Abstract
The aim of this study was to establish a porcine physical map along the chromosome SSC7q by construction of BAC contigs between microsatellites Sw1409 and S0102. The SLA class II contig, located on SSC7q, was lengthened. Four major BAC contigs and 10 short contigs span a region equivalent to 800 cR measured by IMpRH7000 mapping. The BAC contigs were initiated by PCR screening with primers derived from human orthologous segments, extended by chromosome walking, and controlled and oriented by RH mapping with the two available panels, IMpRH7000Rad and IMNpRH12000Rad. The location of 43 genes was revealed by sequenced segments, either from BAC ends or PCR products from BAC clones. The 220 BAC end sequences (BES) were also used to analyze the different marks of evolution. Comparative mapping analysis between pigs and humans demonstrated that the gene organization on HSA6p21 and on SSC7p11 and q11–q14 segments was conserved during evolution, with the exception of long fragments of HSA6p12 which shuffled and spliced the SLA extended class II region. Additional punctual variations (unique gene insertion/deletion) were observed, even within conserved segments, revealing the evolutionary complexity of this region. In addition, 18 new polymorphic microsatellites have been selected in order to cover the entire SSC7p11–q14 region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
From the genomic point of view, interspecies sequencing and gene organization comparison are particularly effective for inferring genome function: Stronger conservation is generally interpreted as indicating more important function, and differences within functionally important regions should reflect adaptive evolution, as observed in the major histocompatibility complex (MHC) (Kulski et al. 2002).
Comparative analyses of human and porcine genomes have been performed at several levels. Cytogenetic studies by chromosome painting showed that porcine Chromosome 7 (SSC7) shares conserved synteny with human Chromosomes 6, 14, and 15 (Goureau et al. 1996). In particular, extensive conservation exists between the short arm of human Chromosome 6 which, along with other genes, harbors the HLA, and a segment covering the short arm of SSC7, where the pig MHC (SLA) class I and class III regions are located, plus a centromeric part of the SSC7 long arm including the SLA class II region (Smith et al. 1995). The porcine ImpRH7000Rad panel characterization confirmed this homology but only roughly localized the synteny breakpoint between human and pig chromosomes: HSA6p–HSA15q/HSA14 and SSC7q (Rink et al. 2002). Recently, we observed that a segment located close to the centromere in humans is located close to the SLA class II region in pigs, 600 cR7000 upstream of the expected position (Demeure et al. 2003). We set out to identify gene order rearrangement between human and pig HSA6p/SSC7q in this particular region on the basis of bacteria artificial chromosome (BAC) library screening and by using a more accurate IMNpRH12000Rad panel.
The importance of this particular genomic segment is associated with the presence of several quantitative trait loci (QTLs) in close vicinity of the SLA, affecting traits like intramuscular fat content (IMF), backfat thickness (BFT), growth, and carcass traits (Bidanel et al. 2001; Milan et al. 2002). The QTL-containing segment spans from the genetic SLA complex to the microsatellite marker S0102, which is located about 10 cM telomerically from the SLA class II region. The selection applied during the last two decades reduced the average carcass fatness with an associated decrease in the IMF, compromising meat quality for consumers. Since the QTLs present seem to act separately although they are located within the same genetic region (Milan et al. 2002; Nezer et al. 2002), we may expect to increase the IMF content without increasing BFT. Mass selection may also reduce the degree of polymorphism in the SLA. A fine map of this region will permit fine selection and, depending on the QTL position, we expect to maintain SLA polymorphism within the herds, possibly conferring a selective advantage over multiple-strain infections (Penn et al. 2002).
In an effort to improve the mapping resolution of this QTL candidate region on pig Chromosome 7, we constructed a porcine physical map that corresponds to the human 6p21.3–6p11.2 region using BAC clone alignment and radiation hybrid (RH) mapping. A detailed picture of gene content and gene order of the segment spanning the SLA class II segment over the q arm of the pig Chromosome 7 up to the S0102 microsatellite is presented.
Materials and methods
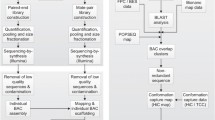
Primer design and pig BAC library screening
The INRA porcine BAC library SBAB-referenced (Rogel–Gaillard et al. 1999) was screened by PCR with primers derived from available human gene sequences. A first screening was performed to select genes located in the human HSA6p21.2–6p21.3 segment, namely, KE2, TAPBP, DAXX, HMGA1, NUDT3, (DIPP) ZNF76, PPARD, SRPK1, MAPK13, STK38 (NDR) CDKN1A (CDKI, WAF1, p21), PIM1, GLO1, TPX, NFYA, CCND3 (Table 1). The BAC library was also screened for a complementary set of genes mapped in HSA6p21.1, 12, and 11, already used to build the fine RH map. These included BMP5, BAG2, LANO (FLJ10775), GCLC, HCRTR2, GSTA3 (Demeure et al. 2003) and one gene, CEBPE, located on HSA14.
Primer design was based on highly conserved segments from the corresponding human and mouse genes and, more recently, on pig sequences directly, using the ICCARE (Interspecific Comparative Clustering and Annotation foR Ests) tool, which makes it possible to compare porcine expressed sequence tags (ESTs) to human gene sequences (http://genopole.toulouse.inra.fr/bioinfo/Iccare/). The PCR-based screening was performed in a total volume of 15 μl with 20 ng of superpool or single pool DNA, and with 20 ng of pig, human, and bacterial genomic DNA as positive and negative controls. PCR amplifications were carried out in PTC thermocyclers. Thermal cycling parameters were defined as follows: denaturation at 95°C for 5 min, followed by 30 cycles of (1) 95°C for 30 sec, (2) annealing temperature of 72°C for 30 sec, and (3) a final step at 72°C for 5 min. PCR products were analyzed on 2% agarose gels, electrophoresed in TBE buffer, and visualized by ethidium bromide staining (Rogel-Gaillard et al. 1999).
