Abstract
Objectives
To evaluate the performance of interpretable machine learning models in predicting breast cancer molecular subtypes.
Methods
We retrospectively enrolled 600 patients with invasive breast carcinoma between 2012 and 2019. The patients were randomly divided into a training (n = 450) and a testing (n = 150) set. The five constructed models were trained based on clinical characteristics and imaging features (mammography and ultrasonography). The model classification performances were evaluated using the area under the receiver operating characteristic (ROC) curve (AUC), accuracy, sensitivity, and specificity. Shapley additive explanation (SHAP) technique was used to interpret the optimal model output. Then we choose the optimal model as the assisted model to evaluate the performance of another four radiologists in predicting the molecular subtype of breast cancer with or without model assistance, according to mammography and ultrasound images.
Results
The decision tree (DT) model performed the best in distinguishing triple-negative breast cancer (TNBC) from other breast cancer subtypes, yielding an AUC of 0.971; accuracy, 0.947; sensitivity, 0.905; and specificity, 0.941. The accuracy, sensitivity, and specificity of all radiologists in distinguishing TNBC from other molecular subtypes and Luminal breast cancer from other molecular subtypes have significantly improved with the assistance of DT model. In the diagnosis of TNBC versus other subtypes, the average sensitivity, average specificity, and average accuracy of less experienced and more experienced radiologists increased by 0.090, 0.125, 0.114, and 0.060, 0.090, 0.083, respectively. In the diagnosis of Luminal versus other subtypes, the average sensitivity, average specificity, and average accuracy of less experienced and more experienced radiologists increased by 0.084, 0.152, 0.159, and 0.020, 0.100, 0.048.
Conclusions
This study established an interpretable machine learning model to differentiate between breast cancer molecular subtypes, providing additional values for radiologists.
Key Points
• Interpretable machine learning model (MLM) could help clinicians and radiologists differentiate between breast cancer molecular subtypes.
• The Shapley additive explanations (SHAP) technique can select important features for predicting the molecular subtypes of breast cancer from a large number of imaging signs.
• Machine learning model can assist radiologists to evaluate the molecular subtype of breast cancer to some extent.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Breast cancer is a major public health problem worldwide as it remains the most common malignancy among women [1]. Its incidence rate of breast cancer is increasing almost annually worldwide, although its mortality rate is declining in many countries, including China [2]. As a heterogeneous disease, it is classified by the St. Gallen guidelines into four molecular subtypes: Luminal A, Luminal B, human epidermal growth factor receptor 2 (HER2), and triple-negative breast cancer (TNBC) [3, 4]. Early recognition of the molecular subtype of breast cancer is essential for timely and specific clinical treatment, improving therapeutic options and patient prognosis [5, 6].
Mammography and ultrasonography (US) are the primary imaging modalities used for breast cancer screening, diagnosis, staging, treatment response assessment, and follow-up [7,8,9]. The imaging manifestations of breast cancer are different with different molecular subtypes. The Luminal subtype shows spiculated margins and posterior acoustic shadow [10, 11], the HER2 subtype shows calcification and a posterior mixed acoustic pattern [11, 12], and the TNBC subtype shows indistinct margins and posterior acoustic enhancement [11]. Therefore, imaging features such as tumor shape, margins, and calcification could be used to differentiate breast cancer molecular subtypes.
Recently, machine learning (ML) methods have become popular. These gain increasing attention in medicine because they can accurately process large amounts of data [13, 14]. However, although significant attention has been paid to improving the predictive performance of ML models, interpreting the predictions currently is an under-investigated area [15, 16]. So the concept of interpretable ML method is gradually introduced into clinical decision support systems and medical research. The interpretable models allow radiologists to focus on rational decisions, ensure proper model operation, and reproduce image-based reasoning, and disease diagnosis [17]. Moreover, rationalizing model decisions would assign priority to meaningful predictions, help to extract important knowledge from ML models, and also increase the acceptance of and confidence in predictions in the molecular subtype of breast cancer research. Currently, simple models such as decision tree (DT), k-nearest neighbor (KNN), logistic regression (LR), and naive Bayes (NB), this decision-making process of models is easy to understand. However, some machine learning algorithms are complex, and users cannot interpret the outcomes of the complex machine learning models, e.g., the random forest (RF) and support vector machine (SVM). Their decision-making process is not as straightforward as the simple models [18].
