Abstract
Objectives
Isocitrate dehydrogenase wild-type (IDHwt) lower-grade gliomas of histologic grades II and III follow heterogeneous clinical outcomes, which necessitates risk stratification. We aimed to evaluate whether radiomics from MRI would allow prediction of overall survival in patients with IDHwt lower-grade gliomas and to investigate the added prognostic value of radiomics over clinical features.
Methods
Preoperative MRIs of 117 patients with IDHwt lower-grade gliomas from January 2007 to February 2018 were retrospectively analyzed. The external validation cohort consisted of 33 patients from The Cancer Genome Atlas. A total of 182 radiomic features were extracted. Radiomics risk scores (RRSs) for overall survival were derived from the least absolute shrinkage and selection operator (LASSO) and elastic net. Multivariable Cox regression analyses, including clinical features and RRSs, were performed. The integrated areas under the receiver operating characteristic curves (iAUCs) from models with and without RRSs were calculated for comparisons. The prognostic value of RRS was assessed in the validation cohort.
Results
The RRS derived from LASSO and elastic net independently predicted survival with hazard ratios of 9.479 (95% confidence interval [CI], 3.220–27.847) and 6.148 (95% CI, 3.009–12.563), respectively. Those RRSs enhanced model performance for predicting overall survival (iAUC increased to 0.780–0.797 from 0.726), which was externally validated. The RRSs stratified IDHwt lower-grade gliomas in the validation cohort with significantly different survival.
Conclusion
Radiomics has the potential for noninvasive risk stratification and can improve prediction of overall survival in patients with IDHwt lower-grade gliomas when integrated with clinical features.
Key Points
• Isocitrate dehydrogenase wild-type lower-grade gliomas with histologic grades II and III follow heterogeneous clinical outcomes, which necessitates further risk stratification.
• Radiomics risk scores derived from MRI independently predict survival even after incorporating strong clinical prognostic features (hazard ratios 6.148–9.479).
• Radiomics risk scores derived from MRI have the potential to improve survival prediction when added to clinical features (integrated areas under the receiver operating characteristic curves increased from 0.726 to 0.780–0.797).
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lower-grade gliomas of World Health Organization (WHO) grades II and III are infiltrative neoplasms with variable clinical outcomes [1]. A mutation in isocitrate dehydrogenase (IDH) is one of the key genetic events which leads to the stratification of gliomas with significantly different survival rates in grade II/III gliomas as well as glioblastomas [1,2,3,4]. Among lower-grade gliomas of histologic grades II and III, IDH wild-type (IDHwt) tumors constitute less than 30% of cases and have been reported to follow worse prognosis than those with IDH mutations [2, 5, 6]. Moreover, a previous study suggested that the majority of IDHwt lower-grade gliomas with grade II/III were considered as the molecular equivalent of conventional glioblastoma [7].
Even though the presence of an IDH mutation is a powerful prognostic factor, heterogeneous clinical outcomes have been reported in IDHwt lower-grade gliomas according to the variable combination of genetic profiles [8,9,10]. Recently, the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy (cIMPACT-NOW) provided the description of a “diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV,” which corresponds to grade II/III IDHwt gliomas with high-level EGFR amplification, the combination of a whole chromosome 7 gain and a whole chromosome 10 loss (+ 7/− 10), or TERT promoter mutations [11]. These specifications highlight that the specific subset of IDHwt lower-grade gliomas follow an aggressive clinical course more closely resembling that of an IDHwt glioblastoma. Thus, it would be beneficial if MRI could stratify IDHwt lower-grade glioma patients according to their risks and identify patients with worse prognosis.
Radiomics exploits MRI data and extracts high-dimensional quantitative imaging features such as intensity distributions, spatial relationships, textural heterogeneity, and shape descriptors [12]. Because radiomics models use high-throughput imaging features, hidden information, which may be visually imperceptible, could be revealed [13]. Radiomics has been applied in many previous studies to predict a specific mutation status, stratify risk, or predict treatment responses in patients with glioblastomas [14,15,16,17]. Several previous studies have also used radiomics in patients with lower-grade gliomas to predict survival [18, 19] or detect molecular subtypes, including IDH mutation status [20,21,22,23]. However, the prognostic significance of radiomics in patients with IDHwt lower-grade gliomas has not been well studied.
We hypothesized that MRI radiomics can independently predict the survival in patients with IDHwt lower-grade gliomas. Therefore, the purpose of this study was to evaluate whether radiomics from MRI would allow prediction of survival in patients with IDHwt lower-grade gliomas and to investigate the added prognostic value of radiomics over clinical features.