When primers derived from human sequences failed to amplify porcine genes like GLO and TPX, human DNA amplicons were used as radiolabeled probes to select BACs by hybridization with high-density nylon filters (Hybond N+, Amersham) covered by all BAC clones of the library.
The positive clones were controlled by PCR on several isolated colonies. DNA amplicons from the selected BAG clones were sequenced on an ABI 373 (Applied Biosystems) after purification (Jetquick columns, GENOMED). To control gene identity, sequences were compared with the GenBank database via the BLAST tool (www.ncbi.nlm. nih.gov/blast/bl2seq/bl2.html).
BAC end sequencing (BES)
Positive BAC clones were grown overnight at 37°C in 100 ml Lurie broth (LB) medium containing 12.5 μg/ml chloramphenicol and purified using a standard alkaline lysis procedure. BAC DNA was digested with EcoRV or PvuI and purified (kit Qiaex II, Qiagen). Approximately 1 μg of BAC DNA was used as template for 5′and 3′ sequencing, using fluorescence-labeled dideoxy-terminators in an ABI 373 automatic sequencer. Sequences were submitted to the EMBL databank (AJ628956–74, AJ629076–108, AJ629125–49, AJ629152–76, AJ629222–46, AJ629414–53).
Chromosome walking
The BAC library was screened with primers based on single-copy BES. Primers were designed with PRIMERS software (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). To confirm BAC overlapping, each primer couple was tested against each BAC belonging to the cluster. BAC clones were also sized by PFGE following NotI digestion as already described (Rogel-Gaillard et al. 1999).
The comparison of BES with the human orthologous working draft sequence, NT_007592_10 (gi:22061075), was performed by BLAST 2 sequences. When none or weak homology was observed, we performed alignments of BES with human and/or mammalian databanks, and we selected precise alignments using FASTA.
RH mapping
In order to control the relative position of the contigs on the porcine genome, BAC end sequences were mapped using ImpRH7000Rad (Yerle et al. 1998) and/or IMNpRH212000Rad (Yerle et al. 2002) RH panels. These panels have an estimated resolution of 26 kb/cR and 12.6 kb/cR, respectively (Genet et al. 2001; Yerle et al. 2002). For each contig, primers were developed in BAC end sequences. Additional primer pairs were used for syntenic breakpoints and when chimeric BAC statement had to be controlled. PCRs were carried out as previously described for BAC screening.
Vectors were submitted to the IMpRH web server at http://impRH.toulouse.inra.fr/ (Milan (et al. 2000) for an initial two-point assignment. The RH map was constructed with CarthaGene software (Schiex et al. 1997, 2001). The RH data from the two panels were merged and a 1000:1 framework map was built under a haploid model, by a stepwise locus-adding strategy using the triplet of markers whose order is the most likely as a starting point. The map was then improved by replacing some markers in order to determine contig order and orientation. The different provisional frameworks were checked using a simulated annealing algorithm to test inversion of map fragments, and a flips algorithm was used to test all local permutations in a window of six markers. The RH map was drawn with MapChart 2.0 (Voorrips 2002).
Identification of new microsatellites
BAC DNA was digested by Sau3AI and subcloned into the BamHI site of pUC18. The plasmid libraries were then screened for the presence of microsatellites with (TG)10 and (TC)10 oligonucleotide probes, labeled with [32P]ATP. Plasmid clones containing microsatellites were sequenced to determine the number of repeats and to obtain their flanking segments needed for primer design for further microsatellite amplification.
Results
General physical and RH maps
The previously described SLA class II contig located on SSC7q11 was the first milestone of the study. In order to design landmarks on the pig Chromosome 7q12–q14 region, we screened the BAC library with primer pairs derived from evolutionarily conserved genes adjacent to the human MHC class II region (HSA6p21 segments), namely, KE2, TAPBP, DAXX, HMGA1, NUDT3 (DIPP), ZNF76, PPARD, SRPK1, MAPK13, STK38 (NDR), CDKNIA (CDKI, WAF1, p21), PIM1, GLO1, TPX, NFYA, and CCND3. We obtained a first set of 11 clusters of porcine BACs that were further organized into overlapping clones and extended by both additional BAC screening with primers derived from additional anchor genes and chromosome walking. Basically, the selected BACs merged in seven contigs covering from 1 Mb to 200 kb (contigs 1–7, Fig. 1).
Comparative mapping between HSA6p and SSC7p11–q 14 using BAC contigs and RH maps. The dotted lines link genes with conserved positions, while genes included in rearranged fragments are linked by solid lines. The centromeres are represented by circles. The HSA6p map is based on human genome assembly data available on the NCBI web site (build version 10, www.ncbi.nlm.nih.gov/mapview/). The IMNpRH12000 map gives BAC end order and contig orientations. The IMpRH7000 map is extracted from Demeure et al. 2003 and gives gene order. It was completed by BAC ends markers located in the contigs.
The BAC library was also screened with specific primers for S0102, SW1409, SW2019, and SW1856 pig microsatellites in order to further integrate the physical and genetic maps. Four clusters of BACs were obtained, organized into overlapping clones (contigs 9–12), and assigned to the physical map draft (Fig. 1). Thus, several landmarks were assigned to the map of pig chromosome 7 within a 12.4-cM-long segment limited by markers SW1409 and S0102, and including the SLA region already described.