Therefore, we established breast cancer molecular subtype models based on Shapley additive explanations (SHAP) [19]. It is a local interpretation method from game theory. This technique can provide an unbiased assessment of each variable’s contribution to the model performance when the variables are not entirely independent, which means that the contribution of every feature is taken into account individually. With the advancements in SHAP value visualization methods, the technique is widely used to analyze data. Here, it is important to note that the SHAP methodology is a unified framework for explaining the output of any machine learning model. Furthermore, it can also interpret the decision-making process of each case.
This study constructed ML models to differentiate between the molecular subtypes based on clinical characteristics and imaging features. The imaging features were derived based on the Breast Imaging Reporting and Data System (BI-RADS) [20]. We also evaluated the performance of more and less experienced radiologists in differentiating between the molecular subtypes with or without model assistance.
Materials and methods
Patients
The institutional review board approved this retrospective study and waived the need for written informed consent. We identified 600 consecutive female patients, from 2012 to 2019, with invasive breast cancer diagnosis and available preoperative mammography and US at our institution. The inclusion criteria were as follows: (1) diagnosed with invasive breast carcinoma; (2) underwent mammography and US before surgery or biopsy; (3) available clinical characteristics; and (4) did not receive irradiation, hormonal therapy, or chemotherapy before surgery. The exclusion criteria were as follows: (1) incomplete imaging data, or only mammography or US examination was performed; (2) missing or incomplete clinical characteristics; (3) received any therapy before mammography and US; and (4) with bilateral breast cancer and occult breast cancer (Fig. 1).
Mammography
The patients were scanned using the Siemens Mammomat Novation (Siemens Healthineers) or Hologic Selenia full-field digital mammography systems (Hologic Gen-Probe). The general imaging conditions were 27–32 kV and 28–68 mAs. Craniocaudal and mediolateral oblique images were obtained for each patient.
Ultrasonography
US images were acquired using an Aixplorer scanner and a Philips iU 22 US system with 7.5–10-MHz linear probes. Bilateral breast US scanning was performed in search of possible lesions. Once a breast lesion was detected, and the recorded data included location and two-dimensional (2D) and color Doppler characteristics. The 2D characteristics included shape, margin, and inner and posterior echoes.
Mammography and ultrasonography imaging signs
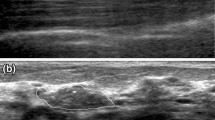
Imaging findings were assessed by three experienced radiologists specializing in breast imaging with 10 years of experience in interpreting breast mammography and the US. Three radiologists independently evaluated the imaging signs and recorded them on the form. Moreover, they were blinded to molecular subtype of the lesion. If the three radiologists agreed on assessing a particular imaging sign, this imaging sign was used as the final result. If the three radiologists were inconsistent in evaluating the imaging sign, the one with the most votes was selected as the final result. All lesions were interpreted based on the BI-RADS lexicon. Recorded mammographic features included masses, calcification, architectural distortion, asymmetries, and other related signs. We define the mass US echo pattern as anechoic, homogeneous, or heterogeneous. Areas with homogeneous echo were defined as isoechoic, hyperechoic, and hypoechoic. Fifty image signs (30 from mammography, 20 from US) were identified (Table 1). An example from one patient is illustrated in Fig. 2.
A 37-year-old woman with TNBC breast cancer. Left craniocaudal (CC) and mediolateral oblique (MLO) views revealed an indistinct mass without calcification in the upper-outer quadrant of breast, which was categorized as BI-RADS category 4B. US image showed an irregular hypoechoic mass with an indistinct margin. Doppler imagery showed the distribution of vessels inside the mass
Clinical information
Clinical information retrieved for each patient included age, menarche age, breast cancer family history, menopausal status, history of nipple discharge, and skin abnormalities (Table 2). The family breast cancer history was defined as occurrence in a first-degree relative [21].
