collin et al. 2015; Rigby et al. 2004). We previously showed that 3A9 TCR:insHEL NOD.H2k-Chr12D are partially protected from diabetes and this locus affects antibody responses (Collin et al. 2015). To determine if Chr12L and Chr12P also influenced diabetes incidence and antibody responses, we crossed 3A9 TCR NOD.H2k-Chr12L and -Chr12P to insHEL NOD.H2k to obtain 3A9 TCR:insHEL NOD.H2k-Chr12L and -Chr12P congenic lines. We monitored the stochastic transition to overt diabetes in 3A9 TCR:insHEL NOD.H2k-Chr12L and -Chr12P female mice. In contrast to the 3A9 TCR:insHEL NOD.H2k-Chr12D strain, diabetes incidence in 3A9 TCR:insHEL NOD.H2k-Chr12L mice was not significantly different to the incidence observed in 3A9 TCR:insHEL NOD.H2k mice (Fig. 5a). Surprisingly, the incidence of diabetes was higher in 3A9 TCR:insHEL NOD.H2k-Chr12P relative to 3A9 TCR:insHEL NOD.H2k mice. Seven of eight 3A9 TCR:insHEL NOD.H2k-Chr12P mice were diabetic at 18 weeks of age, whereas only two of sixteen 3A9 TCR:insHEL NOD.H2k mice were diabetic at this same time point (Fig. 5a). In addition, B10 alleles at the Chr12P locus had a dominant effect as the diabetes incidence of eighteen 3A9 TCR:insHEL NOD.H2k-Chr12PB10/NOD female mice heterozygous at the Chr12P locus paralleled that of 3A9 TCR:insHEL NOD.H2k-Chr12P mice (Fig. 5a). The accelerated diabetes onset and increased diabetes incidence in 3A9 TCR:insHEL NOD.H2k-Chr12P mice relative to 3A9 TCR:insHEL NOD.H2k mice were neither a consequence of more pronounced immune cell infiltration near the pancreatic islets (Fig. 5b, c), nor due to increased anti-HEL IgG autoantibody levels (Fig. 5d). Together, these data suggest that B10 alleles at two juxtaposed loci on chromosome 12 have opposite impacts on diabetes incidence in 3A9 TCR:insHEL NOD.H2k mice.
B10 alleles at the Chr12P locus enhance diabetes incidence. (a) Diabetes incidence is shown for female 3A9 TCR:insHEL mice bearing the following genetic background: B10.BR (n = 8), NOD.H2k (n = 16), NOD.H2k-Chr12L (n = 18), NOD.H2k-Chr12PB/B (n = 8), and NOD.H2k-Chr12PB/N (n = 18), where B/B and B/N denote homozygous B10 and heterozygous B10/NOD Chr12P alleles. Logrank (Mantel-Cox) test. (b) The degree of islet infiltration is shown for non-diabetic 3A9 TCR:insHEL mice of 8–14 weeks of age (B10.BR (176 islets from 7 mice, age 10.4 ± 1.4 weeks), NOD.H2k (174 islets from 11 mice, age 10.0 ± 1.5 weeks), NOD.H2k-Chr12D (102 islets from 4 mice, age 8.6 ± 0.8 weeks), and NOD.H2k-Chr12PB/B (102 islets from 5 mice, age 9.4 ± 0.8). Note that as for the diabetes incidence, the severity of islet infiltration is stochastic, in mouse models of spontaneous autoimmune diabetes. (c) The degree of islet infiltration is shown for mice included in diabetes incidence study; B10.BR (170 islets from 6 mice, age 31.5 ± 8.8 weeks), NOD.H2k (53 islets from 4 mice, age 27.1 ± 4.1 weeks), NOD.H2k-Chr12D (86 islets from 5 mice, age 28.9 ± 9.2 weeks), and NOD.H2k-Chr12PB/B (53 islets from 4 mice, age 15.6 ± 1.0 weeks). The pancreases were harvested within 1 week of detection of high blood glucose levels or when the mice reached 34 weeks of age. Islet infiltration was assessed with one histology slide per mouse, containing 1 to 4 non-consecutive cuts per slide. (d) Anti-HEL IgG serum levels were quantified by ELISA in non-diabetic mice from the indicated strains. All mice were aged between 7.8 and 16.6 weeks, with an average of 9.7 ± 1.8 weeks. Titers are expressed in arbitrary units relative to a reference immune sera set at 1 unit. Each dot represents the data for one mouse. Mean ± SEM are shown. The dashed line represents the detection threshold of the ELISA. The data for the NOD.H2k-Chr12L and Chr12P strains was obtained contemporaneously with previous work that focused on defining the immunogenetic regulation of DN T cell proportion and the impact of B10 alleles on chromosome 12 in modulating antibody production and affinity (Collin et al. 2014; Collin et al. 2015). As such, some of the histology, autoantibody, and diabetes incidence data for B10.BR and NOD.H2k control mice are replicates of those previously published (Collin et al. 2014; Collin et al. 2015)