Screening of the BAC library for additional genes mapped in HSA6p12.1–p11 was performed and further used to build the fine RH map. These genes include BMP5, LANO, BAG2, GCLC, HCRTR, and GSTA3 (Demeure et al. 2003). Contig 8 was lengthened from GSTA3 which is closer to S0102 than GCLC and HCRTR. The contig derived from BMP5 and LANO screening lengthened the SLA class II in inverted orientation in pigs compared to humans and was located far from the orthologous human location. Contig 13, which was derived from the BAC selected by BAG2, is not linked to other segments, while the COL21A1 genes, which belong to the same human segment, have been identified in the centromeric end of contig 1. Contig 14 was derived from BAC selected to enclose SACM2L. This gene is located in HLA class II but we could not find it in either the class II contig or in contig 1 in the pig map.
The relative order of the resulting contigs was defined by RH mapping. The IMpRH7000 panel was tested with 28 markers derived from genes and BAC end sequences (BES). All together, 13 contigs were distributed within 900 cR7000 from the SLA class II contig extremity toward the telomere of the long arm of Chromosome 7. Some HLA class II genes, enclosed in contig 1 in pigs, are separated from SLA class II by a gap of 152.6 cR7000 (about 4 Mb) which included contigs 11–14 (Fig. 1).
The locations of the small contigs 5–9 were determined by building different local framework maps based on the IMpRH panels. The orders are in agreement with the results expected by comparative mapping. Contig 5 is composed of seven BACs where the GLO1 and TPX genes were localized. Contig 6 is composed of four BACs, two of which encode the NFYA gene, and one BES matched with the bovine segment encoding BYA (BoLA class IIb). Contig 7 contains the BYSL and CCND3 genes. Contig 8 is composed of two BACs, one of which encodes the GSTA3 gene. The last contig limited the telomeric part of the region of interest because it was mapped distal from S0102.
An additional primer pair derived from human CEBPE located on HSA14 was used to characterize pig BACs. The relevant clones were mapped on SSC7 at 22 cR7000 from TCRA by RH mapping, confirming previous reports indicating a synteny breakpoint between SSC7 and HSA6/HSA14, but outside the region ending with S0102 (data not shown).
In order to prevent chimeric clone selection and attempting to refine the location and the orientation of the major contigs on the porcine map, the second IMNpRH12000 panel was tested with 16 markers of contigs 1–4 and contig 14 which includes the SACM2L gene from the region II of MHC. As shown in Fig. 1, contigs 1–4, representing the four longer BAC alignments, were distributed onto 357 cR12000, on the basis of mapping 14 markers (BES, gene, or microsatellites) with the ImpRH12000 Contig 14 is separated by 15.2 cR12000 from contig 1 (around 191 kb). The gene content and comparative mapping of the long contigs are detailed below.
A total of 220 BES was used to align the pig physical map on the human draft sequence (NCBI accession NT_007592_10, gi:22061075). BLAST 2 sequences analysis revealed large blocks of conserved homology, although differences do exist. From telomere to centromere, the conserved segments concern contigs 4, 3, 2, and the telomeric part of contig 1, whereas the main differences are located on the centromeric end of contig 1 and in the BAC contigs located between contig 1 and the SLA class II region (Fig. 1).
Comparative mapping of contig 1
Contig 1 comprises 35 BAC clones covering more than 1 Mb (Fig. 2). GenBank accession numbers of BES are AJ629076–AJ629108, AJ629125–AJ629149. Initial BAC screening involved amplification of KE2, DAXX, TABP, HMGA1, and NUDT3 (DIPP) segments. After chromosome walking and BAC alignment, sequencing of clone ends allowed identification of eight additional gene segments. At the centromeric end of contig 1, five BES matched COL21A1 gene segments. Next to COL21A1, the RPS18, KE2, RAB2L, TAPBP, ZNF297, DAXX, SYNGAP1, ITPR3, HMGAl, and NUDT3 genes were sequentially mapped (Fig. 2). The S0665 (MS50) microsatellite was assigned to BACs 697D5 and 198G11 which bridge two sets of overlapping clones but represent a weak link in the contig construction (Fig. 2). RH mapping with the ImpRH12000 panel confirmed the location of S0665 and adjacent sequences (DAXX, 583C11-5, 1045D9-5, DIPP) reinforcing the BAC alignment (Fig. 1).
Representation of contig 1 and the BAC end sequence homologies with the HSA6p. The black small circles represent the sequenced BAC ends. With primers developed in those sequences, we determined the BAC containing those sequences (gray circles). Genes (exons) and microsatellite positions are represented by black and hatched boxes, respectively. The different BAC end sequences, gene, and microsatellite positions are represented on a plane map and the sequence homologies with HSA6p (NT_007592) are illustrated by dotted lines. The human genes’ orientations are represented by arrows. Small black and white arrows localize markers used for RH12000 and RH7000
mapping, respectively.
The distance between RPS18 and NUDT3 seems to be comparable in pigs and humans and there is no major variation in gene content within this region. Nevertheless, divergence between human and pig physical maps can be observed. Human COL21A1 is located on HSA6p12.1, approximately 20 Mb from RPS18 while in pigs, it is located just upstream of RPS18. Conversely, the RXRB, HKE4, HSD17B8, and RING1 genes, which also belong to the HLA class II gene cluster, are not found in contig 1 but are mapped at the end of the SLA class II region in pigs, close to the COL11A2 and DOA genes, like in human, as previously described (Chardon et al. 1999).
Finally, the SAC2ML gene, which belongs to the so-called HLA extended class II gene cluster, was not localized in either the SLA class II contig or in contig 1. Therefore, it appears that an important gene rearrangement in the region located between SLA class II and contig 1 has occurred.