Pathological analysis
Histopathological reports from excisional biopsies, breast-conserving surgeries, or mastectomy specimens were reviewed. Estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) status, and Ki-67 index were used to define the molecular subtypes. These parameters were determined by immunohistochemistry. ER or PR status was considered positive if over 1% of the nuclei stained positive [22]. Positive HER2 expression was defined by an immunohistochemistry score of 3 + or fluorescence in situ hybridization (FISH) amplification with a ratio ≥ 2.0. A Ki-67 of 14% was considered a positive expression [23]. Breast cancer was classified as Luminal A when ER and/or PR was positive, HER2 negative, and Ki-67 < 14%; Luminal B when ER and/or PR was positive, HER2 negative, and Ki-67 ≥ 14% or positive for ER and/or PR and HER2, irrespective of Ki-67 expression; HER2 when ER and PR were negative and HER2 positive; and TNBC when ER, PR, and HER2 were negative.
Model establishment and interpretability analysis
The 600 patients were randomly divided into a training set of 450 patients and a test set of 150 patients. The following prediction binary classification tasks were performed: (1) Luminal versus other subtypes, (2) HER2 versus other subtypes, (3) TNBC versus other subtypes, (4) ER-positive versus ER-negative, (5) PR-positive versus PR-negative, (6) HER2-positive versus HER2-negative, and (7) high Ki-67 (Ki-67 proliferation index ≥ 14%) versus low Ki-67 (Ki-67 proliferation index < 14%).
Before the sample feature data is put into the model for classification, the data was preprocessed first. In the dataset used in the study, age and menarche age is continuous variable, and the rest is classified. For continuous, we adopted standardization for age and menarche age to speed up the training, but for multi-classification variables, we used one-hot encoding. The optimal variables were estimated by t-test, χ2 test, or Fisher’s exact test after excluding errors or inconsistencies. The task-relevant pre-selected features (p < 0.05) were used to train the DT, KNN, LR, NB, and RF models. We used fivefold cross-validation to build the models based on the training set and then validated their performance using the test set. We used SHAP values to evaluate the contribution of each feature to the optimal model and influence of decision for particular cases. Then, we can obtain SHAP values with the SHAP package to interpret model predictions.
Auxiliary reading examination design
A fully crossed multi-radiologist evaluation in two sessions (separated by at least four weeks) was performed to test image reading with and without the optimal model support. Among the 150 test sets, 71 and 64 cases were Luminal subtype breast cancer, and TNBC, respectively. Therefore, this part focused on TNBC and other molecular subtypes, and Luminal subtype and other molecular subtypes.
Two less experienced radiologists (1–3 years) and two more experienced radiologists (5–10 years) aimed to differentiate TNBC and other molecular subtypes, and Luminal subtype and other molecular subtypes in each session based on the mammography and US images, respectively. The radiologists mentioned here were different from those who extracted the signs.
The radiologists read the 150 test images during each session, with or without the support of the optimal model. All radiologists were familiar with the lesion locations on the mammography and US images but were blinded to the patient’s immunohistochemistry results. The patients’ clinical information was provided to the four evaluators. The radiologists were trained individually before each session with 10 examinations that were not included in the trial. The training was intended to familiarize the radiologists with the evaluation workstations and criteria.
The sensitivity, accuracy, and specificity of the judgments of the more and less experienced radiologists, with and without the model support, were calculated.
Statistical analyses
Inter-observer agreement with three radiologists was described using the intra class correlation coefficient (ICC). All ICC values were determined using SPSS [24]. Reproducibility was defined as poor (ICC < 0.40), fair to good (ICC = 0.40–0.75), or excellent (ICC > 0.75).
The predictive discrimination abilities of the models were assessed using the area under the receiver operating characteristic (ROC) curves (AUCs), and the sensitivity, specificity, and accuracy were also calculated. The DeLong test was used to compare the differences in the AUC values of different models. The 95% confidence intervals (CIs) for the AUCs were obtained using the MedCalc software, version 16.2 (MedCalc Software Ltd.).