The Chr12 locus was originally identified by linkage to the proportion of 3A9 DN T cells in lymphoid organs. We thus next quantified the proportion of 3A9 DN T cells in the three different congenic strains. The proportion and absolute numbers of 3A9 DN T cells in the spleen and lymph nodes of all three 3A9 TCR NOD.H2k-Chr12 congenic strains was comparable to that of 3A9 TCR NOD.H2k mice (Fig. 6a, b), except for the absolute number of 3A9 DN T cells, which was marginally higher for 3A9 TCR NOD.H2k-Chr12L mice relative to the 3A9 TCR NOD.H2k parental strain (Fig. 6b). Although statistically significant, this difference is primarily driven by two out of eighteen 3A9 TCR NOD.H2k-Chr12L mice for which the proportion of 3A9 DN T cells exceeded 5%, well above the 2.8 ± 0.5% average. The statistical difference is lost when these outliers are removed. These results suggest that B10 alleles at the Chr12 locus do not significantly increase 3A9 DN T cell proportion in lymphoid organs.
The Chr12 locus does not impact the proportion and number of 3A9 DN T cells in the spleen and lymph nodes. (a) The proportion and (b) absolute number of 3A9 DN T cells are shown for the spleen (left), skin-draining lymph nodes (middle), and pancreatic draining lymph nodes (right) of 3A9 TCR NOD.H2k, 3A9 TCR NOD.H2k-Chr12L, 3A9 TCR NOD.H2k-Chr12D, and 3A9 TCR NOD.H2k-Chr12P mice. Each dot represents data from one mouse. The percentage of 3A9 DN T cells for both male and female mice is pooled as it is comparable for both sexes in all four strains and does not vary within the age range tested (not shown). The age range and percentage of female mice for each strain are as follows: spleen, 3A9 TCR NOD.H2k (n = 12, 8.9 to 13.1 week old, 42% female), 3A9 TCR NOD.H2k-Chr12L (n = 19, 7.7 to 11.7 week old, 68% female), 3A9 TCR NOD.H2k-Chr12D (n = 13, 7.7 to 11.7 week old, 53% female), and 3A9 TCR NOD.H2k-Chr12P (n = 16, 8 to 9.6 week old, 50% female); skin-draining lymph nodes, 3A9 TCR NOD.H2k (n = 10, 8.9 to 13.1 week old, 50% female), 3A9 TCR NOD.H2k-Chr12L (n = 18, 7.7 to 11.7 week old, 67% female), 3A9 TCR NOD.H2k-Chr12D (n = 12, 7.7. to 11.1 week old, 50% female), and 3A9 TCR NOD.H2k-Chr12P (n = 14, 8 to 9.6 week old, 57% female); and pancreatic lymph nodes, 3A9 TCR NOD.H2k (n = 5, 8.9 to 13.1 week old, 40% female), 3A9 TCR NOD.H2k-Chr12L (n = 5, 8 to 11 week old, 80% female), 3A9 TCR NOD.H2k-Chr12D (n = 5, 7.7. to 9.4 week old, 20% female), and 3A9 TCR NOD.H2k-Chr12P (n = 7, 8 to 9.3 week old, 42% female). *P < 0.05 based on an ANOVA with Bonferroni
As the proportion of 3A9 DN T cells is a multigenic trait, we decided to revisit the data from the 3A9 TCR transgenic F2 mice to determine if B10 alleles at the Chr12 locus interacted with other loci to determine the proportion of 3A9 DN T cells in lymphoid organs. The two-dimensional LOD scores revealed clear interactions between Idd2 (Chr9) and Idd13 (Chr2) (Fig. 7). This is in line with our previously published work (Collin et al. 2018). Based on these two dimensional plots, the Chr12 locus did not significantly interact with the Idd2 locus, while it interacted modestly with the Idd13 locus linked to the proportion of 3A9 DN T cells in the skin draining lymph nodes (Fig. 7b). To test this