Filling in the gap between SLA class II andcontig 1
Extension of the SLA class II contig
In order to fill in the gap and bridge contig 1 to RING1, the most telomeric gene of the SLA class II contig, we attempted to extend the SLA class II contig already available. Unfortunately, all the BES of the relevant SLA class II contig extremity contained repeats and were not suitable for BAC screening and chromosome walking. We therefore screened the BAC library with primers designed from genes located close to COL21A1 in humans. BMP5 and LANO genes were good candidates since RH mapping with the IMNpRH12000 panel assigned these loci to the SLA class II–COL21A1 interval (Fig. 1). Library screening with BMP5 and LANO primer pairs and further chromosome walking contributed to lengthen the SLA class II contig by adding 10 additional BACs that represented an extension of about 500 kb (Fig. 3). From BES derived from these new BACs, a series of BLAST hits was found sequentially on the HSA6p12.1 sequence (Fig. 3). The distance between LANO and BMP5 is about 200 kb in pigs instead of the 1.8 Mb in humans. These two genes are reversed in order in the pig map compared with the human map.
Representation of the lengthened SLA class II contig and the BAC end sequence homologies with the HSA6p. The black small circles represent the sequenced BAC ends. With primers developed in those sequences, we determined the BAC containing those sequences (gray circles). Genes (exons) and microsatellite positions are represented by black and hatched boxes, respectively. The different BAC end sequences, gene, and microsatellite positions are represented on a plane map and the sequence homologies with HSA6p (NT_007592) are illustrated by dotted lines. The human genes’ orientations are represented by arrows. Small black and white arrows localize markers used for RH12000 and RH7000 mapping, respectively.
Recovering additional clones in the SLA class II–contig 1 interval
A major disruption of 152.6 cR7000 (about 4 Mb) appeared between the LANO and COL21A1 genes. As we can observe, Sw2019 and Sw1856 microsatellites were mapped in this interval. Additional genes like BAG2 and SAC2ML, located close to COL21A1 and the HLA class II cluster in humans, respectively, were also RH mapped in this interval in pigs.
Several clones containing BAG2 gene sequences were recovered (contig 13), but none of them was suitable to bridge the surrounding contigs. Two of the three derived BES provided BLAST hits on human sequence flanking one side of the COL21A1 gene and the third one on the other side of the gene.
Screening of the BAC library with SACM2L primers resulted in the isolation of five BACs (contig 14), none of which overlapped with either RING1 or RPS18 containing BACs. All the derived BES contained repetitive elements, except one which-matched a short human sequence located 300 kb from the human BAG2 sequence.
Thus, a segment of HSA6p12.1 seems to be spliced and shuffled in pig. The gene order LANO–BMP5–COL21A1–BAG2 in humans is converted to BMP5–LANO–BAG2–COL21A1 in pigs, with an insertion of SACM2L (from class II MHC region) between BAG2 and COL21A1. RH mapping with the ImpRH12000 panel confirms this alignment in pigs (Fig. 1).
The SW2019 and SW1856 microsatellites were used to build two independent small contigs (contigs 11 and 12, respectively) composed of 19 additional BACs. Most of the BES of these two contigs contained repetitive elements with the exception of six BES isolated from contig 12 which presented BLAST hits with different parts of the HSA6 chromosome sequence, including the COL21A1 segment, the pericentromeric region, and segments farther away on the long arm.
No other gene primers issued from the human genomic segment HSA6p12 allowed us to fill the gap until now. The comparison of BES by BLASTN with other available mammalian sequences revealed similarities: lower with rodents than with human and higher on short segments with equine and bovine sequences. In the two last species, the map of this region is not achieved and this comparison did not help to fill the gap.
The study of the “SLA class II–contig 1” interval revealed at least three hotspots of rearrangements between species. The first one is adjacent to the SACM2L gene (HSAp21.3) in the middle of extended class II. In rodent, this gene seems to belong to the ancient duplication unit including class I gene (MMU17 or RNO20). The second hotspot includes COL21A1 (HSA6p12.1), absent in rodents. This gene is located between BMP5 and BPAG1 which both limit homology disruption between human, pig, and rodent at least. In rodent, a first homologous segment limited by GST and BMP5 is located on MMU9 or RNO8. The second homologous segment, which includes BPAG1, BAG2, and RAB2L genes, is located on other chromosomes (MMU1 or RNO9). The gene cluster GST1, 5, 3, 4 in human is located near the third breakpoint at the limits MMU1/MMU9 and RNO9/RNO8 in rodent (www.ensembl.org/musculus/ or /Rattus_norvegicus/).
Comparative mapping of contig 2
Contig 2 included 26 BAC clones, spanning about 1 Mb. Altogether, eight genes were identified within this contig (Fig. 4). GenBank accession numbers of BES are AJ629152–AJ629176 and AJ629414–AJ629428. The four primer pairs derived from human PPAKD, CLPS, SRPKl, and MAPK13 genes were used to screen the BAC library. They sorted several clones that merged into contig 2 after chromosome walking. Sequencing of the amplification products confirmed that there was no misassignment. Two derived BES, 572A10 and 382D11, the first in 5′ (AJ629158) and the second in 3′ end sequences (AJ629163), contained segments of the ZNF76 gene. The 5′ and 3′ end sequences of BAC 60B11 (AJ629422 and AJ629423) contained two exons of the SOCS5 and MAPK14 genes, respectively. Finally the 3′ end of both 219A3 and 1007A4 BACs (AJ629425 and AJ629426) matched the SLC26A8 gene sequence. Moreover, 33 out of 39 BES presented BLAST hits and could be sequentially aligned onto the human draft sequence. Although several genes which might be orthologous to human genes have not yet been located in contig 2, the length of this segment appears to be conserved in both species and genome organization of the HSA6p21.31 segment is well preserved in pigs. Thus, a robust conservation exists between pigs and humans in this region, except for the SOCS5 gene.
