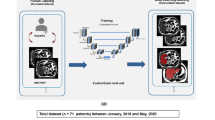
Abstract
Objective
To automate the segmentation of whole liver parenchyma on multi-echo chemical shift encoded (MECSE) MR examinations using convolutional neural networks (CNNs) to seamlessly quantify precise organ-related imaging biomarkers such as the fat fraction and iron load.
Methods
A retrospective multicenter collection of 183 MECSE liver MR examinations was conducted. An encoder-decoder CNN was trained (107 studies) following a 5-fold cross-validation strategy to improve the model performance and ensure lack of overfitting. Proton density fat fraction (PDFF) and R2* were quantified on both manual and CNN segmentation masks. Different metrics were used to evaluate the CNN performance over both unseen internal (46 studies) and external (29 studies) validation datasets to analyze reproducibility.
Results
The internal test showed excellent results for the automatic segmentation with a dice coefficient (DC) of 0.93 ± 0.03 and high correlation between the quantification done with the predicted mask and the manual segmentation (rPDFF = 1 and rR2* = 1; p values < 0.001). The external validation was also excellent with a different vendor but the same magnetic field strength, proving the generalization of the model to other manufacturers with DC of 0.94 ± 0.02. Results were lower for the 1.5-T MR same vendor scanner with DC of 0.87 ± 0.06. Both external validations showed high correlation in the quantification (rPDFF = 1 and rR2* = 1; p values < 0.001). In both internal and external validation datasets, the relative error for the PDFF and R2* quantification was below 4% and 1% respectively.
Conclusion
Liver parenchyma can be accurately segmented with CNN in a vendor-neutral virtual approach, allowing to obtain reproducible automatic whole organ virtual biopsies.
Key points
• Whole liver parenchyma can be automatically segmented using convolutional neural networks.
• Deep learning allows the creation of automatic pipelines for the precise quantification of liver-related imaging biomarkers such as PDFF and R2*.
• MR “virtual biopsy” can become a fast and automatic procedure for the assessment of chronic diffuse liver diseases in clinical practice.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
On the assessment of chronic diffuse liver diseases, such as non-alcoholic fatty liver disease (NAFLD), non-alcoholic steatohepatitis (NASH), or iron overload, magnetic resonance (MR) imaging has an important role in the evaluation of parenchymal deposits, including fat and iron [1]. NAFLD and NASH may progress towards increasing stages of hepatic fibrosis and, finally, to cirrhosis and HCC development [2]. Proton density fat fraction (PDFF) and R2* relaxation rate are used as reproducible quantitative metrics, obtained from MR images, for steatosis and iron concentration estimations, respectively. MR imaging biomarkers take an important role in the diagnosis and treatment monitoring of patients suffering from chronic diffuse liver diseases [3,4,5,6], offering additional information to the qualitative diagnosis made by radiologists.
Liver biopsy has important limitations related to the invasiveness of the procedure, the sampling bias due to the heterogeneous distribution of different histological features within the liver parenchyma, the intra- and inter-observer variability of pathological grading system, and the large patients’ unacceptance and withdrawal [7,8,9]. On the other hand, the so-called virtual biopsies, carried out through MR images and computational methods, can be performed multiple times and evaluate the heterogeneous distributions with high patients’ acceptance.
Multi-echo chemical shift–encoded gradient-echo (MECSE) MR images allow for the simultaneous and precise quantification of fat and iron within the liver parenchyma [4, 5, 10]. PDFF and R2* measurements are highly related to the histopathological grading systems, allowing MR virtual biopsy to become a common procedure performed in clinical practice for the assessment of chronic diffuse liver diseases [4, 5].
Liver PDFF and R2* quantification is usually obtained from small regions of interest (ROI); however, there is a lack of standardization when placing the ROIs across the liver (different sizes, locations, and number), introducing subsampling biases and subjective region selection, increasing variability across sites and studies, and reducing the reproducibility and repeatability of the imaging biomarkers quantified [11, 12]. To obtain an estimation of the patient liver status and to grade the heterogeneity of fat or iron distribution, MR virtual biopsies should evaluate the imaging biomarkers across the whole liver in a voxel-wise approach. Liver segmentation is nowadays done in a manual or semi-automatic way, hindering radiologists’ workflow.