Materials and methods
This retrospective study was approved by the institutional review board of our hospital, and the requirement for obtaining informed patient consent was waived. The NIH/NCI-approved Cancer Genome Atlas (TCGA) and The Cancer Imaging Archive (TCIA) databases are publicly available datasets in which all data are anonymized [24]. Thus, individual institutional approval was not required for the external validation set.
Patients
Institutional cohort
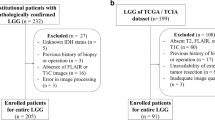
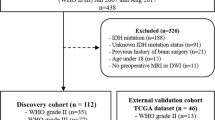
From January 2007 to February 2018, 459 patients with pathologically confirmed lower-grade gliomas were identified. Patients with IDHwt lower-grade gliomas who underwent preoperative MRI were included. Patients were excluded if they presented any of the following: (1) IDH mutant tumors (n = 202); (2) unknown IDH mutation status (n = 91); (3) previous history of brain surgery or treatment (i.e., radiation therapy or chemotherapy) (n = 21); (4) age under 18 years old (n = 16); (5) no preoperative MRI performed (n = 12). Thus, 117 patients with IDHwt lower-grade gliomas were enrolled in this study (Fig. 1). These included 35 (29.9%) WHO grade II gliomas and 82 (70.1%) grade III gliomas.
The extent of tumor resection was determined by visually comparing the preoperative and postoperative lesion volume on both T2-weighted images (T2WI) and contrast-enhanced T1-weighted imaging (T1C), in conjunction with intraoperative impressions of the surgeons, classified as one of three categories: gross total, subtotal (< 100% and ≥ 75% of gross tumor removal)/partial (< 75% of gross tumor removal) resection, or biopsy only. Karnofsky Performance Status (KPS) scores which were determined preoperatively and postoperative treatment (i.e., chemotherapy, radiation therapy) were retrieved from their electronic medical records.
Overall survival (OS) was used as a clinical outcome. OS was defined as the number of days from the initial surgery, i.e. tumor resection or biopsy, to either patient death or the date of the last follow-up if the patient did not die.
External validation cohort
TCGA (http://cancergenome.nih.gov) provides 265 lower-grade gliomas of histologic grades II and III, within which 46 were IDHwt. Among them, 33 patients (10 grade II and 23 grade III) with preoperative MRI containing both T2WI and T1C images were retrieved and constituted the external validation cohort. Information including age, extent of resection, WHO grade, and death were available for these 33 patients. KPS was known for 22 patients.
MR image acquisition
A 3.0-T MRI scanner (Achieva, Philips Healthcare) and an 8-channel SENSE head coil were used for all preoperative MRI scans. The preoperative MRI protocol included T2WI (TR/TE, 2800–3000/80–100 ms; FOV, 240 mm; section thickness, 5 mm; matrix, 256 × 256) and three-dimensional T1C (TR/TE, 6.3–8.3/3.1–4 ms; FOV, 240 mm; section thickness, 1 mm; matrix, 192 × 192). T1C images were acquired after administering 0.1 mL/kg gadolinium-based contrast material (Gadovist, Bayer).
The preoperative T2WI and T1C of 33 patients from the TCGA datasets were downloaded from TCIA (http://www.cancerimagingarchive.net).
Image pre-processing and radiomic feature extraction
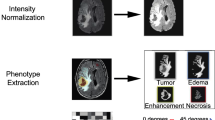
First, T2WI and T1C images were resampled to an identical spatial resolution of 1 × 1 × 1 mm using nilearn (https://nilearn.github.io). Next, those images were subjected to N4 bias correction to remove low-frequency intensity and non-uniformity from the images [25, 26]. After resampling and N4 bias correction, T1C images were registered to identical spatial coordinates using T2WI as a template using SimpleITK (http://www.simpleitk.org). Signal intensity was normalized using the WhiteStripe R package [27], implemented in R version 3.4.0 (R Foundation for Statistical Computing). Tumor segmentation was then performed by C.J.P., a neuroradiologist with 6 years of experience, with a semi-automatic method using 3D slicer (www.slicer.org) [28]. The software automatically selected the T2 hyperintense lesion where the signal is above the specific threshold, which corresponds to the infiltrative tumor and surrounding edema. Then, C.J.P. revised the automatically selected region of interest (ROI) by excluding erroneously detected cerebrospinal fluid or correcting the tumor border, as the periphery of the tumor could not be exactly determined by the software due to the infiltrative nature of the gliomas. T1C was not referred for the tumor segmentation as it is difficult to determine the tumor border in the T1C. Another neuroradiologist (S.S.A.) with 13 years of experience subsequently re-evaluated and confirmed the ROIs.