interaction, we selected F2 mice homozygous for B10 alleles at the Idd13 locus and segregated these mice according to their B10 or NOD genotype at the SNP providing the strongest LOD score within the Chr12 locus. In skin-draining lymph nodes, 3A9 TCR F2 mice homozygous for B10 alleles at both Idd13 and Chr12 have more 3A9 DN T cells than those bearing homozygous NOD alleles at the Chr12 locus (Fig. 7b). In contrast, the genotype at Chr12 did not impact the proportion of 3A9 DN T cells in F2 mice bearing B10 alleles at the Idd2 locus (Fig. 7b, c). Based on these results, we can conclude that B10 alleles at the Idd13 and Chr12 loci influence the proportion of 3A9 DN T cells, at least in the skin-draining lymph nodes. However, due to the low statistical power of our analyses, we cannot exclude the possibility of additional genetic interactions in defining 3A9 DN T cell proportion. Still, together, these results suggest a complex allelic regulation of 3A9 DN T cell proportion in lymphoid organs.
Complex gene interactions contribute to defining the proportion of 3A9 DN T cells. Left, Genome-wide two-dimensional LOD score analysis of F2 mice for 3A9 DN T cell proportion in (a) the spleen, (b) the skin-draining lymph nodes, and (c) the pancreatic lymph nodes. Middle, The proportion of 3A9 DN T cells from F2 mice bearing homozygous B10 alleles at the rs3674264 (spleen) or the rs6193859 (lymph nodes) SNP on chromosome 2 and segregated according to their genotype on chromosome 12 at the rs3718992 (spleen and pancreatic lymph nodes)or the rs13481365 SNP (skin-draining lymph nodes). Right, The proportion of 3A9 DN T cells from F2 mice bearing homozygous B10 alleles at the rs3088463 (spleen) or at the CEL-9_49183636 (lymph nodes) SNP on chromosome 9 and segregated according to their genotype on chromosome 12 at rs3718992 (spleen and pancreatic lymph nodes) or the rs13481365 (skin-draining lymph node) SNP. **P < 0.01 Mann Whitney test
Discussion
DN T cells contribute to immune tolerance in various pathologies, including autoimmune diabetes (Dugas et al. 2010; Duncan et al. 2010; Ford et al. 2007; Hillhouse et al. 2013; Hillhouse and Lesage 2013; Juvet and Zhang 2012; Liu et al. 2016; Martina et al. 2015; Zhang et al. 2011). We had previously shown that the proportion of DN T cells in lymph nodes was significantly linked to Idd2 and suggestively linked to Idd13 (Collin et al. 2014). We had also found that the proportion of DN T cells in lymphoid organs of NOD congenic mice bearing diabetes resistance alleles at Idd2 and Idd13 was increased relative to NOD mice (Collin et al. 2018; Collin et al. 2014; Dugas et al. 2014). Following from these studies, we provide further evidence of the complex genetic regulation of DN T cells in lymphoid organs. First, we find that the distribution of 3A9 DN T cell proportion in spleen and pancreatic lymph nodes of F2 mice follows a Gaussian distribution. Second, we identify multiple additional genetic loci suggestively linked to 3A9 DN T cell proportion. Third, among these loci, we find that B10 alleles either positively or negatively impact the proportion of 3A9 DN T cells relative to NOD alleles. Fourth, the proportion of 3A9 DN T cells in NOD mice was not influenced by B10 alleles on chromosome 12. This was surprising considering that this locus showed suggestive linkage to 3A9 DN T cell proportion and that the F2 mice bearing B10 alleles at the corresponding suggestive SNP had a higher proportion of 3A9 DN T cells relative to F2 mice homozygous for NOD alleles at this SNP. Finally, joint LOD score analyses provide evidence of additional interactions between genetic loci determining the proportion of 3A9 DN T cells in lymphoid organs.