We used t-test, χ2 test, or Fisher’s exact test to select statistically significant variables to build the model. The sensitivity, specificity, and accuracy of each radiologist with and without model assistance were calculated with McNemar’s test. ICC, t-test, χ2 test, Fisher’s exact test, and McNemar’s test were conducted using IBM SPSS Statistics for Windows, version 20.0 (IBM Corp.). Differences with p < 0.05 were considered statistically significant.
Results
Our study population consisted of 600 patients (age: mean ± SD, 47.59 ± 11.37 years; range, 22–87 years). Overall, the radiologists showed excellent agreement in their feature assessment in the mammography and US images (ICC = 0.748–0.962; p < 0.001). Exact ICC values for each sign are presented in Table S1. Among the 56 variables included 50 imaging features and 6 clinical information; those with p < 0.05 were used to build the machine models. The pre-selected feature sets for each category are shown in Table S2. The results from the multivariate models are presented in Table 3. The ROC curves of the machine learning models established based on the pre-selected feature set are shown in the supplementary information Fig. S1.
The pre-selected feature set of the optimal model was evaluated by the SHAP method, and some of the feature sets are shown in Fig. 3 and Fig. S2. The most important features for the prediction of the models were identified. Moreover, it can also explain the decision-making process of each case, as shown in Fig. 4.
A 51-year-old woman with invasive ductal carcinoma in the left breast. Left CC (a) and MLO (b) views revealed fine pleomorphic, fine linear branching calcifications with linear/segmental. US image (c) showed an irregular hypoechoic mass with spiculated margin, shadowing posterior feature, calcification. Doppler imagery showed no blood supply to the lesion. Immunohistochemistry showed ER ( −), PR ( −), HER2 (3 +), Ki-67 (+ , 25%), so the lesion was HER2 subtype (d). The pre-selected feature set of the optimal model was further explained by the SHAP technique (e). Tumor with calcification, including morphology and distribution on mammography, was the top features for predicting the molecular subtype
Differentiation of one molecular subtype from all the others
The DT model showed the best performance in distinguishing TNBC from the other subtypes with an AUC, accuracy, sensitivity, and specificity of 0.971 (95% CI, 0.947–0.995), 0.947, 0.905, and 0.941, respectively. The DT and NB models differed significantly in differentiating TNBC from the other subtypes (p = 0.016). The RF model had the highest AUC (0.900; 95% CI, 0.842–0.958) for differentiating the Luminal from the other subtypes. Its accuracy, sensitivity, and specificity were 0.860, 0.871, and 0.886, respectively. The AUCs of RF and KNN models differed significantly in differentiating between the Luminal and the other subtypes (p = 0.024). The RF model also can achieve AUC of 0.855 (95% CI: 0.766–0.945) for differentiating between HER2 and the other subtypes. Its accuracy, sensitivity, and specificity were 0.893, 0.900, and 0.724, respectively.
Differentiation based on individual molecular markers
The LR model was the best in distinguishing PR-positive from PR-negative and ER-positive from ER-negative, with AUC, accuracy, sensitivity, and specificity of 0.879 (95% CI, 0.813–0.945), 0.860, 0.902, and 0.809 and of 0.878 (95% CI, 0.816–0.941), 0.867, 0.940, and 0.788, respectively. The LR and KNN models differed significantly in their ability to differentiate between PR-positive and PR-negative (p = 0.024) and between ER-positive and ER-negative (p = 0.006). The NB model achieved the highest AUC (0.739, 95% CI: 0.636–0.842) for differentiating between high and low Ki-67. Its accuracy, sensitivity, and specificity were 0.733, 0.647, and 0.742, respectively. A significant difference was observed between NB and KNN (p < 0.001).