The main difficulties for automatic segmentation of the liver in these MR images are related to its similarities with the surrounding organs, aggravated by the low image spatial resolution and large slice thickness, which causes partial volume effects. Moreover, diseased livers have different shapes, morphologies, and sizes among patients.
Traditionally, model-based [13], atlas-based [14,15,16], and level set–based [17, 18] methodologies have been used to segment the liver on both MR and CT images. Unfortunately, these methods entail high computation costs and long execution times, being a challenge to generate an atlas model able to comprehend the huge variability in liver morphology within the population, failing in the generalization to all liver signal intensities and shapes.
Learning-based methodologies [19,20,21,22,23,24,25] have been proposed to automate some tasks traditionally done by radiologists [26]. Regarding organ segmentation, artificial intelligence (AI) and mainly convolutional neural networks (CNNs) are able to model all variations found on a training dataset and to perform an automatic segmentation in some seconds without high computational needs. Up to now, most of the developed learning-based CNN methods for liver segmentation are built using computed tomography (CT) images [19,20,21,22,23]. The ones developed over MR examinations [22,23,24,25] used different sequences including diffusion-weighted [24], T2-weighted [23, 24], and dynamic contrast-enhanced [23, 25] exams.
Our hypothesis is that MECSE-MR images, needed to precisely quantify liver fat and iron deposits, can be used for whole liver segmentation by using a CNN solution to automatically extract imaging biomarkers. The aim of this study is to develop and validate, with both internal and external independent cases, a novel CNN-based model trained to generate a whole liver virtual dissection and quantification.
Materials and methods
A retrospective multicenter and international collection of 182 patients with suspected diffuse liver disease and a 2D-MECSE-MR examination was conducted (Table 1), only patients without intrahepatic lesions were included. A first cohort of 153 studies, from two different centers, was selected to create and test the model. A second cohort included 29 studies from two independent centers in different countries for external validation purposes.
Dataset preparation
All the studies were manually segmented by a radiologist with more than 5 years’ experience on abdominal MR imaging. The first echo time was selected as the reference image for manual segmentation due to its higher signal-to-noise ratio [23]. The liver parenchyma was delineated, avoiding the gallbladder and large vessels.
The different splits of the whole dataset required for the development and validation of the model are illustrated in Fig. 1.
Convolutional neural network architecture
The proposed architecture was an encoder-decoder CNN [27] with four convolutional blocks on each branch.
To increase the network generalization and reduce overfitting, a normalization of the activations was applied after each convolutional layer including a batch normalization layer [28].
Furthermore, to achieve an acceptable boundary detection and a faster convergence of the network, a deep supervision [29, 30] path was included. 1 × 1 convolutional layers were added at the output of each decoder level and summed to achieve the final activation map.
Preprocessing and data augmentation
All the images were resampled to a shape of 192 × 192 by applying a bicubic interpolation and normalized to (0–1) range.
To avoid overfitting and to increase the network generalization, on-the-fly data augmentation was applied during the training process. The transformations applied include rotations between ± 5° and Gaussian noise addition (μ = 0; σ є [0.001, 0.01]) to the training images.
Model training and validation/tuning
To analyze the generalization and robustness of the designed architecture to new unseen data and to tune some CNN hyperparameters, a 5-fold cross-validation strategy was followed.
The training was performed along 300 epochs with a batch size of 40 images. The loss function to optimize on each iteration was based on the Dice coefficient (DC) [31]. An ADAM optimizer [32] was used during the training process. The initial learning rate was set to 1e-5 and the remaining hyperparameters were kept with their default values [32].
PDFF and R2* quantification
Both PDFF and R2* were quantified voxel-wise in all MR studies using QUIBIM Precision – Liver fat and iron v1.0.0 analysis module (QUIBIM), approved as medical device with CE mark class IIa. The median liver value was used for each patient. Different ranges to differentiate steatosis and siderosis grades were defined following the thresholds introduced in [4]:
-
Steatosis (PDFF): none (< 4.8%), low (4.8–8.5%), mild (8.6–12.9%), high (> 12.9%).