Representation of contigs 2–4 and the BAC end sequence homologies with the HSA6p. The black small circles represent the sequenced BAC ends. With primers developed in those sequences, we determined the BAC containing those sequences (gray circles). Genes (exons) and microsatellite positions are represented by black and hatched boxes, respectively. The different BAC end sequences, gene, and microsatellite positions are represented on a plane map and the sequence homologies with HSA6p (NT_007592) are illustrated by dotted lines. The human genes’ orientations are represented by arrows. Small black and white arrows localize markers used for RH12000 and RH7000 mapping, respectively.
BLAST alignment of two 5′ BES from the 521G2 and 60B11 BACs (AJ629418 and AJ629422) revealed 83% and 76% similarity with human SOCS5 cDNA sequences, respectively, suggesting that the SOCS5 gene is close to the SRPK1 gene in pigs but is encoded on the opposite DNA strand (Fig. 4). We verified that the two sequenced gene segments are also present on the 439C12 BAC, which overlaps both 521G2 and 60B11 BACs. In humans, the SOCS5 locus is located on HSA2 and not on HSA6. Interestingly, there is no similarity between the human SOCS5 cDNA sequences and the working draft HSA6p sequence, which excludes the possible location of any SOCS5-like paralogous gene in this human genomic segment. Thus, we have possibly identified a micro-reshuffling in this area.
The gap lengths are estimated in pigs by the RH distance between markers and converted into DNA length by resolution averaged from the IMNpRH12000 panel study (12.6 kb/cR). These values are compared to the distance which separates the homologous point on the human working draft sequence (version 10). The gaps flanking contig 2 have the same estimated lengths in pigs and in humans. It is remarkable to observe that, on Fig. 1, the RH12000 interval between contigs 1 and 2 is 78.4 cR (990 kb) and is 50.9 cR (640 kb) between contigs 2 and 3. It corresponds to 990 kb (Figs. 2 and 4: 26.05–25.06 Mb) and to 580 kb (Figs. 4 and 5: 27.28–26.7 Mb), respectively, on the human sequence.
Comparative mapping of contigs 3 and 4
Contigs 3 and 4 comprise 14 and 13 overlapping BACs, respectively, covering about 600 kb each. GenBank accession numbers of BES are AJ629222–AJ629246 (contig 3) and AJ629429–AJ629453 (contig 4). Contig 3 contains STK38 (NDR) and CDKN1A genes. The majority of the BES matches the human sequence except for those located on both contig ends, rich in SINE and LINE repeats. Gene order is conserved. In contig 4, we detected two additional genes next to the PIM1 gene used for the first screening round, namely, MITCH1 and RNF8, and we found sequence similarity between 14 BES and the orthologous human sequence (Fig. 5).
The gap between contigs 3 and 4 is 58.5 cR long measured by IMNpRH12000. This corresponds to 740 kb in pigs as opposed to 360 kb in the human segment (Fig. 4: 27.8–27.44 Mb). This discrepancy might be due to irregularity of the retention value along the IMNpRH map.
It should be noted that the genes present in contigs 3 and 4 have the same organization in rat RNO20 and mouse MMU17, indicating that the segment seems to be highly conserved in mammals.
Interspersed repeat density and GC percentage
All together, the BES located on the SLA class II – contig 1 interval represent a total of 9887 bp. This region is characterized by a low percentage of GC content (42% on the average), and a density of 0.5 SINE/kb, similar to the segments which flank the gap (Fig. 5). The LINE density is about 0.4, not significantly higher than the one determined for the flanking segments (0.2, 0.3), whereas it reaches 0.6 LINE/kb in contig 3. The DNA-MER and LTR fragments are rare since, they are in the flanking segments as well (Table 2).
The GC percentage of BES located on contig 1 varies along the contig. The level is low (40%) along the sequence covering COL21A1, just like in its human counterpart. The GC percentage average jumps to 53% along the gene dense segment which mainly corresponds to the extended SLA/HLA class II region and drops to 45% at the end of the contig where gene density decreases. The maximum is greater than 60% along the extended class II segment and lower than 60% between the ITPR3 and NUDT3 genes. This variation corresponds to the transition between the human segment, HSA6p21.32 and HSA6p21.31 (Fig. 5). The SINE density clearly increases between the ITPR3 and NUDT3 genes, compared to the extended class II region.
The GC percentage and type of interspersed repeat vary along contig 2 (Fig. 5). The GC percentage is high (54%) on the segment encoding ZNF76 and PPARD genes and low (43%) on the following segment which encodes several kinases, in spite of apparently equivalent gene density. We observed the same GC percentage (43%) in the first part of contig 3 which encodes other kinases, whereas we found about 50% CG in the second part of contig 3 and in the beginning of contig 4 encoding other genes. The human counterpart of contig 3 covers the junction between HSA6p21.31 and HSA6p21.2. The GC percentage of BES recovered from contig 3 may reflect the HSA6p21.31 and HSA6p21.2 junction. Contig 4 represents part of the HSA6p21.2 segment. Alltogether, BES represented about 10,000 nucleotides for both contigs 3 and 4.
The GC percentage seems to depend on the gene family encoded by the DNA segment and the data derived from BES seem to correspond to full-length sequence results as well.
Microsatellite markers
A new set of 18 polymorphic microsatellite markers is available as a result of this study (Table 3). Their exact locations are indicated in the figures which describe the contigs. Four are located in the class II region, three mark the contig 1 gene-rich region, four are polymorphic markers of the two ends of contig 2, two are mapped in contig 3, and one microsatellite limits the telomeric part of contig 4. Two additional microsatellites are located in the vicinity of the NFYA gene (contig 6), the last one being associated with the GSTA3 gene (contig 8).
Discussion
We constructed a physical map by generating BAC contigs covering the region adjacent to the histocompatibility complex class II on the long arm of pig Chromosome 7. A total of 284 BACs were assigned and 220 STSs were generated by BAC end sequencing, 54 of which corresponded to gene segments (exons: 12.8%, introns: 44.7%).