Radiologists assessment with and without the selected model support
As the DT model achieved the highest AUC in differentiating between TNBC and the other subtypes, it was selected as the optimal model to help radiologists diagnose the molecular subtype of breast cancer. We compared the diagnostic performances of the four radiologists in distinguishing TNBC from the other subtypes, Luminal breast cancer from the other subtypes with and without the DT model support (Tables 4 and 5). The accuracy, sensitivity, and specificity of all radiologists have improved with the help of the DT model.
In the diagnosis of TNBC versus other subtypes, the average sensitivity, specificity, and accuracy among the less experienced radiologists were 0.630, 0.780, and 0.730 unaided and 0.720, 0.905, and 0.844 aided, respectively. The parallel values for the more experienced radiologists were 0.760, 0.775, 0.767 unaided and 0.820, 0.865, and 0.850 aided, respectively. The average sensitivity, specificity, and accuracy of less experienced and more experienced radiologists increased by 0.090, 0.125, and 0.114 and by 0.060, 0.090, and 0.083, respectively.
In the diagnosis of Luminal versus other subtypes, the average sensitivity, average specificity, and average accuracy among the less experienced radiologists were 0.553, 0.573, and 0.617 unaided and 0.637, 0.725, and 0.776 aided, respectively. The parallel values for the more experienced radiologists were 0.722, 0.652, 0.767 unaided and 0.740, 0.752, and 0.815 aided, respectively. The average sensitivity, average specificity, and average accuracy of less experienced and more experienced radiologists increased by 0.084, 0.152, and 0.159 and by 0.020, 0.100, and 0.048, respectively.
Discussion
Identification of the subtype-specific molecular characteristics of human breast cancer has important implications for clinical treatment options, disease progression, and, ultimately, patient prognosis. Our study developed and established ML models based on clinical and imaging signs to differentiate between breast cancer molecular subtypes and reproduce image-based reasoning and diagnosis of the disease. The imaging signs extracted based on the BI-RADS lexicon ensured that the study could be easily replicated and established in other medical environments. Furthermore, the ML models are expected to help radiologists, and other clinicians recognize imaging signs that are important for breast cancer molecular subtype classification.
Among the five models, the DT model was best in distinguishing between TNBC and the other subtypes, with an AUC of 0.971. The pre-selected feature set of the DT model was further explained by the SHAP method. The two most important features for differentiating TNBC from the other subtypes in the DT model were indistinct margins and calcification on mammography, as in previous studies [25, 26]. Ko et al [27] have emphasized pointed that TNBC is less frequently associated with calcifications on mammography. Masses with indistinct margins and fine linear calcification branching on mammography were the top features of the optimal model that differentiated between Luminal and non-Luminal and ER-positive and ER-negative. However, Wang et al [26] have recently reported that ER-positive tumors might also manifest as spiculated masses. Huang et al [28] have also found that the Luminal subtypes were positively associated with spiculated margins. The mass margin of the ER-positive and Luminal subtypes in our study contrasted with that in the study by Wang et al [26]. The RF model achieved the highest AUC of 0.855 than other models for distinguishing HER2 from the other subtypes. Its best predictive features were related to calcification, including morphology and distribution. Similar to our results, others reported that HER2 breast cancer exhibited typical calcification with mostly fine pleomorphic or fine linear branching [29, 30]. These results indicated that the model we established was reliable.
Nowadays, many studies [31, 32] on breast cancer molecular subtypes are based on radiomics. In our study, an interpretable ML model for the prediction of molecular subtype of breast cancer was established by using imaging signs and clinical information. It can help radiologists better understand how the models work and present a process that reproduces image-based reasoning and disease diagnosis. Some of the results of our studies were slightly higher than those that were beneficial to the prediction of molecular subtype of breast cancer by radiomics. In this study, the DT model showed the best performance in distinguishing TNBC from the other subtypes. A study by Ma et al has extracted radiomics features from mammography for determining TNBC status, with an AUC value of 0.865, which was far less than that of our model [33]. Additionally, the RF model had the best AUCs of 0.900 and 0.855 for Luminal and non-Luminal breast cancers, and for HER2 and non-HER2 breast cancers, respectively. These results were higher than the AUCs of 0.796 and 0.784 reported by Ma et al [33]. In this study, the LR model was the best of ER-positive and ER-negative breast cancer, and PR-positive and PR-negative breast cancer, with an AUC of 0.878 and 0.879, respectively. The AUC predicted by this model for both ER-positive and ER-negative breast cancers was close to that predicted by Li et al [34] using radiomics model (0.890) and higher than that predicted by this model for PR-positive and PR-negative breast cancers (0.690). This may be because radiomics is easily affected by various factors such as image processing and feature extraction techniques. It is also possible that current radiomics is mainly based on a single imaging modality.