-
Siderosis (R2*): none (< 42 s-1), mild (42–91 s-1), high (> 91 s-1).
To evaluate automatic segmentation performance, PDFF and R2* were quantified over both CNN-based and manual liver masks.
Heterogeneity assessment
The test dataset was used to analyze if the trained model was able to generalize to cases in which fat and/or iron were distributed heterogeneously. Four circular ROIs (7cm2) were drawn across the liver (two ROIs within the left lobe and two ROIs within the right lobe) avoiding non-liver parenchyma regions. The median value of each ROI was calculated and the difference between the lowest and highest values was extracted (ΔhDFF, ΔR2*).
Testing and performance evaluation
Seventy-six previously unseen cases were used to evaluate the network performance. After image normalization, all MR series were segmented with a 2D slice-by-slice approach using the trained network and stacked to finally obtain the whole liver mask. At the CNN output, a probability liver map was obtained. To increase robustness in the quantification, a more conservative segmentation was chosen ensuring that all the segmented voxels belong to liver parenchyma, without worrying if some peripheral voxels corresponding to the liver were missed. For that, all the voxels with a probability higher than 90% were defined as liver.
After segmentation, along the 3D mask, some small isolated segmented regions occasionally appeared. Therefore, all regions with a volume lower than the biggest component were removed, leaving a single large liver region.
Finally, to evaluate the performance and generalization of the trained network, different parameters were calculated to measure the differences between the CNN-based and the manual (ground truth) masks: dice coefficient (DC), volume overlap error (VOE), relative volume difference (RVD), average symmetric surface distance (ASSD), maximum surface distance (MSD), false discovery rate (FDR), Spearman correlation index (r) for liver volume, PDFF and R2*, and relative error in the quantification of PDFF and R2*.
Results
PDFF and R2* quantification
For each subset of data, the percentage of patients belonging to each steatosis and siderosis grade was analyzed using the median values obtained with the manual masks and the defined thresholds. Different PDFF and R2* distributions are found along the subsets (Fig. 2); therefore, the reproducibility of the model and its generalization to different steatosis and siderosis grades were analyzed.
Heterogeneity assessment
When analyzing PDFF and R2* on four different ROIs across the liver, a ΔPDFF of 4.01 ± 1.75% (mean ± SD) and a ΔR2* of 11.13 ± 7.45s-1 were obtained, being the maximum difference 8.57% for the PDFF and 38.56s-1 for the R2*. Whether the steatosis and/or siderosis grades are associated with each patient changed depending on the ROI value used was analyzed. Fifty-five of the cases suffered a steatosis grade change, while 33% had a change in the siderosis grade. Therefore, there are some cases in the test dataset where PDFF and/or R2* were distributed heterogeneously and the generalization to these cases was proved.
Model validation
The network robustness to different datasets was evaluated comparing the DC calculated over the training and tuning-validation datasets on each fold of the 5-fold cross-validation. Table 2 shows, on each fold, the highest mean DC obtained over the validation dataset and its corresponding value over the training dataset. The DC showed similar values along the different folds in both datasets, while maintaining similar results on each fold independently.
Model testing
The mean, standard deviation, and median values of the different metrics are summarized in Table 3. The median DC was 94% with a FDR of 4%, meaning that the number of non-liver segmented voxels was minimized while maintaining a good segmentation performance.
Furthermore, a low relative error in both PDFF and R2* quantification was obtained, with mean ± SD values of 2.8 ± 3.25% and 0.59 ± 0.52% and median values of 1.4% and 0.47%, respectively.
PDFF and R2* correlations between the results obtained when analyzing the liver using the manual segmentation and the CNN mask are shown in Fig. 3 (upper row), showing a perfect correlation on both components (PDFF and R2*: r = 1, p < 0.001). The correlation between the liver volumes is also represented, obtaining a correlation value of r = 0.98 (p < 0.001).