We performed a linkage analysis for 3A9 DN T cells in three different lymphoid organs, namely, the spleen, skin-draining lymph nodes, and pancreatic lymph nodes. Of note, 3A9 TCR transgenic F2 mice never progress to diabetes, as they do not carry the insHEL transgene. In this context, one could assume that skin-draining lymph nodes and pancreatic lymph nodes are somewhat equivalent; in the absence of insulitis or diabetes, the pancreatic lymph nodes may resemble any other lymph node. Still, each lymph node has its unique properties by draining specific tissues (Esterhazy et al. 2019; Thomson and McCoy 2019). Also, one cannot exclude that some F2 mice may exhibit some degree of pancreatic inflammation driven by NOD alleles, independently of the TCR transgene. Regardless, the linkage analysis in skin-draining and pancreatic lymph nodes both reveal that Idd2 is most tightly linked to 3A9 DN T cell proportion, with additional linkage to Idd13 and a locus on mouse chromosome 4. In contrast, the linkage analysis in the spleen revealed a single locus on chromosome 6, barely reaching the significance threshold. This same locus is also suggestively linked to 3A9 DN T cell proportion in lymph nodes. The lower sensitivity in detecting loci in the spleen could be due to many factors. For one, the lower proportion of T cells in the spleen relative to the lymph nodes may decrease the sensitivity in separating the influence of B10 and NOD alleles on the phenotype. Second, spleen and lymph node tissue homeostasis are not guided by the same factors. Third, the immune cell composition of the spleen and lymph nodes differ such that the spleen holds a higher proportion of NK cells, dendritic cells, and other myeloid cells than the lymph nodes, which could influence the distribution of 3A9 DN T cells. Performing concurrent linkage analyses in the spleen and lymph nodes can inform on the distinct regulation in each of these lymphoid tissues.
Beyond the complex genetic regulation of DN T cells, we also observed epistasis with the chromosome 12 locus in regard to autoimmune diabetes susceptibility. Of interest, this locus does not coincide with any Idd loci. While the Chr12L locus did not significantly affect diabetes incidence, Chr12D locus conferred protection to the 3A9 TCR:insHEL NOD.H2k mice, the Chr12P locus enhanced susceptibility to disease. Although it is uncommon that non-NOD alleles enhance susceptibility to diabetes, it is not without precedence. In the 3A9 TCR:insHEL NOD.H2k transgenic mouse model, we found that congenic mice bearing resistance alleles at either the Idd9 or the Idd13 locus both exhibited enhanced diabetes susceptibility (Hillhouse et al. 2016). Similar observations have also been reported in non-TCR transgenic NOD mice, where the Idd14 locus on chromosome 13 displays a similar behavior to the Chr12 locus (Wang et al. 2014). Indeed, generation of Idd14 subcongenic lines revealed the adjacent Idd31 locus. NOD.B6-Idd31 congenic mice show a lower incidence of diabetes than NOD mice only if the Idd14 locus bears NOD alleles, whereas NOD.B6-Idd14 mice have a higher incidence of diabetes than the parental NOD strain (Wang et al. 2014). Complex genetic interactions have also been observed in other congenic sublines, where some non-NOD alleles mask the effect of other diabetes-protective alleles on the same chromosomes. However, these masking loci have not been reported to enhance susceptibility in NOD mice (Hamilton-Williams et al. 2013; Hollis-Moffatt et al. 2005; Morin et al. 2006; Rogner et al. 2001). Hence, genetic epistasis, although challenging to unravel, likely contributes significantly to defining complex traits, such as autoimmune disease susceptibility (Carlborg and Haley 2004).