Our study aimed to study comparative performances of different supervised ML models in predicting the molecular subtype of breast cancer, and the results showed that it is not necessarily that the complex ML models (RF) can always outperform simpler models (e.g., DT, LR, NB). In our study, we found that in the prediction of TNBC and non-TNBC, ER + and ER − , PR + and PR − , HER2 + and HER2 − , and low Ki-67 and high Ki-67, the prediction performance of the simple model was higher than the complex machine learning model (RF). Despite the RF model in predicting Luminal and non-Luminal breast cancer can achieve the highest AUC (0.900), only 0.004 higher than the LR model. Similarly, the RF model predicted the highest AUC of breast cancer with HER2 and non-HER2, but its AUC was only slightly higher than other DT model of 0.017. There are many reasons for this phenomenon. Just as the no-free lunch (NFL) theorem states [35] that all learning algorithms perform just as well when averaged over all possible data sets. Our experimental results also support the idea that no single algorithm can maintain the best performance in all data classification. However, Occam’s razor principle highlights that the most simple model or hypothesis should be selected preferentially if simple models can draw a similar conclusion as complex models. Therefore, in this study, the DT model was chosen as the best auxiliary diagnostic model to assist radiologists in identifying the molecular subtype of breast cancer. This not only ensures the diagnostic performance of the model but also simplifies the complexity of the model. Indeed, additional studies will be required to confirm this finding in the future.
Besides, little was reported on the performance of artificial intelligence systems in differentiating between breast cancer molecular subtypes. To date, published studies [36, 37] have mainly evaluated the stand-alone performance of artificial intelligence. In our study, with and without the assisted model diagnosis, the sensitivity, specificity, and accuracy of the model were lower than those of the DT model in the diagnosis of TNBC and non-TNBC breast cancer, and the Luminal subtype and non-Luminal subtype breast cancer, regardless of radiologists’ experience level. It further indicates that it is difficult for radiologists to identify the breast cancer molecular subtype based on mammographic and US images. As expected, the evaluation indicators of all radiologists were significantly improved in predicting the molecular subtype of breast cancer with the help of the DT model. Among them, the performance of the less experienced radiologists showed greater improvement than more experienced radiologists. This finding suggests that the less experienced radiologists benefit more from the DT models than the more experienced ones. Our results suggest that it might be feasible to help radiologists differentiate between breast cancer molecular subtypes with model support. In particular, the model could provide more information for less experienced radiologists. Larger-scale studies are needed to confirm these findings.
This study had some limitations. First, this retrospective study was conducted in a single institution, possibly causing selection bias. A future multicenter study could help address this issue. Second, we compared the performance of only five ML algorithms. Comparing all available algorithms was beyond the scope of this study. Third, this study only focused on the dichotomy of the molecular subtype of breast cancer and failed to explore the four classifications of the molecular subtype of breast cancer.
In conclusion, this study showed the potential of interpretable ML models for differentiating between breast cancer molecular subtypes. Although not a replacement for invasive immunohistochemical detection methods, it provides a process that allows reproducing image-based reasoning and disease diagnosis. Moreover, applying the optimal model in clinical practice could help breast radiologists better identify the molecular subtype in breast cancer patients. Further studies involving large patient populations, multiple centers, and multi-modal imaging methods are needed to verify this conclusion.
