Abstract
Objectives
Despite the robust diagnostic performance of MRI-based radiomic features for differentiating between glioblastoma (GBM) and primary central nervous system lymphoma (PCNSL) reported on prior studies, the best sequence or a combination of sequences and model performance across various machine learning pipelines remain undefined. Herein, we compare the diagnostic performance of multiple radiomics-based models to differentiate GBM from PCNSL.
Methods
Our retrospective study included 94 patients (34 with PCNSL and 60 with GBM). Model performance was assessed using various MRI sequences across 45 possible model and feature selection combinations for nine different sequence permutations. Predictive performance was assessed using fivefold repeated cross-validation with five repeats. The best and worst performing models were compared to assess differences in performance.
Results
The predictive performance, both using individual and a combination of sequences, was fairly robust across multiple top performing models (AUC: 0.961–0.977) but did show considerable variation between the best and worst performing models. The top performing individual sequences had comparable performance to multiparametric models. The best prediction model in our study used a combination of ADC, FLAIR, and T1-CE achieving the highest AUC of 0.977, while the second ranked model used T1-CE and ADC, achieving a cross-validated AUC of 0.975.
Conclusion
Radiomics-based predictive accuracy can vary considerably, based on the model and feature selection methods as well as the combination of sequences used. Also, models derived from limited sequences show performance comparable to those derived from all five sequences.
Key Points
• Radiomics-based diagnostic performance of various machine learning models for differentiating glioblastoma and PCNSL varies considerably.
• ML models using limited or multiple MRI sequences can provide comparable performance, based on the chosen model.
• Embedded feature selection models perform better than models using a priori feature reduction.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Glioblastoma (GBM) and primary central nervous system lymphoma (PCNSL) together comprise the two most common primary malignant brain tumors [1]. Whereas GBM accounts for 14.6% of all brain neoplasms, PCNSL accounts for about 1.9% [2]. Even though the treatment strategies are vastly different, they both share overlapping clinical and imaging characteristics, which makes accurate pre-operative identification critical but challenging [3,4,5]. The utility of conventional and more advance imaging sequences (including diffusion and perfusion studies) has previously been assessed with modest success [6,7,8,9,10]. However, these may not be widely available.
More recently, a number of studies (Table 1) have attempted a radiomic-based differentiation between GBM and PCNSL with good success [1, 3, 5, 11,12,13,14,15,16,17,18,19,20,21,22]. A number of these were performed using machine learning (ML), which includes a wide variety of statistical analysis algorithms [23]. The success of a ML technique depends considerably on the amount, type and completeness of data, type of feature selection/reduction technique, and the problem to be addressed. As such, the predictive performance of various ML models for a specific problem can vary and remains largely unaddressed when differentiating GBM from PCNSL. This is, therefore, a need to compare the predictive performance of various models to determine the best performing models for this two-class problem. Similarly, given the heterogeneity of prior studies, it is important to determine if individual sequences or a combination of sequences have equivalent or superior performance when compared to all sequences combined. This will help guide the selection of best performing models for future studies and facilitate model selection for larger studies using multi-institutional datasets.
In this study, we compared the predictive performance of various ML techniques for differentiating between PCNSL and GBM using a combination of various feature selection and ML algorithms. The aims were to identify the best and worst performing models, as well as to determine if accurate distinction between these entities could be achieved using a single sequence or required a combination of sequences for best results.
Methods
This is a single institution retrospective study, performed post approval of the local institutional review board. Patients were identified using a combination of institutional cancer registries and electronic medical records. Inclusion criteria were a pathologically proven diagnosis of GBM or PCNSL. Exclusion criteria included imaging studies with motion artifacts, absence of available index MRI scan, or absence of all required sequences (axial T1WI, T2WI, diffusion-weighted imaging [DWI], fluid-attenuated inversion recovery [FLAIR], and contrast-enhanced T1WI [T1-CE]). Additionally, patients were excluded where the image pre-processing (see below) or feature extraction was unsuccessful. This yielded a total of 94 patients, 34 with PCNSL and 60 with GBM, who were eventually assessed.