Considering gene expression in various immune cell types in the context of predictive SNPs reveals that most genetic variants linked to type 1 diabetes susceptibility in humans map to the humoral arm of the immune response (Farh et al. 2015). This observation is in line with the fact that islet autoantibodies serve to predict type 1 diabetes onset (Orban et al. 2009; Steck et al. 2011; Ziegler et al. 2013). Interestingly, the humoral response also significantly contributes to diabetes onset in the 3A9 TCR:insHEL NOD.H2k transgenic model (Silva et al. 2011). In line with the impact of a humoral response in this autoimmune model, we had observed that injection of 3A9 DN T cells results in a decreased diabetes incidence coupled with a decrease in anti-HEL autoantibody levels (Dugas et al. 2010). In 3A9 TCR:insHEL NOD.H2k mice, B10 resistance alleles at the Idd2 locus lead to an increase in 3A9 DN T cells, a decrease in diabetes susceptibility, and a decrease in autoantibody levels (Collin et al. 2014). As 3A9 DN T cells are able to eliminate B cells (Hillhouse et al. 2010), increasing the number of 3A9 DN T cells likely impedes autoantibody production, which consequently results in a reduced diabetes incidence in Idd2 congenic mice. In contrast, this association in phenotypes is not observed in Chr12 congenic mice. Indeed, although the Chr12 locus was suggestively linked to the proportion of 3A9 DN T cells in an interim analysis, we did not observe an increase in 3A9 DN T cells in 3A9 TCR NOD.H2k mice congenic for B10 alleles at the Chr12 locus or subloci. However, we previously reported that 3A9 TCR:insHEL NOD.H2k-Chr12D mice exhibit a lower incidence of diabetes and produce fewer autoantibodies than 3A9 TCR:insHEL NOD.H2k mice (Collin et al. 2015). Antibody affinity maturation is also affected in the Chr12D congenic mice (Collin et al. 2015). Yet, autoantibody production is not enhanced in 3A9 TCR:insHEL NOD.H2k-Chr12P mice. Altogether, these results uncouple the genetic regulation of autoantibody production and DN T cell proportion in the context of this transgenic model.
The Chr12 congenic mice were generated based on an interim LOD score analysis, which linked the Chr12 locus to DN T cell proportion in the spleen and showed suggestive linkage in both skin-draining and pancreatic lymph nodes. Although interim analyses have low statistical power, we still opted to undertake the time-consuming generation of these congenic mice. Indeed, interim analyses can reveal unexpected loci that cannot be perceived due to complex genetic epistasis (Keller et al. 2019). In addition, we had found that it coincided with the Nbwa1 locus, linked to autoantibody production in the context of lupus (Rigby et al. 2004), suggesting that this locus could impact autoantibody production in our setting as well. Although the Chr12 congenic line and sublines did not show variations in 3A9 DN T cell proportion, the Chr12D subline influences autoantibody production and both Chr12D and Chr12P subline had opposite impacts on autoimmune diabetes incidence, revealing genetic epistasis in the autoimmune diabetes susceptibility. This study highlights the strength of congenic sublines to unravel genetic epistasis.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Allen PM, Matsueda GR, Evans RJ, Dunbar JB Jr, Marshall GR, Unanue ER (1987) Identification of the T-cell and Ia contact residues of a T-cell antigenic epitope. Nature 327:713–5
Anderson MS, Bluestone JA (2005) THE NOD MOUSE: a model of immune dysregulation. Annu Rev Immunol 23:447–85
Carlborg O, Haley CS (2004) Epistasis: too often neglected in complex trait studies? Nat Rev Genet 5:618–25
Chen YG, Mathews CE, Driver JP (2018) The role of NOD mice in type 1 diabetes research: lessons from the past and recommendations for the future. Front Endocrinol (Lausanne) 9:51