Correlation analysis over the testing dataset for the liver volume (left), PDFF (middle), and R2* (right) quantification when comparing the results obtained when using the ground truth and the predicted segmentation mask. In the upper rows, the correlation between manual-based and automatic-based quantitative data is represented. In the bottom rows, Bland-Altman plots to compare both variables of each parameter are included. Within the correlation images, in x- and y-axes respectively, the histogram of the variables obtained with the ground truth and the CNN predicted mask is represented
Bland-Altman plots (Fig. 3, lower row) show the difference in the quantification of liver volume, PDFF, and R2*. These parameters show a low bias (volume: 0.07L, PDFF: 0.12% and R2*: 0.27s-1) and narrow limits of agreement (volume: [- 0.11 to 0.26] L, PDFF: [- 0.12 to 0.36]% and R2*: [- 0.18 to 0.71]s-1). Similar differences are obtained along the whole spectrum for each variable (no differences are seen between low and high volume, PDFF, and R2* values).
Figure 4 illustrates three different test studies. As seen, large vessels, such as the inferior vena cava, were excluded from the predicted segmentation mask.
Segmentations obtained on three different test cases at different anatomical levels. In yellow the CNN predicted segmentation mask is shown and in red the contour of the ground truth is represented. a Results in a study from one of the centers used for the model development. b Results in a study from the other center used for the model development. c Performance on a study which belongs to a patient with partial hepatectomy
External validation
The reproducibility of the trained model was evaluated using an additional MR dataset from different centers and scanners (Table 4). The results obtained with a 1.5-T scanner are lower than those obtained with a 3-T scanner (DC: 0.93 ± 0.03 Philips 3T, 0.87 ± 0.06 Philips 1.5T). However, although the model was trained with Philips MR studies, the results with the Siemens scanner were higher (DC: 0.93 ± 0.03 Philips, 0.94 ± 0.02 Siemens).
In the assessment of PDFF and R2* quantification, a low relative error was seen. At institution 1, the mean ± SD relative error was 3.24 ± 3.93% for the PDFF and 0.80 ± 0.85% for the R2*, while the median values were 1.99% for the PDFF and 0.53% for the R2*. At institution 2, the mean ± SD values were 3.42 ± 3.99% for the PDFF and 0.86 ± 0.76% for the R2* and the median values were 1.96% for the PDFF and 0.73% for the R2*.
Figure 5 shows the correlation between the results obtained in the liver volume, PDFF, and R2* quantification, showing a nearly perfect correlation between the results obtained with both masks. At institution 1, the correlation between volume values was r = 0.96 (p < 0.001), while at institution 2, it was r = 0.88 (p < 0.001). At both institutions, both PDFF and R2* showed a perfect correlation (r = 1; p < 0.001). A Bland-Altman analysis was also conducted to analyze the differences in the quantification of the three parameters. For all the cases, the results showed a low bias value and narrow limits of agreement. In addition, similar values are obtained in the whole spectrum for each parameter.
Differences between the liver volume (left), PDFF (middle), and R2* (right) values obtained using the ground truth and the CNN predicted mask at institution 1 and institution 2. Within each institution, the upper row shows the correlation analysis between both values and the bottom row contains the Bland-Altman plots. In the correlation images, in x- and y-axes respectively, the histogram of the variables obtained with the ground truth and the CNN predicted mask is represented
The mask obtained on a study from each center is represented in Fig. 6.
Segmentation obtained on two studies from the external validation at different anatomical levels. In yellow the predicted segmentation mask is shown and in red the contour of the ground truth is represented. a Study acquired on a Siemens 3-T MR scanner. b Study acquired on a Philips 1.5-T MR scanner
Processing time
The overall processing time for the whole 3D liver CNN segmentation and quantification was 26 ± 6 s (mean ± SD).
Discussion
A novel methodology for an automatic vendor and field strength–neutral segmentation of the liver parenchyma on low-contrast MECSE-MR images based on CNN has been proposed. This automatic segmentation allows the quantification of fat (PDFF) and iron (R2*) in the assessment of diffuse liver diseases.