Abbreviations
- AUC:
-
Area under curve
- BI-RADS:
-
Breast Imaging Reporting and Data System
- CC:
-
Craniocaudal
- DT:
-
Decision tree
- ER:
-
Estrogen receptor
- FISH:
-
Fluorescence in situ hybridization
- HER2:
-
Human epidermal growth factor receptor 2
- ICC:
-
Intraclass correlation coefficient
- IHC:
-
Immunohistochemistry
- KNN:
-
k-Nearest neighbor
- LR:
-
Logistic regression
- ML:
-
Machine learning
- MLM:
-
Machine learning model
- MLO:
-
Mediolateral oblique
- NB:
-
Naive Bayes
- NFL:
-
No free lunch
- PR:
-
Progesterone receptor
- RF:
-
Random forest
- ROC:
-
Receiver operating characteristic
- SHAP:
-
Shapley additive explanations
- SVM:
-
Support vector machine
- TNBC:
-
Triple-negative breast cancer
- US:
-
Ultrasonography
References
Vogell A, Evans ML (2019) Cancer screening in women. Obstet Gynecol Clin N Am 46(3):485–499
Harbeck N, Gnant M (2017) Breast cancer. Lancet 389(10074):1134–1150
Zardavas D, Irrthum A, Swanton C et al (2015) Clinical management of breast cancer heterogeneity. Nat Rev Clin Oncol 12(7):381–394
Goldhirsch A, Wood WC, Coates AS et al (2011) Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 22(8):1736–1747
Toss A, Cristofanilli M (2015) Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res 17(1):60
Huber KE, Carey LA, Wazer DE (2009) Breast cancer molecular subtypes in patients with locally advanced disease: impact on prognosis, patterns of recurrence, and response to therapy. Semin Radiat Oncol 19(4):204–210
McDonald ES, Clark AS, Tchou J et al (2016) Clinical diagnosis and management of breast cancer. J Nucl Med 57(Suppl 1):9S-16S
Seely JM, Alhassan T (2018) Screening for breast cancer in 2018-what should we be doing today? Curr Oncol 25(Suppl 1):S115–S124
Niell BL, Freer PE, Weinfurtner RJ et al (2017) Screening for breast cancer. Radiol Clin N Am 55(6):1145–1162
Taneja S, Evans AJ, Rakha EA et al (2008) The mammographic correlations of a new immunohistochemical classification of invasive breast cancer. Clin Radiol 63(11):1228–1235
Rashmi S, Kamala S, Murthy SS et al (2018) Predicting the molecular subtype of breast cancer based on mammography and ultrasound findings. Indian J Radiol Imaging 28(3):354–361
An YY, Kim SH, Kang BJ et al (2015) Breast cancer in very young women (<30 years): Correlation of imaging features with clinicopathological features and immunohistochemical subtypes. Eur J Radiol 84(10):1894–1902
Deo RC (2015) Machine learning in medicine. Circulation 132(20):1920–1930
Choy G, Khalilzadeh O, Michalski M et al (2018) Current applications and future impact of machine learning in radiology. Radiology 288(2):318–328
Guo Y, Hu Y, Qiao M et al (2018) Radiomics analysis on ultrasound for prediction of biologic behavior in breast invasive ductal carcinoma. Clin Breast Cancer 18(3):e335–e344
Son J, Lee SE, Kim EK et al (2020) Prediction of breast cancer molecular subtypes using radiomics signatures of synthetic mammography from digital breast tomosynthesis. Sci Rep 10(1):21566
Elshawi R, Al-Mallah MH, Sakr S (2019) On the interpretability of machine learning-based model for predicting hypertension. BMC Medical Inform Decis Mak 19(1):146
Lebedev AV, Westman E, Van Westen GJP et al (2014) Random forest ensembles for detection and prediction of Alzheimer’s disease with a good between-cohort robustness. Neuroimage Clin 6:115–125
Rodríguez-Pérez R, Bajorath J (2020) Interpretation of compound activity predictions from complex machine learning models using local approximations and Shapley values. J Med Chem 63(16):8761–8777
Rao AA, Feneis J, Lalonde C et al (2016) A pictorial review of changes in the BI-RADS Fifth Edition. Radiographics 36(3):623–639
Elkin EB, Klem ML, Gonzales AM et al (2011) Characteristics and outcomes of breast cancer in women with and without a history of radiation for Hodgkin’s lymphoma: a multi-institutional, matched cohort study. J Clin Oncol 29(18):2466–2473