Chen YG, Scheuplein F, Osborne MA, Tsaih SW, Chapman HD, Serreze DV (2008) Idd9/11 genetic locus regulates diabetogenic activity of CD4 T-cells in nonobese diabetic (NOD) mice. Diabetes 57:3273–80
Collin R, Doyon K, Mullins-Dansereau V, Karam M, Chabot-Roy G, Hillhouse EE, Orthwein A, Lesage S (2018) Genetic interaction between two insulin-dependent diabetes susceptibility loci, Idd2 and Idd13, in determining immunoregulatory DN T cell proportion. Immunogenetics 70:495–509
Collin R, Dugas V, Chabot-Roy G, Salem D, Zahn A, Di Noia JM, Rauch J, Lesage S (2015) Autoimmunity and antibody affinity maturation are modulated by genetic variants on mouse chromosome 12. J Autoimmun 58:90–9
Collin R, Dugas V, Pelletier AN, Chabot-Roy G, Lesage S (2014) The mouse Idd2 locus is linked to the proportion of immunoregulatory double-negative T cells, a trait associated with autoimmune diabetes resistance. J Immunol 193:3503–12
Driver JP, Serreze DV, Chen YG (2011) Mouse models for the study of autoimmune type 1 diabetes: a NOD to similarities and differences to human disease. Semin Immunopathol 33:67–87
Dugas V, Beauchamp C, Chabot-Roy G, Hillhouse EE, Lesage S (2010) Implication of the CD47 pathway in autoimmune diabetes. J Autoimmun 35:23–32
Dugas V, Liston A, Hillhouse EE, Collin R, Chabot-Roy G, Pelletier AN, Beauchamp C, Hardy K, Lesage S (2014) Idd13 is involved in determining immunoregulatory DN T-cell number in NOD mice. Genes Immun 15:82–7
Duncan B, Nazarov-Stoica C, Surls J, Kehl M, Bona C, Casares S, Brumeanu TD (2010) Double negative (CD3+ 4- 8-) TCR alphabeta splenic cells from young NOD mice provide long-lasting protection against type 1 diabetes. PLoS One 5:e11427
Esteban LM, Tsoutsman T, Jordan MA, Roach D, Poulton LD, Brooks A, Naidenko OV, Sidobre S, Godfrey DI, Baxter AG (2003) Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol 171:2873–8
Esterhazy D, Canesso MCC, Mesin L, Muller PA, de Castro TBR, Lockhart A, ElJalby M, Faria AMC, Mucida D (2019) Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature 569:126–130
Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA, Bernstein BE (2015) Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518:337–43
Ford MS, Chen W, Wong S, Li C, Vanama R, Elford AR, Asa SL, Ohashi PS, Zhang L (2007) Peptide-activated double-negative T cells can prevent autoimmune type-1 diabetes development. Eur J Immunol 37:2234–41
Fox CJ, Paterson AD, Mortin-Toth SM, Danska JS (2000) Two genetic loci regulate T cell-dependent islet inflammation and drive autoimmune diabetes pathogenesis. Am J Hum Genet 67:67–81
Hamilton-Williams EE, Bergot AS, Reeves PL, Steptoe RJ (2016) Maintenance of peripheral tolerance to islet antigens. J Autoimmun 72:118–25
Hamilton-Williams EE, Rainbow DB, Cheung J, Christensen M, Lyons PA, Peterson LB, Steward CA, Sherman LA, Wicker LS (2013) Fine mapping of type 1 diabetes regions Idd9.1 and Idd9.2 reveals genetic complexity. Mamm Genome 24:358–75
Hill NJ, Lyons PA, Armitage N, Todd JA, Wicker LS, Peterson LB (2000) NOD Idd5 locus controls insulitis and diabetes and overlaps the orthologous CTLA4/IDDM12 and NRAMP1 loci in humans. Diabetes 49:1744–7
Hillhouse EE, Beauchamp C, Chabot-Roy G, Dugas V, Lesage S (2010) Interleukin-10 limits the expansion of immunoregulatory CD4-CD8- T cells in autoimmune-prone non-obese diabetic mice. Immunol Cell Biol 88:771–80
Hillhouse EE, Delisle JS, Lesage S (2013) Immunoregulatory CD4(-)CD8(-) T cells as a potential therapeutic tool for transplantation, autoimmunity, and cancer. Front Immunol 4:6
Hillhouse EE, Lesage S (2013) A comprehensive review of the phenotype and function of antigen-specific immunoregulatory double negative T cells. J Autoimmun 40:58–65