Different learning-based models have been reported for liver segmentation on a variety of MR series [22,23,24,25]. However, only one publication used a MECSE-MR sequence [22] with a DC of 0.93 ± 0.04, the same as us in our testing dataset. Although an external validation with different MR scanners was also performed, no results per scanner were provided. The mean DC reported in [22] for the external validation was 0.92 ± 0.05, while in our external validation, we achieved a mean DC of 0.87 ± 0.06 in a Philips 1.5-T scanner and 0.94 ± 0.02 in a Siemens 3-T scanner.
Previous studies focused on the maximization of the segmentation metrics, not focusing on using the mask to extract imaging biomarkers. The main objective of our method was achieving the highest accuracy in liver segmentation reducing false-positive areas to avoid errors and obtain the highest precision on PDFF and R2* quantification. Accordingly, the obtained liver mask has to be accurate but conservative, without including neighbor structures such as gallbladder or large vessels, even at the cost of losing small peripheral liver areas. The results obtained in this study show a high correlation and a quite low relative error when comparing the PDFF and R2* liver values quantified using the manual and the CNN-based mask.
A study by Stocker et al [33] shows the differences obtained when quantifying PDFF and R2* using an in-house automatic tool for liver segmentation and quantification. When comparing the Bland-Altman plots, narrower limits of agreement were obtained for both PDFF and R2* in our study, meaning that the differences between the results from the manual and the automatic tools were lower in our solution.
Noteworthily, the reproducibility of the developed model was analyzed using MECSE-MR studies from external centers with different scanners from the ones used during training-tunning. CNN-based models are known to offer different results when small changes are applied to the input images [34, 35], being very sensitive to training data properties. For this reason, we constructed a dataset covering a large population spectrum, from different centers and scanners, applying different preprocessing techniques such as data augmentation and image normalization to improve the reproducibility of the model.
The lower precision obtained with the 1.5-T MR scanner can be related to the fact that these opposed-phase images were acquired with a lower in-plane resolution (2.3 × 2.3 mm2) and a higher slice thickness (10-10.5 mm), aggravating partial volume effects and, therefore, making difficult tissue differentiation. To increase the model generalization to these acquisitions, the CNN model should be further tuned and retrained with images from scanners with lower field strength magnets and lower image resolution. Another option would be the application of recently published solutions to perform a manufacturer shift using different CNN-based style transfer applications based on generative adversarial networks (GANs) [36]. However, when comparing the PDFF and R2* values quantified from the manual ground truth and the predicted mask, the high correlation between values and the low relative error in the quantification foster its applicability in clinical practice. Additionally, the CNN solution showed generalization to other manufacturers; the results obtained with Siemens images were better than those obtained with Philips scanner. It is also relevant to note that the training and internal validation processes were conducted using images from Spain and Portugal (Europe), while the external validation faced studies from Argentina and Chile (South America), fostering the international generalization of the tool.
In this way, the automatic quantification of PDFF and R2* allows the generation of virtual biopsies, facing the main problems that pathological biopsies entail, offering a huge advance on the assessment of diffuse liver diseases. Moreover, any error obtained in the CNN-based segmentation can be manually edited by an expert before performing the quantitative analysis over the new corrected mask, ensuring the validity of the results.
It is well-known that other 3D-MR sequences are commonly used. Since the proposed algorithm is based on a 2D CNN, the MR images are segmented slice-by-slice. Therefore, since the CNN has learned to segment independent slices from different anatomical levels, the expansion of this solution to 3D acquisitions is direct.
The main limitation of this study relates to the limited sample size, with a lack of studies coming from some other manufacturers and from scanners with different MECSE-MR acquisition protocols. Other next steps include liver parcellation in left-right lobes, and in the different hepatic segments. A complete liver volumetry together with a separate PDFF and R2* quantification will improve the heterogeneity evaluation of fat and iron distribution. Furthermore, the segmentation of the lesions will be of interest to exclude them from the liver parenchyma quantification.