Data derived from our human/pig comparative mapping revealed overall gene conservation and gene content, except for the segment located between the RING1–SACM2L–RPS18 genes in the end of SLA class II. In humans, the distance between RING 1 and RPS18 is about 80 kb. In pigs, the corresponding segment is about 4 Mb long. It may be due to double insertions of the BMP5–LANO–BAG2 and COL21A1 genes on both sides of SACM2L, intermingled by repeat-rich segments. Our physical map indicated that the shifted fragments LANO–BMP5–COL21A1– BAG2, located on HSA6p12.1 and not on HSA6p21.3 like the RING1–SACM2L–RPS18 gene cluster, were inserted in a rearranged fashion since gene order and orientation were not conserved. These precise results are in agreement with fluorescene in situ hybridization (FISH) and RH mapping data reported by Demeure et al. (2002) as well as with the genetic map. The selected BACs enclose the homology breakpoints: the full-length sequence of BAC encoding RING1 confirms the direct linkage between RING1 and BMP5 surrounding segment (BX640585, manuscript in preparation). It is interesting to note that, in cattle, a chromosomal rearrangement disrupted the BoLA class II region into two partially duplicated segments (Hess et al. 1999). In mice, the location of SACM2L-like fragments within two evolutionarily variable regions (Kumanovics et al. 2002) demonstrates that the segment is sensitive to evolution of post-species divergence. Thus, it seems that the region contiguous to the MHC class II region is prone to frequent chromosomal rearrangements in mammals.
Our current map still has a gap within the LANO–COL21A1 region, which has proved particularly difficult to cover because of the abundance of repeats present in the BAC ends at the border of contigs. Until now, four small contigs, including the Sw2019 and Sw1856 markers and the BAG2 and SACM2L genes, were assigned to this region. The length of the gaps LANO–Sw2019, Sw2019–Sw1856, Sw1856–BAG2, and BAG2–RAB2L based on the IMpRH7000 panel (26 kb/cR on average) might be 550, 990, 9901, and 660 kb, respectively. The distance between SACM2L and COL21A1 could be approximately 200 kb based on the IMNpRH12000 panel, which has a resolution of 12.6 kb/cR (Yerle et al. 2002).
The insertion of the 230-kb COL21A1 gene segment in pig clearly indicates a homology disruption between mammals. Noteworthy, this disruption also concerned rodents. The human segment HSA6p12.1, which comprises the gene clusters GST...LANO...BMP5–COL21A1–BPAGl...BAG2...R AB23..., corresponds to mice Chromosomes 1 and 9 and rat Chromosomes 8 and 9 segments except for the COL21A1 segment. This gene is absent at the point that separates the two synthenic gene clusters.
Extensive comparative studies by BLASTN between the pig BES and the available mammalian sequences failed to help fill in the gaps which still exist in our physical map.
The length of the gaps between the large contigs 1, 2, 3, and 4 was estimated by the IMNpRH12000 panel. The three gaps (DIPP–ZNF76, 439C12–358G3, and 797A4–703H8) are supposed to extend over 980, 630, and 730 kb, respectively. These distances are overestimated since the BACs are not always located at the contig ends. Nevertheless, one can estimate possible rearrangements upon comparison with distances in humans. The distance between DIPP and ZNF76 genes is 980 kb in humans, suggesting a complete conservation of the region in both species. The second and third gaps (SRPK1–WAF and PPIL1 – MTCH1 intervals) are 850 and 210 kb long, respectively, in humans. In pigs, the second gap is shorter (630 kb) while the third one is much longer (730 kb). Those results suggest the existence of more rearrangements than those observed in this segment, possibly associated with the variable length of repeat sequences and/or insertion/deletion of genes.
The contig lengthening and gap filling by fingerprinting of the whole BAC library will help to increase gene density and will better define the pig gene order.
In a previous study, the rearrangement was observed by the use of IMpRH7000 and IMNpRH12000 panels and confirmed by FISH. Here we determined the gene order. This map confirms and defines more precisely the results obtained by RH mapping and FISH techniques on long porcine genomic segments.
Moreover, the BAC end sequencing provided a global idea concerning the overall GC content and permitted precise comparisons with the human genome. Our results definitively demonstrate that rearrangements have occurred during either human or pig evolution, on HSA6p or SSC7q11-ql4, although previous studies had failed to identify them (Genet et al. 2001; Hawken et al. 2002; Tanaka et al. 2003). Genes that are conserved in humans, pigs, mice, and rats seem to be linked to essential functions (cell cycle and apoptosis regulation, hematopoiesis) or tissue-specific expression (in testes or blood vessels). This degree of conservation is also observed in genomes of inferior species (Kulski et al. 2003). The variation between species seems to have occurred within particular locations, often at the transition point between high and low GC levels (Kanaya et al. 2000). COL21A1 and SACM2L flanking segments illustrated this observation. However, even in long genomic DNA segments conserved between species during evolution, we observed some “gene INDEL” such as SOCS5 (flanked by SRPK1 and SLC26A8 within contig 2), while its counterpart seems to be unmodified in mice and rats. When we compared the genome of various species, conserved and variable segments were observed at each scale at which we performed the studies.