Image pre-processing
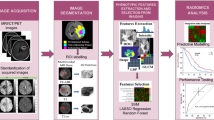
Post de-identification of images, the DICOM images were converted to nifti format prior to pre-processing. Figure 1 provides an overview of the study workflow. The data were initially resampled to voxel size 1 × 1 × 5 mm3 using the AFNI package (https://afni.nimh.nih.gov/) [24]. All sequences were registered to T1WI using Advanced Normalization Tools (ANTs) (http://stnava.github.io/ANTs/.) [25]. Following the resampling and co-registration, the image intensities were normalized to [0,255] using the feature scaling method available in the ANT registration suite.
Tumor segmentation was performed on axial T1W CE and FLAIR images by two radiologists (S.P., G.B.) in consensus using an in-house developed semi-automatic tool Layered Optimal Graph Image Segmentation for Multiple Objects and Surfaces (LOGISMOS) that first automatically identifies the tumor surfaces followed by an efficient “just-enough interaction” with an optional surface editing step which may be invoked if needed [26]. The T1-CE images were used to generate the masks for the enhancing disease (including internal necrosis where present). The FLAIR images were used to generate a mask for the entire lesion (tumor and surrounding edema). Figure 2 shows representative examples of ROI segmentation for both tumor types. The T1-CE-derived mask was subsequently subtracted from this mask to generate the mask for the FLAIR signal abnormality surrounding the tumor. This way, two masks—one for the tumor and the other for the surrounding FLAIR signal abnormality—were expert-identified.
Representative examples of the two tumor types (GBM: a–c; PCNSL: d–f) along with ROI segmentation for whole tumor and FLIAR signal abnormality. The edema mask was created through subtraction of the T1-CE mask from the FLAIR mask. The dotted red line surrounding the segmented volume (b, c, e, and f) represents the volume of interest as defined by the user
Feature extraction
For each tumor, features were extracted using two masks, one for the tumor component (including enhancing and necrotic tumor) and the other for the surrounding FLAIR component. Features were extracted using PyRadiomics v3.0 [27]. Since there were ten possible mask and sequence combinations (five MRI sequences and two masks), on each of which 107 radiomic features were obtained, there were a total of 1070 features. For each sequence-specific model, the feature set included 214 (2 masks × 1 sequence × 107 features) radiomic features. Additionally, 3 limited sequence combinations were also evaluated and included: T1-CE/ADC, T1-CE/ADC/FLAIR, and ADC/FLAIR.
Each set of 107 features included 3D shape features (n = 14), first-order features (n = 18), gray level co-occurrence matrix features (n = 24), gray level dependency matrix features (n = 14), gray level run length matrix features (n = 16), gray level size zone matrix features (n = 16), and neighboring gray tone difference matrix features (n = 5). The default value for the number of bins was fixed by bin width of 25 gray levels. In rare cases where the edema was minimal, leading to absence of a corresponding mask, the value of the corresponding feature was set to − 9999.
Feature selection
Due to the large size of the possible feature sets to be used relative to the sample size and highly correlated variables, feature selection is generally considered a critical piece of the model building process. Three feature selection methods were considered: a linear combination filter, a high correlation filter, and principal component analysis (PCA). The linear combination (lincomb) filter addresses both collinearity and dimension reduction. The high correlation (corr) filter removes variables which have a large absolute correlation. For the models using all sequences, the highest allowable correlation was set to 0.4 and for the models using each sequence separately, the threshold was set to 0.6. These thresholds were chosen to sufficiently reduce the dimensionality of the feature set for model fitting while retaining many of the important variables. The number of components retained in the PCA transformation was determined by specifying the fraction of the total variance that should be covered by the components. For the models using all sequences, this threshold was set to 80% and for the models using each sequence separately, the threshold was set to 85%, again with the goal of sufficiently retaining as much information as possible with enough dimension reduction to allow model fitting. Finally, models were also run using the entire feature set without any a priori feature reduction. These feature selection methods were implemented using the recipes package in R version 4.0.2 [28, 29]. Prior to any feature selection, all variables were standardized and missing values were imputed using mean imputation.