In conclusion, whole liver parenchyma can be automatically and accurately segmented using CNN before imaging biomarker extraction. Deep learning liver virtual dissection allows the creation of automatic pipelines to characterize diffuse liver diseases through the quantification of fat fraction and iron concentration by calculating the PDFF and R2* in a voxel-wise manner. This model allows MR virtual biopsy to become a fast and automatic procedure for the assessment of chronic diffuse liver diseases in clinical practice.
Abbreviations
- AI:
-
Artificial intelligence
- ASSD:
-
Average symmetric surface distance
- CNN:
-
Convolutional neural network
- DC:
-
Dice coefficient
- FDR:
-
False discovery rate
- MECSE:
-
Multi-echo chemical shift encoded
- MSD:
-
Maximum surface distance
- NAFLD:
-
Non-alcoholic fatty liver disease
- NASH:
-
Non-alcoholic steatohepatitis
- PDFF:
-
Proton density fat fraction
- RVD:
-
Relative volume difference
- VOE:
-
Volumetric overlap error
References
Donato H, França M, Candelária I, Caseiro-Alves F (2017) Liver MRI: from basic protocol to advanced techniques. Eur J Radiol 93:30–39
Farrell GC, Larter CZ (2006) Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 43(2 Suppl 2):S99–S112
Kinner S, Reeder SB, Yokoo T (2016) Quantitative imaging biomarkers of NAFLD. Dig Dis Sci 61(5):1337–1347
França M, Alberich-Bayarri Á, Martí-Bonmatí L et al (2017) Accurate simultaneous quantification of liver steatosis and iron overload in diffuse liver diseases with MRI. Abdom Radiol (NY) 42(5):1434–1443
Kühn JP, Hernando D, Muñoz del Rio A et al (2012) Effect of multipeak spectral modeling of fat for liver iron and fat quantification: correlation of biopsy with MR imaging results. Radiology. 265(1):133–142
Rostoker G, Laroudie M, Blanc R et al (2017) Signal-intensity-ratio MRI accurately estimates hepatic iron load in hemodialysis patients. Heliyon. 3(1):e00226
Ratziu V, Charlotte F, Heurtier A et al (2005) Sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology. 128(7):1898–1906
Butensky E, Fischer R, Hudes M et al (2005) Variability in hepatic iron concentration in percutaneous needle biopsy specimens from patients with transfusional hemosiderosis. Am J Clin Pathol 123(1):146–152
Deugnier Y, Turlin B (2007) Pathology of hepatic iron overload. World J Gastroenterol 13(35):4755–4760
Martí-Bonmatí L, Alberich-Bayarri A, Sánchez-González J (2012) Overload hepatitides: quanti-qualitative analysis. Abdom Imaging 37(2):180–187
McCarville MB, Hillenbrand CM, Loeffler RB et al (2010) Comparison of whole liver and small region-of-interest measurements of MRI liver R2* in children with iron overload. Pediatr Radiol 40(8):1360–1367
Campo CA, Hernando D, Schubert T, Bookwalter CA, Pay AJV, Reeder SB (2017) Standardized approach for ROI-based measurements of proton density fat fraction and R2* in the liver. AJR Am J Roentgenol 209(3):592–603
Esfandiarkhani M, Foruzan AH (2017) A generalized active shape model for segmentation of liver in low-contrast CT volumes. Comput Biol Med 82:59–70
Yan Z, Zhang S, Tan C et al (2015) Atlas-based liver segmentation and hepatic fat-fraction assessment for clinical trials. Comput Med Imaging Graph 41:80–92
Dura E, Domingo J, Göçeri E, Martí-Bonmatí L (2017) A method for liver segmentation in perfusion MR images using probabilistic atlases and viscous reconstruction. Pattern Anal Appl 21(4):1083–1095
Xu Y, Xu C, Kuang X et al (2016) 3D-SIFT-Flow for atlas-based CT liver image segmentation. Med Phys 43(5):2229
Yuan Z, Wang Y, Yang J, Liu Y (2010). A novel automatic liver segmentation technique for MR images. 2010 3rd International Congress on Image and Signal Processing, Yantai. 1282–1286
Göçeri E (2016) Fully automated liver segmentation using Sobolev gradient-based level set evolution. Int J Numer Method Biomed Eng 32(11)