In addition, to evaluate the reproducibility of tumor segmentations, we randomly selected 50 patients in whom we obtained two different ROIs from two different readers (C.J.P., and S.S.A.). Then, we calculated the Dice coefficient (DICE), which is the most commonly used metric in validating medical volume segmentations [29, 30]. The DSC measure represents the relative overlap between two binary volume data and is expressed in the equation below.
V denotes the volume of binary data; R1 and R2 represent the ROIs from reader 1 and reader 2, respectively. The DICE score is 1.0 when the two volumes match perfectly.
Radiomic features were extracted from the ROIs on T2WI and T1C images by using Pyradiomics 1.2.0 (http://www.radiomics.io/pyradiomics.html) [31].
In total, 13 shape, 18 first-order, 23 gray-level co-occurrence matrix (GLCM), 16 gray-level run length matrix (GLRLM), 16 gray-level size zone matrix (GLSZM), and 5 neighborhood gray tone difference matrix (NGTDM) were extracted from the ROIs on T2WI and T1C images, constituting a total of 182 radiomic features. A schematic workflow of image pre-processing and radiomic feature extraction is shown in Fig. 2.
Pathological evaluation and molecular subtyping
All surgical specimens were histopathologically diagnosed according to the 2016 WHO classification. Both peptide nucleic acid–mediated clamping polymerase chain reaction and immunohistochemical analyses were performed to detect the presence of a IDH1-R132H mutation [2]. Monoclonal antibody H09 was used for immunohistochemical analysis. The degree of IDH1-R132H staining was determined positive if there were any stained cells. Cases without IDH1-R132H staining were determined to be negative [32, 33]. In IDH1-negative cases, IDH1/2 status was confirmed by a peptide nucleic acid–mediated clamping polymerase chain reaction.
Statistical analysis
Statistical analysis was performed in R software (version 3.5.1; R Foundation for Statistical Computing) (www.R-project.org). Because of the relatively large number of imaging variables compared with the number of events, the least absolute shrinkage and selection operator (LASSO) and elastic net were used to select important features, to minimize the potential risk for overfitting, by shrinking the regression coefficients of irrelevant variables toward zero [34]. The performances of these methods were tested by 10-fold cross-validation with 100 replications to enhance the generalizability of the results. The “glmnet” with “coxnet” and the “glmnetUtils” R packages were used for the LASSO and elastic net, respectively [35, 36]. A radiomics risk score (RRS) was calculated for each patient using a linear combination of features frequently selected from the LASSO and elastic net, weighted according to their regression coefficients. Univariable analysis of RRS and clinical features—age, KPS, extent of resection, WHO grade, and postoperative treatment—for OS prediction was performed. Subsequently, the variables which were significantly associated with OS were included for multivariable Cox regression analyses to create prognostic models: (1) model 1, clinical features only; (2) model 2, clinical features and RRS. To assess the discriminative ability, we obtained time-dependent receiver operating characteristic (ROC) curves and calculated the integral of the area under the curves (iAUC) [37]. The difference in the iAUC of the two models in the institutional cohort was tested based on a 95% confidence interval (CI) from a bootstrap with 1000 resampling, and internal validation for the developed model was performed using a bootstrap on the same resampled data. The iAUC difference was considered statistically significant if the 95% CI of the iAUC difference did not contain a zero value. Furthermore, the Akaike information criterion (AIC) was used to compare competing prognostic models, with a smaller AIC indicating a better model fit [38]. Additionally, likelihood ratio chi-square tests were performed to compare model fitness in terms of the log likelihood (LL) between two nested models, which was presented as minus 2LL (− 2LL).
With regard to the external validation, the prognostic value of RRS was assessed by comparing the iAUCs of model 1 (clinical features only) and model 2 (clinical features and RRS), which was tested based on a 95% CI from a bootstrap with 1000 resampling. In this analysis, 11 patients who lacked the KPS in the TCGA dataset were excluded.