Data analysis
Twelve different predictive models were fit to determine the best classifier for each feature set. These models can be categorized into three broad groups: linear classifiers, non-linear classifiers, and ensemble classifiers. The linear classifiers used were linear, logistic, ridge, elastic net, and LASSO regression. The non-linear classifiers used were neural network, support vector machine (SVM) with a polynomial kernel, SVM with a radial kernel, and multi-layer perceptron (MLP). Finally, the ensemble classifiers used were random forest, generalized boosted regression model (GBRM), and boosting of classification trees with AdaBoost.
Each model was fit using the three feature selection techniques as well as the entire feature set (full), except for the linear regression, logistic regression, and the neural network which cannot be fitted with the full feature set. This is because the model parameters cannot be uniquely estimated in linear and logistic regression models when the number of features is much larger than the sample size. For neural network, however, the problem is more of excessive computational requirement.
This yielded 45 possible model/feature selection combinations to be fit to each of the possible feature sets. These combinations were evaluated for individual MRI sequences (n = 5), a combination of sequences (T1-CE + ADC + FLAIR, T1-CE + ADC, and ADC+ FLAIR; n = 3), and all sequences combined (n = 1). Overall, a total of 405 different models were assessed. Predictive performance of each model was evaluated using fivefold repeated cross-validation with five repeats. For models with tuning parameters, important parameters were tuned using nested cross-validation to avoid bias. The feature selection techniques were carried out within each cross-validated split of the data, so as not to bias the estimate of predictive performance. Model fitting and cross-validated predictive performance were implemented using the MachineShop and RSNNS packages in R version 4.0.2 [29,30,31]. Predictive performance was measured with the area under the receiver operating characteristic curve (ROC AUC) for interpretability. As models were formulated to predict GBM, ROC AUC estimates the probability that a randomly selected subject that had GBM will have a greater predicted value than a randomly selected subject that had PCNSL. Higher ROC AUC values indicate better predictive performance.
To compare predictive performance between different model/feature selection techniques and between models fit to features from all sequences, the combination of sequences, and individual sequences, the corrected resampled t test was used on the resampled ROC AUC values to properly account for the correlation arising from overlapping observations in the training and test sets from the repeated cross-validation procedure [32, 33]. To assess the variability in the performance of different model/feature selection techniques, model performance was compared between the top performing model and the top and bottom five models in each sequence category (Table 2; also Supplementary Table 3). All p values were adjusted for multiple comparisons using the false discovery rate adjustment [34].
Results
A total of 94 patients were studied, 34 with PCNSL (36.2%) and 60 with GBM (63.8%). Further details on patient demographics, scanner types, and typical scanning parameters at authors’ institute are provided in Supplementary Table 1. The mean cross-validated ROC AUC for the best and worst five models, using all sequences, individual sequences, and a combination of sequences is provided in Table 2. Figure 3 shows the mean ROC AUC for all models across all feature combinations, when built using data from all five MRI sequences.
In general, most of the high performing models, regardless of whether they were derived from all sequences, individual sequences, or a combination of limited sequences, were the ones using full feature set without any a priori feature reduction. On the other hand, most of the worst performing models used the linear combination feature reduction strategy. The model performance among the top 5 models was comparable across categories, and the corrected resampled t test with false discovery rate adjustment did not indicate any differences between the top models and the top performing model (AUC 0.977). The top performing model was however significantly better when compared against the bottom five models in each category. The adjusted p values and the overall performance of the 45 models when using all sequences, T1-CE, and T1-CE/FLAIR/ADC-derived models are provided in Supplementary Data (Supplementary Tables 3–6).