Yang D, Xu D, Zhou SK et al (2017). Automatic liver segmentation using an adversarial image-to-image network. Medical Image Computing and Computer-Assisted. MICCAI 2017. Lecture Notes in Computer Science, vol 10435. Springer, Cham
Han X (2017). Automatic liver lesion segmentation using a deep convolutional neural network method. arXiv: 1704.07239
Qin W, Wu J, Han F et al (2018) Superpixel-based and boundary-sensitive convolutional neural network for automated liver segmentation. Phys Med Biol 63(9):095017
Christ PF, Ettlinger F, Grün F et al (2017). Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. arXiv: 1702.05970
Wang K, Mamidipalli A, Retson T et al (2019) Automated CT and MRI liver segmentation and biometry using a generalized convolutional neural network. Radiol Artif Intell 1(2):180022
Lavdas I, Glocker B, Kamnitsas K et al (2017) Fully automatic, multiorgan segmentation in normal whole body magnetic resonance imaging (MRI), using classification forests (CFs), convolutional neural networks (CNNs), and a multi-atlas (MA) approach. Med Phys 44(10):5210–5220
Jansen MJA, Kuijf HJ, Niekel M et al (2019) Liver segmentation and metastases detection in MR images using convolutional neural networks. J Med Imaging (Bellingham) 6(4):044003
Montagnon E, Cerny M, Cadrin-Chênevert A et al (2020) Deep learning workflow in radiology: a primer. Insights Imaging 11(1)
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham
Wang L, Yang Y, Min R, Chakradhar S (2017) Accelerating deep neural network training with inconsistent stochastic gradient descent. Neural Netw 93:219–229
Lee C, Xie S, Gallagher P, Zhang Z, Tu Z (2015). Deeply-supervised nets. Artif Intell Stat 562–570
Dou Q, Chen H, Jin Y, Yu L, Qin J, Heng P (2016) 3D deeply supervised network for automatic liver segmentation from CT volumes. Medical Image Computing and Computer-Assisted. MICCAI 2016. Springer, Cham
Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ (2017). Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA 2017, ML-CDS 2017. Lecture Notes in Computer Science, vol 10553. Springer, Cham
Kingma DP, Ba J (2015) Adam: a method for stochastic optimization. international conference on learning representations. 1–13. arXiv: 1412.6980
Stocker D, Bashir MR, Kannengiesser SAR, Reiner CS (2018) Accuracy of automated liver contouring, fat fraction, and R2* measurement on gradient multiecho magnetic resonance images. J Comput Assist Tomogr 42(5):697–706
Fezza SA, Bakhti Y, Hamidouche W, Deforges O (2019) Perceptual evaluation of adversarial attacks for CNN-based image classification. 2019 Eleventh International Conference on Quality of Multimedia Experience (QoMEX). 1–6
Chen L, Bentley P, Mori K, Misawa K, Fujiwara M, Rueckert D (2019) Intelligent image synthesis to attack a segmentation CNN using adversarial learning. Simulation and Synthesis in Medical Imaging. SASHIMI 2019. Lecture Notes in Computer Science, vol 11827. Springer, Cham
Yan W, Huang L, Xia L et al (2020) MRI manufacturer shift and adaptation: increasing the generalizability of deep learning segmentation for MR images acquired with different scanners. Radiol Artif Intell 2(4):e190195
Acknowledgements
DMA is recipient of a Río Hortega award (CM19/00212), Instituto de Salud Carlos III.
Funding
This study was partially funded by the Spanish Ministry of Science and innovation, Instituto de Salud Carlos III (PI19/0380) and GILEAD Sciences (Grant Number: GLD19/00050). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Luis Marti-Bonmati.
Conflict of interest
The authors of this manuscript declare relationships with the following companies: QUIBIM SL.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• Observational
• Multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jimenez-Pastor, A., Alberich-Bayarri, A., Lopez-Gonzalez, R. et al. Precise whole liver automatic segmentation and quantification of PDFF and R2* on MR images. Eur Radiol 31, 7876–7887 (2021). https://doi.org/10.1007/s00330-021-07838-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-07838-5