Hillhouse EE, Liston A, Collin R, Desautels E, Goodnow CC, Lesage S (2016) TCR transgenic mice reveal the impact of type 1 diabetes loci on early and late disease checkpoints. Immunol Cell Biol 94:709–13
Hollis-Moffatt JE, Hook SM, Merriman TR (2005) Colocalization of mouse autoimmune diabetes loci Idd21.1 and Idd21.2 with IDDM6 (human) and Iddm3 (rat). Diabetes 54:2820–5
Hunter K, Rainbow D, Plagnol V, Todd JA, Peterson LB, Wicker LS (2007) Interactions between Idd5.1/Ctla4 and Other Type 1 Diabetes Genes. J Immunol 179:8341–9
Juvet SC, Zhang L (2012) Double negative regulatory T cells in transplantation and autoimmunity: recent progress and future directions. J Mol Cell Biol 4:48–58
Kachapati K, Adams DE, Wu Y, Steward CA, Rainbow DB, Wicker LS, Mittler RS, Ridgway WM (2012) The B10 Idd9.3 locus mediates accumulation of functionally superior CD137(+) regulatory T cells in the nonobese diabetic type 1 diabetes model. J Immunol 189:5001–15
Keller MP, Rabaglia ME, Schueler KL, Stapleton DS, Gatti DM, Vincent M, Mitok KA, Wang Z, Ishimura T, Simonett SP, Emfinger CH, Das R, Beck T, Kendziorski C, Broman KW, Yandell BS, Churchill GA, Attie AD (2019) Gene loci associated with insulin secretion in islets from non-diabetic mice. J Clin Invest 130:4419–4432
Lesage S, Hartley SB, Akkaraju S, Wilson J, Townsend M, Goodnow CC (2002) Failure to censor forbidden clones of CD4 T cells in autoimmune diabetes. J Exp Med 196:1175–88
Liston A, Lesage S, Gray DH, O’Reilly LA, Strasser A, Fahrer AM, Boyd RL, Wilson J, Baxter AG, Gallo EM, Crabtree GR, Peng K, Wilson SR, Goodnow CC (2004) Generalized resistance to thymic deletion in the NOD mouse: a polygenic trait characterized by defective induction of Bim. Immunity 21:817–30
Liu T, Cong M, Sun G, Wang P, Tian Y, Shi W, Li X, You H, Zhang D (2016) Combination of double negative T cells and anti-thymocyte serum reverses type 1 diabetes in NOD mice. J Transl Med 14:57
Maier LM, Wicker LS (2005) Genetic susceptibility to type 1 diabetes. Curr Opin Immunol 17:601–8
Martina MN, Noel S, Saxena A, Rabb H, Hamad AR (2015) Double Negative (DN) alphabeta T Cells: misperception and overdue recognition. Immunol Cell Biol 93:305–310
Morin J, Boitard C, Vallois D, Avner P, Rogner UC (2006) Mapping of the murine type 1 diabetes locus Idd20 by genetic interaction. Mamm Genome 17:1105–12
Mullen Y (2017) Development of the nonobese diabetic mouse and contribution of animal models for understanding type 1 diabetes. Pancreas 46:455–466
Orban T, Sosenko JM, Cuthbertson D, Krischer JP, Skyler JS, Jackson R, Yu L, Palmer JP, Schatz D, Eisenbarth G, Diabetes Prevention Trial-Type 1 Study G (2009) Pancreatic islet autoantibodies as predictors of type 1 diabetes in the Diabetes Prevention Trial-Type 1. Diabetes Care 32:2269–2274
Podolin PL, Pressey A, DeLarato NH, Fischer PA, Peterson LB, Wicker LS (1993) I-E+ nonobese diabetic mice develop insulitis and diabetes. J Exp Med 178:793–803
Polychronakos C, Li Q (2011) Understanding type 1 diabetes through genetics: advances and prospects. Nat Rev Genet 12:781–92
Ridgway WM, Peterson LB, Todd JA, Rainbow DB, Healy B, Burren OS, Wicker LS (2008) Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv Immunol 100:151–75
Rigby RJ, Rozzo SJ, Boyle JJ, Lewis M, Kotzin BL, Vyse TJ (2004) New loci from New Zealand Black and New Zealand White mice on chromosomes 4 and 12 contribute to lupus-like disease in the context of BALB/c. J Immunol 172:4609–17
Rogner UC, Boitard C, Morin J, Melanitou E, Avner P (2001) Three loci on mouse chromosome 6 influence onset and final incidence of type I diabetes in NOD.C3H congenic strains. Genomics 74:163–71
Sgouroudis E, Albanese A, Piccirillo CA (2008) Impact of protective IL-2 allelic variants on CD4+ Foxp3+ regulatory T cell function in situ and resistance to autoimmune diabetes in NOD mice. J Immunol 181:6283–92