The best performing model used only a limited combination of sequences (T1-CE/FLAIR/ADC) and achieved an AUC of 0.977. This was comparable to the model using all five sequences as input, which had the best cross-validated AUC of 0.972 (p value of 0.790 when compared with best performing model). Similarly, the best performing models using only T1-CE/ADC (mean AUC of 0.975) and using only FLAIR/ADC (mean AUC of 0.971) also had comparable performance to the best performing model. Table 3 lists the overall top five models of the analysis, along with the performance metrics.
Discussion
Our study suggests that the model performance for a radiomics-based differentiation between GBM and PCNSL can vary significantly, based on the chosen model, although several best predictive models have comparable performance. Interestingly, the best performing models were those that performed embedded feature selection fit to the full texture feature set, as opposed to models using any of the a priori feature reduction strategies. Another important observation is that models derived from a limited combination of sequences may perform as well as the models using data from all five sequences. These observations are relevant not only in terms of highlighting the variability across machine learning models for the same problem but also in terms of evaluating the necessity of a priori feature reduction which is almost routinely performed. Finally, the excellent performance of a limited sequence combination may allow for a less computationally intensive workflow that may be easier to integrate in clinical practice.
In the current study, of the three feature selection/reduction strategies, models using PCA overall performed better while the lincomb-based models performed the worst. None of these however performed as well as models using full feature set with embedded feature selection. It is pertinent to note here that PCA leads to the fewest number of features included in the models and the linear combination filter leads to the largest. As the number of tumors in the data is small relative to the number of features, it makes sense that feature selection methods leading to smaller feature sets would perform better, particularly for those models which do not do any embedded feature selection, such as the linear, logistic, neural network, and SVM models. However, any feature selection filter would result in some loss of information. The impact of loss of information is evident when evaluating performance of embedded feature selection methods of the elastic net, LASSO, random forest, AdaBoost, and GBRM models which show poor predictive performance with feature reduction (AUC: 0.666–0.738) and considerably improved performance when using full feature set (AUC: 0.961–0.972). Of note, both the ensemble models and penalized regression models like GBRM or LASSO do perform feature selection. However, the feature selection in such cases is part of the model fitting process, unlike other models which require a priori feature reduction using some form of feature selection strategy.
In the analysis using all sequences, the GBRM classifier had an observed ROC AUC of 1.000 and mean cross-validated ROC AUC of 0.972, while the LASSO classifier had an observed ROC AUC of 1.000 and mean cross-validated ROC AUC of 0.967. The absence of a compelling drop in performance and the use of nested cross-validation for tuning parameters would argue against over-fitting of models in our analysis. The low Brier scores of multiple top performing models also attest to the robustness of results in our case. Similarly, the models using a combination of sequences also had excellent cross-validated performance, when using full features without any specified feature reduction strategy. This is an interesting observation since most of the prior studies have consistently used some form of feature reduction strategy, varying between principal component analysis, recursive random forest, and minimum redundancy, maximum relevance [14, 17, 22]. Our findings suggest that feature reduction, though meant to reduce redundant and highly correlated features, may not always be ideal and can negatively impact model performance. Embedded feature selection in the model, on the other hand, may perform better.
Another important observation from our study is that information derived from a limited combination of sequences may suffice to differentiate GBM from PCNSL. In fact, none of the overall top five performing models used the full feature set (Table 3). It is pertinent to note here that all five models, at the very least, consistently used post-contrast imaging and ADC map–derived texture features. This is relevant since it could help reduce the required computational effort and time. We found the model using T1-CE, ADC, and FLAIR to have the highest mean cross-validated AUC of 0.977, though other combinations (all sequences, T1-CE + ADC, ADC + FLAIR) also had comparable performance. In this regard, the model performance using only the ADC and FLAIR images is worth noting since it implies that excellent accuracy could be obtained without contrast administration, a finding that may be useful for patients who are unable to get a contrast-enhanced study. Almost all prior studies dealing with the same two-class problem, except for Wang et al, have used a contrast-enhanced sequence for analysis [18]. The top radiomics features and their relative importance for the best overall performing model are provided in Supplementary Table 2. Most of the top-ranking features were second-order features and derived from the T1-CE images. A combination of sequences, in general, did better than individual sequences alone, with the maximal AUC achieved for T1-CE and FLAIR, both with mean cross-validated AUC of 0.968.