In addition, the optimal cutoff values of RRSs were defined by the log-rank test, using the Contal and O’Quigley’s method, which was performed using the “cutp” function of “survMisc” in R [39]. The patients in the institutional and external validation cohort were then classified into low-risk and high-risk groups according to fixed cutoff values which were derived from institutional cohort. Differences in clinical characteristics between the low-risk and high-risk group were evaluated using Student’s t test and chi-square test. The Kaplan-Meier curves of those low-risk and high-risk groups were compared to evaluate the relationship between the RRS and OS. A p value < 0.05 was considered to be statistically significant.
Results
Characteristics of the 150 enrolled patients are summarized in Table 1. In the institutional cohort and external validation set, the median OS was 693 days (interquartile range, 384–1168 days) and 269 days (interquartile range, 155–582 days), respectively. The extent of resection was significantly different between the two groups (p < 0.001), but other clinical features were not significantly different.
The DICE scores which were calculated to assess the reproducibility of segmentations from two different readers ranged from 0.809 to 0.992 (mean ± standard deviation, 0.966 ± 0.035), which represents high reproducibility.
LASSO and elastic net selected 13 and 80 radiomic features for OS prediction, respectively, that were the most important for predicting tumor outcome. These radiomic features ranked according to their regression coefficients are listed in Supplementary Tables 1 and 2. RRS_LASSO and RRS_elasticnet were derived from the sum of features selected by LASSO and elastic net multiplied by each regression coefficient, respectively (Supplementary Tables 1 and 2). Thirteen features that were selected from both LASSO and elastic net included 12 texture features from GLSZM, NGTDM, and GLCM and one shape feature. The texture feature zone percentage from T1C was the parameter with the highest negative coefficient, associated with a favorable prognosis.
Univariable analysis of clinical features and RRS revealed that age, KPS, extent of resection, WHO grade, and RRS showed significant association with OS. Those selected features were included in the multivariable Cox models. Multivariable Cox models with and without RRS for OS prediction are summarized in Table 2. Both RRS_LASSO and RRS_elasticnet were independent predictors of OS with hazard ratios of 9.479 (95% CI, 3.220–27.847) and 6.148 (95% CI, 3.009–12.563), respectively. Time-dependent ROC curves from model 1 (clinical features only) and model 2 (clinical features and RRSs) are presented in Fig. 3. The iAUC of model 1 for OS prediction was 0.726 (95% CI, 0.678–0.814). When RRS_LASSO and RRS_elasticnet were added (model 2), the iAUC significantly increased to 0.780 (95% CI, 0.737–0.855) and 0.797 (95% CI, 0.752–0.872), respectively. On bootstrap testing, the increase in iAUC was statistically significant, with the differences in iAUC being 0.054 (95% CI, 0.013–0.104) and 0.040 (95% CI, 0.009–0.091), respectively. Smaller AIC values were noted in the models with RRS (392.2 and 381.0 for models with RRS_LASSO and RRS_elasticnet, respectively) compared with those without RRS (409.9), which indicate better prognostic models. In addition, smaller − 2LL values were also noted in the models with RRS (378.2 and 367.0 for models with RRS_LASSO and RRS_elasticnet, respectively) compared with those without RRS (397.9) with significant differences (p < 0.005 for all), which indicates better fitness of models.
Time-dependent receiver operating characteristic curves from models with and without radiomics risk scores (RRSs). Model 2 with both RRS_LASSO (a) and RRS_elasticnet (b) yielded significantly higher integrated area under the ROC curves (iAUC) of 0.758 (confidence interval [CI], 0.697–0.821) and 0.770 (CI, 0.706–0.833), respectively, compared with that of model 1 with only clinical features (0.701 [CI, 0.631–0.772])
The performance of survival prediction models was tested through internal validation (Table 3). Models with RRS derived from both LASSO and elastic net demonstrated significantly superior performance, with higher iAUC values (LASSO: 0.758, 95% CI, 0.697–0.821; elastic net: 0.770, 95% CI, 0.706–0.833) than in the model without RRS (0.701, 95% CI, 0.631–0.772) for OS prediction. On bootstrap testing, the increase in iAUC was statistically significant, with the differences in iAUC being 0.058 (95% CI, 0.008–0.090) for LASSO and 0.069 (95% CI, 0.001–0.108) for elastic net.
The performance of survival prediction models was also tested in the external validation cohort (Table 4). Models with RRS derived from both LASSO and elastic net yielded higher performance (LASSO: 0.754, 95% CI, 0.586–0.901; elastic net: 0.787, 95% CI, 0.619–0.931) than in the model without RRS (0.746, 95% CI, 0.543–0.921); however, the increase in iAUC was not statistically significant.