References
JP Bidanel D Milan N Iannuccelli Y Amigues MY Boscher et al. (2001) ArticleTitleDetection of quantitative trait loci for growth and fatness in pigs Genet Sel Evol 33 289–309 Occurrence Handle10.1051/gse:2001120 Occurrence Handle1:CAS:528:DC%2BD3MXltFWksLw%3D Occurrence Handle11403749
P Chardon C Rogel–Gaillard L Cattolico S Duprat M Vaiman et al. (2001) ArticleTitleSequence of the swine major histocompatibility complex region containing all non-classical class I genes Tissue Antigens 57 55–65 Occurrence Handle10.1034/j.1399-0039.2001.057001055.x Occurrence Handle1:CAS:528:DC%2BD3MXhsFaju74%3D Occurrence Handle11169259
O Demeure C Renard M Yerle T Faraut J Riquet et al. (2003) ArticleTitleRearranged gene order between pig and human in a QTL region on SSC 7 Mamm Genome 14 71–80 Occurrence Handle10.1007/s00335-002-3034-1
C Genet C Renard C Cabau C Rogel–Gaillard J Gellin et al. (2001) ArticleTitleIn the QTL region surrounding porcine MHC, gene order is conserved with human genome Mamm Genome 12 246–249 Occurrence Handle10.1007/s003350010261
A Goureau M Yerle A Schmitz J Riquet D Milan et al. (1996) ArticleTitleHuman and porcine correspondence of chromosome segments using bidirectional chromosome painting Genomics 36 252–262 Occurrence Handle10.1006/geno.1996.0460 Occurrence Handle1:CAS:528:DyaK28XlslWiurs%3D Occurrence Handle8812451
RJ Hawken J Murtaugh GH Flickinger M Yerle A Robic et al. (1999) ArticleTitleA first-generation porcine whole-genome radiation hybrid map Mamm Genome 10 824–830 Occurrence Handle10.1007/s003359901097 Occurrence Handle1:CAS:528:DyaK1MXkvV2msLY%3D Occurrence Handle10430669
M Hess T Goldammer A Gelhaus K Ried G Rappold et al. (1999) ArticleTitlePhysical assignment of the bovine MHC class IIa and class IIb genes Cytogenet Cell Genet 85 244–247 Occurrence Handle1:CAS:528:DyaK1MXlvFShsrw%3D Occurrence Handle10449907
CB Jorgensen AK Wintero M Yerle M Fredholm (1997) ArticleTitleMapping of 22 expressed sequence tags isolated from a porcine small intestine cDNA library Mamm Genome 8 423–427 Occurrence Handle10.1007/s003359900460
S Kanaya T Fukagawa A Ando H Inoko Y Kudo et al. (2000) Evolution, structure and function M Kasahara (Eds) Major Histocompatibility Complex Springer-Verlag Tokyo 131–145
JK Kulski T Shina T Anzai S Kohara H Inoko (2002) ArticleTitleComparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man Immunol Rev 190 95–122 Occurrence Handle10.1034/j.1600-065X.2002.19008.x Occurrence Handle1:CAS:528:DC%2BD3sXnt12ksQ%3D%3D Occurrence Handle12493009
A Kumanovics A Madan S Qin L Rowen L Hood et al. (2002) ArticleTitleQUOD ERAT FACIENDUM: sequence analysis of the H2-D and H2-Q regions of 129/SvJ mice Immunogenetics 54 479–489 Occurrence Handle10.1007/s00251-002-0494-0 Occurrence Handle1:CAS:528:DC%2BD38XnvFOhurs%3D Occurrence Handle12389096
D Milan R Hawken C Cabau S Leroux C Genet et al. (2000) ArticleTitleIMpRH server: an RH mapping server available on the Web Bioinformatics 16 558–559 Occurrence Handle10.1093/bioinformatics/16.6.558
D Milan JP Bidanel N lannuccelli J Riquet Y Amigues et al. (2002) ArticleTitleDetection of quantitative trait loci for carcass composition traits in pigs Genet Sel Evol 34 705–728 Occurrence Handle10.1051/gse:2002026
C Nezer L Moreau D Wagenaar M Georges (2002) ArticleTitleResults of a whole genome scan targeting QTL for growth and carcass traits in a Pietrain x Large White intercross Genet Sel Evol 34 371–387 Occurrence Handle10.1051/gse:2002013 Occurrence Handle1:CAS:528:DC%2BD38Xmt1Gmurg%3D Occurrence Handle12081803
DJ Penn K Damjanovich WK Potts (2002) ArticleTitleMHC heterozygosity confers a selective advantage against multiple-strain infections Proc Natl Acad Sci USA 99 11260–11264 Occurrence Handle10.1073/pnas.162006499 Occurrence Handle1:CAS:528:DC%2BD38XmslSmtbw%3D Occurrence Handle12177415
C Renard M Vaiman N Chiannilkulchai L Cattolico C Robert et al. (2001) ArticleTitleSequence of the pig major histocompatibility region containing the classical class I genes Immunogenetics 53 490–500 Occurrence Handle10.1007/s002510100348 Occurrence Handle1:CAS:528:DC%2BD3MXosFShurY%3D Occurrence Handle11685460
A Rink EM Santschi KM Eyer B Roelofs M Hess et al. (2002) ArticleTitleA first-generation EST RH comparative map of the porcine and human genome Mamm Genome 13 578–587 Occurrence Handle10.1007/s00335-002-2192-5 Occurrence Handle1:CAS:528:DC%2BD38Xos1amu7c%3D Occurrence Handle12420136
C Rogel–Gaillard N Bourgeaux A Billault M Vaiman P Chardon (1999) ArticleTitleConstruction of a swine BAC library: application to the characterization and mapping of porcine type C endoviral elements Cytogenet Cell Genet 85 205–211 Occurrence Handle10.1159/000015294
T Schiex C Gaspin (1997) ArticleTitleCARTHAGENE: constructing and joining maximum likelihood genetic maps Proc Int Conf Intell Syst Mol Biol 5 258–267 Occurrence Handle1:STN:280:ByiH28zis1c%3D Occurrence Handle9322047