Overall, multiple machine learning–based models in our study achieved excellent accuracy which was better than multiple previously reported studies where the AUC varied between 0.877 and 0.956, and comparable to other studies by Yun et al, Nakagawa et al, and Chen et al (see Table 1). Of the multiple previously reported studies, only Yun et al reported the model performance across three different models, and using three feature reduction techniques. [22]. Their best performing machine learning model used generalized linear model boosting with backward feature elimination as a feature reduction technique, achieving an AUC of 0.943 which is inferior to the best performing model in the current study (AUC of 0.977). However, the best overall performing model in their study was MLP (AUC of 0.991). In our analysis, even though MLP-based models showed high performance (mean AUC: 0.913–0.933), they were not among the top 5 performing models in any category. This could possibly be secondary to the imaging data used. For example, all MRI studies in their cohort were done on a 3-T magnet with a slice thickness of 1 mm for the post-contrast images. The magnet strength and image resolution are known to affect texture features and could account for some of these differences.
Chen et al also achieved excellent results (AUC of 0.982–0.991) for the same two-class problem using features derived from T1-CE images [3]. However, unlike general radiomic features, which are derived from image features such as intensity, shape, wavelet, or texture parameters, they extracted radiomic features using scale invariant feature transform (SIFT), which may explain the slight variability in results compared to our current study. Nakagawa et al were also able to achieve a similar model performance (AUC of 0.980) in their study using eXtreme gradient boosting (XGBoost), and a combination of T1-CE, T2WI, ADC, and rCBV maps derived from dynamic susceptibility contrast (DSC)–enhanced perfusion MRI [16]. However, DSC imaging may not be widely available. Our results, which use conventional and routinely available sequences, are more pragmatic.
Limitations of our study include its retrospective nature and a relatively small sample size. Given this limitation, it is quite possible that the model performance may vary with additional training data. Another limitation would be the absence of external validation cohort which would help determine the generalizability of our findings. Unlike GBM, there are no freely available data repositories for PCNSL patients to our knowledge which could be readily used. We also did not compare our results to expert human readers. However, multiple prior studies have compared machine learning models with human readers and noted the AUC of the expert human readers to vary between 0.79 and 0.94, indicating that the top performing machine learning models would likely outperform expert human readers [11, 16, 20]. Future studies should evaluate the combined accuracy of human readers and best performing machine learning models. Finally, despite the number of different models that were evaluated, we did not assess deep neural networks since this technique is more computationally demanding and data hungry. Nevertheless, our study evaluates the performance of 45 different feature machine learning techniques and provides additional insights into factors that may determine final model performance. Other relative strengths of the study include a documented imaging protocol, use of feature selection techniques, discrimination and calibration statistics, pathological confirmation in all cases, nested cross-validation, and potential clinical utility, all of this contribute to the radiomic quality score as proposed previously [35].
Conclusion
Our findings suggest that excellent discrimination between PCNSL and GBM can be achieved through machine learning. Additionally, features derived from a limited number of sequences may perform as well as features derived from multiple sequences. Another pertinent observation is that models without any a priori feature reduction strategy generally outperform models derived post-feature reduction. Finally, the model performance varies considerably, based on the feature selection technique and the model used. Future studies should focus on a more extensive evaluation of different machine learning models to determine the best possible combination of feature selection and learning model for a more optimized model performance.