Silva DG, Daley SR, Hogan J, Lee SK, Teh CE, Hu DY, Lam KP, Goodnow CC, Vinuesa CG (2011) Anti-islet autoantibodies trigger autoimmune diabetes in the presence of an increased frequency of islet-reactive CD4 T cells. Diabetes 60:2102–11
Steck AK, Johnson K, Barriga KJ, Miao D, Yu L, Hutton JC, Eisenbarth GS, Rewers MJ (2011) Age of islet autoantibody appearance and mean levels of insulin, but not GAD or IA-2 autoantibodies, predict age of diagnosis of type 1 diabetes: diabetes autoimmunity study in the young. Diabetes Care 34:1397–9
Tellier J, van Meerwijk JP, Romagnoli P (2006) An MHC-linked locus modulates thymic differentiation of CD4+CD25+Foxp3+ regulatory T lymphocytes. Int Immunol 18:1509–19
Theofilopoulos AN, Kono DH, Baccala R (2017) The multiple pathways to autoimmunity. Nat Immunol 18:716–724
Thomson CA, McCoy KD (2019) Not all lymph nodes are created equal. Immunity 51:12–14
Tsaih SW, Khaja S, Ciecko AE, MacKinney E, Chen YG (2013) Genetic control of murine invariant natural killer T cells maps to multiple type 1 diabetes regions. Genes Immun 14:380–6
Wang N, Elso CM, Mackin L, Mannering SI, Strugnell RA, Wijburg OL, Brodnicki TC (2014) Congenic mice reveal genetic epistasis and overlapping disease loci for autoimmune diabetes and listeriosis. Immunogenetics 66:501–6
Wicker LS, Todd JA, Peterson LB (1995) Genetic control of autoimmune diabetes in the NOD mouse. Annu Rev Immunol 13:179–200
Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, Chen SL, Rosa R, Cumiskey AM, Serreze DV, Gregory S, Rogers J, Lyons PA, Healy B, Smink LJ, Todd JA, Peterson LB, Wicker LS, Santamaria P (2007) Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet 39:329–37
Zhang D, Zhang W, Ng TW, Wang Y, Liu Q, Gorantla V, Lakkis F, Zheng XX (2011) Adoptive cell therapy using antigen-specific CD4-CD8- T regulatory cells to prevent autoimmune diabetes and promote islet allograft survival in NOD mice. Diabetologia 54:2082–92
Ziegler AG, Rewers M, Simell O, Simell T, Lempainen J, Steck A, Winkler C, Ilonen J, Veijola R, Knip M, Bonifacio E, Eisenbarth GS (2013) Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 309:2473–9
Acknowledgements
We thank all of the animal house staff for aid in maintaining the mouse colonies used in this study.
Funding
This work was supported by the Canadian Diabetes Association (Grant OG-3-13-4018) and the Canadian Institutes of Health Research (Grant PJT 159603) to SL. RC and ANP received scholarships from Diabète Québec and l’Université de Montréal. RC and VD received scholarships from the Fonds de recherche Québec-Santé. SL is a Research Scholars Emeritus from the Fonds de recherche Québec -Santé.
Author information
Authors and Affiliations
Contributions
RC acquired data, analyzed data, participated in the elaboration of the experimental plan, prepared the figures, and corrected and approved the manuscript. VD acquired data, generated the congenic mice, participated in the elaboration of the experimental plan, and corrected and approved the manuscript. ANP acquired data, analyzed data, participated in the elaboration of the experimental plan, and corrected and approved the manuscript. GCR acquired data and corrected and approved the manuscript. SL participated in the elaboration of the experimental plan, supervised the study, and wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The Maisonneuve-Rosemont Hospital ethics committee, overseen by the Canadian Council for Animal Protection, approved the experimental procedures.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Collin, R., Dugas, V., Pelletier, AN. et al. Evidence of genetic epistasis in autoimmune diabetes susceptibility revealed by mouse congenic sublines. Immunogenetics 73, 307–319 (2021). https://doi.org/10.1007/s00251-021-01214-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-021-01214-9