Schiex T, Chabrier P, Bouchez M, Milan D (2001) Boosting EM for Radiation Hybrid and Genetic Mapping. Algorithms in bioinformatics: first international workshop, WABI 2001, Aarhus, Denmark, August 28–31, 2001, Proceedings. www.inra.fr/bia/T/schiex/Doc/publis.html
TP Smith GA Rohrer LJ Alexander DL Troyer KR Kirby–Dobbels et al. (1995) ArticleTitleDirected integration of the physical and genetic linkage maps of swine chromosome 7 reveals that the SLA spans the centromere Genome Res 5 259–271 Occurrence Handle1:CAS:528:DyaK2MXpsFaqt78%3D Occurrence Handle8593613
M Tanaka T Matsumoto S Yanai M Domukai D Toki et al. (2003) ArticleTitleConservation of the syntenies between porcine chromosome 7 and human chromosome 6, 14 and 15 demonstrated by radiation hybrid mapping and linkage analysis Anim Genet 34 255–263 Occurrence Handle10.1046/j.1365-2052.2003.00999.x
RE Voorrips (2002) ArticleTitleMapChart: Software for the Graphical Presentation of Linkage Maps and QTLs J Hered 93 77–78 Occurrence Handle10.1093/jhered/93.1.77 Occurrence Handle1:CAS:528:DC%2BD38XktlOntrw%3D Occurrence Handle12011185
M Yerle P Pinton A Robic A Alfonso Y Palvadeau et al. (1998) ArticleTitleConstruction of a whole-genome radiation hybrid panel for high-resolution gene mapping in pigs Cytogenet Cell Genet 82 182–188 Occurrence Handle10.1159/000015095 Occurrence Handle1:CAS:528:DyaK1MXkslyqtQ%3D%3D Occurrence Handle9858812
M Yerle P Pinton C Delcros N Arnal D Milan et al. (2002) ArticleTitleGeneration and characterization of a 12,000-rad radiation hybrid panel for fine mapping in pig Cytogenet Genome Res 97 219–228 Occurrence Handle10.1159/000066616 Occurrence Handle1:CAS:528:DC%2BD38XovVeisr8%3D Occurrence Handle12438716
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported in this article have been submitted to EMBL/GenBank and have been assigned the following accession numbers: gene amplicons (AJ629177–AJ629187), microsatellites (AJ544106–AJ544111, AJ544188–AJ544192, and AJ544212–AJ5442217), BAC-end sequences (AJ628956–974, AJ629076–9108, AJ629125–149, AJ629152–176, AJ629222–46, AJ629414–453, AJ630314–355, and AJ634055. Locus name, gene name (EMBL accession number): SLA class II telomeric end: COL11A2, collagen type XI alpha2 (BX324144); RXRB, retinoid X receptor beta; KE4, HLA class II region expressed gene KE4; KE6 (HSD17B8), estradiol 17 beta-dehydrogenase; and RING1, ring finger protein 1 (BX640585), BMP5, bone morphogenetic protein 6-like id EST SSC2B02 (F14943); LANO (FLJ10775, TINAG), LAP (leucine-rich repeats and PDZ) and no PDZ protein (NM_018214)
GAP: SACM2I, suppressor of actin mutations 2-like (BI360056); BAG2, BCL2-associated athanogene 2 (NM_004282)
Contig1: RPS18 (KE3), ribosomal protein S18 (AJ629092); HKE2, HLA class II region expressed gene KE2 (AJ629178); RAB2L, RAB2, member RAS oncogene family-like (NM_00476 l); TAPBP, tapasin binding protein (BE234046); ZNF297 (BING1), Zn finger protein with POZ motif (AJ629088); DAXX (Fas binding protein) death-associated protein 6 (AJ629179); SYNGAP (RASA5, KIAA1938), synaptic Ras GTPase activating protein 1 homolog (AJ629106); BAK1, BCL2 antagonist killer 1 (AJ629098); ITPR3, inosotol triphosphate receptor 3 (AJ634055); HMGA1(HMGY), high mobility group protein 1 (AJ62180); NUDT3(DIPP), nucleoside diphosphate linked moiety X or diphosphoinositol polyphosphate (BE749729)
Contig 2: ZNF76, Zn finger protein 76 (expressed in testis) (AJ629181, AJ629163); PPARD, peroxisome proliferative activated receptor (AJ629182, AY188501); CLPS, colipase (AJ306320, AF012819); SRPK1, serine/arginine protein kinase (BP172161); SOCS5 (CISH6, KIAA0671), suppressor of cytokine signaling 5 (HSA2p21), or cytokine-inductible SH2 containing element (AJ629418, AJ629422); SLC26A8, solute carrier family 26, member 8 (AJ629426); MAPK14, mitogen-activated protein kinase 14 (AJ629427); MAPK13, mitogen-activated protein kinase 13 (BF713204)
Contig 3: STK38 (NDR), serine/threonine kinase 38 (AJ629183); CDKN1A (p21, WAF), cyclin-dependent kinase inhibitor 1A (AJ629184); PPIL1 (CYPL1), peptidylprolyl isomerase (cyclophilin)-like 1 (AJ629228)
Contig4: MTCH1 (CGI-64, PSAP), mitochondrial carrier homolog 1 (AJ620434); PIM1, protein kinase 1 related oncogen (AJ629185); RNF8, ring finger protein (C3HC4 type) (AJ629445)
Small contigs: GLO1, glyoxalase 1 (NM_006708); TPX, testis-specific protein 1 (AJ006634); CCND3, cyclin D3 (AJ629186); NFYA, nuclear transcription factor Y alpha (AJ629187); GSTA3, gluthation S-transferase (BP447660); CEBPE (CRP1), CCAAT/enhancer binding protein (C/EBP) epsilon (BF652076)
Rights and permissions
About this article
Cite this article
Barbosa, A., Demeure, O., Urien, C. et al. A physical map of large segments of pig Chromosome 7q11–q14: comparative analysis with human Chromosome 6p21. Mamm Genome 15, 982–995 (2004). https://doi.org/10.1007/s00335-004-3008-6
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00335-004-3008-6