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- AFNI:
-
Analysis of Functional NeuroImages
- ANT:
-
Advanced normalization tools
- AUC:
-
Area under the curve
- DSC:
-
Dynamic susceptibility contrast
- DWI:
-
Diffusion weighted imaging
- FLAIR:
-
Fluid-attenuated inversion recovery
- GBM:
-
Glioblastoma
- GBRM:
-
Generalized boosted regression model
- LASSO:
-
Least absolute shrinkage and selection operator
- LOGISMOS:
-
Layered Optimal Graph Image Segmentation for Multiple Objects and Surfaces
- ML:
-
Machine learning
- MLP:
-
Multi-layer perceptron
- MRI:
-
Magnetic resonance imaging
- PCA:
-
Principal component analysis
- PCNSL:
-
Primary central nervous system lymphoma
- r-CBV:
-
Relative cerebral blood volume
- ROC:
-
Receiver operating characteristic
- SIFT:
-
Scale invariant feature transform
- SVM:
-
Support vector machine
- T1-CE:
-
T1 contrast enhanced
References
Alcaide-Leon P, Dufort P, Geraldo AF et al (2017) Differentiation of enhancing glioma and primary central nervous system lymphoma by texture-based machine learning. AJNR Am J Neuroradiol 38(6):1145–1150
Ostrom QT, Cioffi G, Gittleman H et al (2019) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro Oncol 21(Suppl 5):v1–v100
Chen Y, Li Z, Wu G et al (2018) Primary central nervous system lymphoma and glioblastoma differentiation based on conventional magnetic resonance imaging by high-throughput SIFT features. Int J Neurosci 128(7):608–618
Bathla G, Hegde A (2016) Lymphomatous involvement of the central nervous system. Clin Radiol 71(6):602–609
Yang Z, Feng P, Wen T, Wan M, Hong X (2017) Differentiation of glioblastoma and lymphoma using feature extraction and support vector machine. CNS Neurol Disord Drug Targets 16(2):160–168
Choi YS, Lee HJ, Ahn SS et al (2017) Primary central nervous system lymphoma and atypical glioblastoma: differentiation using the initial area under the curve derived from dynamic contrast-enhanced MR and the apparent diffusion coefficient. Eur Radiol 27(4):1344–1351
Kickingereder P, Sahm F, Wiestler B et al (2014) Evaluation of microvascular permeability with dynamic contrast-enhanced MRI for the differentiation of primary CNS lymphoma and glioblastoma: radiologic-pathologic correlation. AJNR Am J Neuroradiol 35(8):1503–1508
Kickingereder P, Wiestler B, Sahm F et al (2014) Primary central nervous system lymphoma and atypical glioblastoma: multiparametric differentiation by using diffusion-, perfusion-, and susceptibility-weighted MR imaging. Radiology 272(3):843–850
Nakajima S, Okada T, Yamamoto A et al (2015) Differentiation between primary central nervous system lymphoma and glioblastoma: a comparative study of parameters derived from dynamic susceptibility contrast-enhanced perfusion-weighted MRI. Clin Radiol 70(12):1393–1399
Toh CH, Castillo M, Wong AM et al (2008) Primary cerebral lymphoma and glioblastoma multiforme: differences in diffusion characteristics evaluated with diffusion tensor imaging. AJNR Am J Neuroradiol 29(3):471–475
Kang D, Park JE, Kim YH et al (2018) Diffusion radiomics as a diagnostic model for atypical manifestation of primary central nervous system lymphoma: development and multicenter external validation. Neuro Oncol 20(9):1251–1261
Kim Y, Cho HH, Kim ST, Park H, Nam D, Kong DS (2018) Radiomics features to distinguish glioblastoma from primary central nervous system lymphoma on multi-parametric MRI. Neuroradiology 60(12):1297–1305
Kunimatsu A, Kunimatsu N, Kamiya K, Watadani T, Mori H, Abe O (2018) Comparison between glioblastoma and primary central nervous system lymphoma using MR image-based texture analysis. Magn Reson Med Sci 17(1):50–57
Kunimatsu A, Kunimatsu N, Yasaka K et al (2019) Machine learning-based texture analysis of contrast-enhanced MR imaging to differentiate between glioblastoma and primary central nervous system lymphoma. Magn Reson Med Sci 18(1):44–52
Liu S, Fan X, Zhang C et al (2019) MR imaging based fractal analysis for differentiating primary CNS lymphoma and glioblastoma. Eur Radiol 29(3):1348–1354