Rakha EA, Green AR (2017) Molecular classification of breast cancer: what the pathologist needs to know. Pathology 49(2):111–119
Zhang K, Zhu Q, Sheng D et al (2020) A new model incorporating axillary ultrasound after neoadjuvant chemotherapy to predict non-sentinel lymph node metastasis in invasive breast cancer. Cancer Manag Res 12:965–972
Prieto L, Lamarca R, Casado A et al (1997) The evaluation of agreement on continuous variables by the intraclass correlation coefficient. J Epidemiol Community Health 51(5):579–581
Boisserie-Lacroix M, Mac GG, Debled M et al (2012) Radiological features of triple-negative breast cancers (73 cases). Diagn Interv Imaging 93(3):183–190
Wang Y, Ikeda DM, Narasimhan B et al (2008) Estrogen receptor-negative invasive breast cancer: imaging features of tumors with and without human epidermal growth factor receptor type 2 overexpression. Radiology 246(2):367–375
Ko ES, Lee BH, Kim HA et al (2010) Triple-negative breast cancer: correlation between imaging and pathological findings. Eur Radiol 20(5):1111–1117
Huang J, Lin Q, Cui C et al (2020) Correlation between imaging features and molecular subtypes of breast cancer in young women (≤30 years old). Jpn J Radiol 38(11):1062–1074
Lee SH, Chang JM, Shin SU et al (2017) Imaging features of breast cancers on digital breast tomosynthesis according to molecular subtype: association with breast cancer detection. Br J Radiol 90(1080):20170470
Killelea BK, Chagpar AB, Bishop J et al (2013) Is there a correlation between breast cancer molecular subtype using receptors as surrogates and mammographic appearance? Ann Surg Oncol 20(10):3247–3253
Zhou J, Tan H, Bai Y et al (2019) Evaluating the HER-2 status of breast cancer using mammography radiomics features. Eur J Radiol 121:108718
Fleury E, Marcomini K (2019) Performance of machine learning software to classify breast lesions using BI-RADS radiomic features on ultrasound images. Eur Radiol Exp 3(1):34
Ma W, Zhao Y, Ji Y et al (2019) Breast cancer molecular subtype prediction by mammographicradiomic features. Acad Radiol 26(2):196–201
Li H, Zhu Y, Burnside ES et al (2016) Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2:16012
Zhang Y, Xin Y, Li Q et al (2017) Empirical study of seven data mining algorithms on different characteristics of datasets for biomedical classification applications. Biomed Eng Online 16(1):125
Wu M, Zhong X, Peng Q et al (2019) Prediction of molecular subtypes of breast cancer using BI-RADS features based on a “white box” machine learning approach in a multi-modal imaging setting. Eur J Radiol 114:175–184
Tagliafico AS, Bignotti B, Rossi F et al (2019) Breast cancer Ki-67 expression prediction by digital breast tomosynthesis radiomics features. Eur Radiol Exp 3(1):36
Funding
This work was supported by the National Natural Science Foundation of China (82171929), National Key R&D Program of China (2019YFC0121903, 2019YFC0117301) and National Natural Science Foundation of Guangdong Province, China (2019A1515011168, 2018A0303130215).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Weiguo Chen.
Conflict of interest
The authors declare no competing interests.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Retrospective study and waived the need for written informed consent.
Ethical approval
The institutional review board of Nanfang Hospital, Southern Medical University approval was obtained.
Methodology
• retrospective
• observational
• performed at one institution
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, M., Liu, R., Wen, C. et al. Predicting the molecular subtype of breast cancer and identifying interpretable imaging features using machine learning algorithms. Eur Radiol 32, 1652–1662 (2022). https://doi.org/10.1007/s00330-021-08271-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-08271-4