Nakagawa M, Nakaura T, Namimoto T et al (2018) Machine learning based on multi-parametric magnetic resonance imaging to differentiate glioblastoma multiforme from primary cerebral nervous system lymphoma. Eur J Radiol 108:147–154
Suh HB, Choi YS, Bae S et al (2018) Primary central nervous system lymphoma and atypical glioblastoma: differentiation using radiomics approach. Eur Radiol 28(9):3832–3839
Wang BT, Liu MX, Chen ZY (2019) Differential diagnostic value of texture feature analysis of magnetic resonance T2 weighted imaging between glioblastoma and primary central neural system lymphoma. Chin Med Sci J 34(1):10–17
Wu G, Chen Y, Wang Y et al (2018) Sparse representation-based radiomics for the diagnosis of brain tumors. IEEE Trans Med Imaging 37(4):893–905
Xia W, Hu B, Li H et al (2020) Multiparametric-MRI-based radiomics model for differentiating primary central nervous system lymphoma from glioblastoma: development and cross-vendor validation. J Magn Reson Imaging 53(1):242–50
Xiao DD, Yan PF, Wang YX, Osman MS, Zhao HY (2018) Glioblastoma and primary central nervous system lymphoma: preoperative differentiation by using MRI-based 3D texture analysis. Clin Neurol Neurosurg 173:84–90
Yun J, Park JE, Lee H, Ham S, Kim N, Kim HS (2019) Radiomic features and multilayer perceptron network classifier: a robust MRI classification strategy for distinguishing glioblastoma from primary central nervous system lymphoma. Sci Rep 9(1):5746
Wang H, Zhou Y, Li L, Hou W, Ma X, Tian R (2020) Current status and quality of radiomics studies in lymphoma: a systematic review. Eur Radiol 30:6228–40
Cox RW (1996) AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29(3):162–173
Avants BB, Tustison N, Song G (2009) Advanced normalization tools (ANTS). Insight J 2(365):1–35
Zhang H, Lee K, Chen Z, Kashyap S, Sonka M (2020) LOGISMOS-JEI: Segmentation using optimal graph search and just-enough interaction. In: Handbook of medical image computing and computer assisted intervention. Academic Press, Cambridge, p 249–272
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77(21):e104–e107
Kuhn M, Wickham H (2018) RStudio. recipes: Preprocessing Tools to Create Design Matrices
Team RC (2013) R: a language and environment for statistical computing
Smith BJ (2020) MachineShop: machine learning models and tools. R package version 2.5.0
Bergmeir CN, Benítez Sánchez JM (2012) Neural networks in R using the Stuttgart neural network simulator: RSNNS. American Statistical Association
Bouckaert RR, Frank E (2004) Evaluating the replicability of significance tests for comparing learning algorithms. In: Dai H, Srikant R, Zhang C (eds) Advances in Knowledge Discovery and Data Mining. PAKDD 2004. Lecture Notes in Computer Science, vol 3056. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-24775-3_3
Nadeau C, Bengio Y (2003) Inference for the generalization error. Mach Learn 52(3):239–281
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol 57(1):289–300
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14(12):749–762
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Girish Bathla.
Conflict of interest
Girish Bathla has a research grant from Siemens AG, Forchheim, Germany, as well as American Cancer Society, that is unrelated to the submitted work. The rest of the authors report no relationships that could be construed as a conflict of interest.
Statistics and biometry
Caitlin ward (co-author) kindly provided statistical advice for this manuscript.
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 96 kb)
Rights and permissions
About this article
Cite this article
Bathla, G., Priya, S., Liu, Y. et al. Radiomics-based differentiation between glioblastoma and primary central nervous system lymphoma: a comparison of diagnostic performance across different MRI sequences and machine learning techniques. Eur Radiol 31, 8703–8713 (2021). https://doi.org/10.1007/s00330-021-07845-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-07845-6