The optimal cutoff values of RRS_LASSO and RRS_elasticnet were derived from the institutional cohort, which were − 11.53 and − 14.86, respectively. These RRSs stratified institutional cohort into low-risk and high-risk groups with significantly different OS (p < 0.001, both, Supplementary figure and Supplementary Table 3). There was significant difference in the extent of resection between the two groups and total resection of the tumor was more frequently performed in the low-risk group. Those optimal cutoff values of RRS also stratified external validation cohort into two groups with significantly different OS (p = 0.001 and 0.020, respectively, Fig. 4). The detailed clinical characteristics of the low-risk and high-risk groups in the external validation cohort are demonstrated in Supplementary Table 4.
Discussion
We identified a subset of radiomic features that was significantly associated with OS in patients with IDHwt lower-grade gliomas of histologic grades II and III. When the RRS which was derived from the selected radiomic features was integrated with the clinical features, a superior model performance was achieved in predicting prognosis. There was also a trend toward better performance when RRS was added to a model with clinical features in the external validation cohort. In addition, the optimal cutoff of the RRS derived from the institutional cohort divided the external validation cohort into two groups with significantly different survival outcomes, demonstrating the prognostic value of the RRS. Our study therefore suggests that the radiomic features may allow noninvasive risk stratification of patients with IDHwt lower-grade gliomas and can be used as a potential imaging biomarker.
Several previous studies have adapted radiomics to predict survival in patients with lower-grade gliomas. Zhou et al utilized various combinations of clinical features, a standardized feature set named Visually AccesSAble Rembrandt Images features, and radiomic features to predict tumor progression, revealing that the radiomic features alone achieved the highest performance compared with the other combinations tested in patients with grade II/III gliomas [18]. Another recent study identified a significant association between radiomic features and progression-free survival in patients with grade II/III gliomas, and the model integrating both clinicopathologic features and radiomic features demonstrated high accuracy for progression-free survival prediction [19]. These studies have pointed to the prognostic potential of radiomic features. Similar results were observed in our study, demonstrating that the radiomic features also have prognostic value in patients with the IDHwt subgroup of lower-grade gliomas. In this study, we identified that radiomics not only is an independent factor for survival but also has added prognostic value when integrated with clinical features. Even though the absolute increase of iAUC was not considerably big when the radiomics was added to a model with clinical features (iAUC increased to 0.780–0.797 from 0.726), still it was statistically significant. Furthermore, the prognostic value of radiomics was proved in the external validation set. In addition, after incorporating strong prognostic clinical features such as extent of resection or WHO grade, RRSs were revealed to have significant association with survival. Therefore, radiomics has the potential for accurate noninvasive risk stratification in patients with IDHwt lower-grade gliomas.
The majority of radiomic features that were selected by LASSO and elastic net for survival prediction were texture features. Other studies also reported that texture features among radiomic features showed the prognostic values in patients with gliomas [15, 17, 40, 41]. Texture features quantify the image pattern on the basis of the spatial relationship or co-occurrence of the pixel value [42], which provide information on intratumoral heterogeneity [12]. Specifically, the GLSZM feature named “zone percentage” derived from T1C, which was consistently selected from both LASSO and elastic net, presented the strongest association with overall survival. The feature is a measurement of coarseness of the texture [29], which was reported to have strong discriminative ability between lower- and high-grade gliomas according to a recent study [43]. Therefore, in our study, tumors with higher values of GLSZM zone percentage could follow more favorable outcomes based on more homogenous textures. In addition, intratumoral heterogeneity in gliomas has been reported to be associated with aggressive tumor behavior and drug resistance [44, 45]. Therefore, texture features may play a key role in predicting prognosis.
In our study, we extracted the radiomic features from T2WI and T1C images. FLAIR, T1WI, and advanced imaging such as DWI or perfusion images were not used because many patients from the external validation cohort lack these data. Extracting features from more sequences may increase the model performance for survival prediction but would inevitably decrease the generalizability of the model. Our study results showed that the radiomic features only obtained with conventional MR images (T2WI and T1C) had prognostic value, which was validated in the independent cohort, and therefore these findings could be applied to clinical situations in which advanced imaging cannot be routinely performed. In addition, a previous study had demonstrated that the T1C alone performed best for the accurate survival stratification of patients with glioblastoma, which was comparable to the performance of a combination of other conventional MR images (T1WI, T1C, T2WI, and FLAIR) [45]. Similarly, another study reported that texture features from T1C images most accurately predicted OS in patients with glioblastomas compared with those from T1WI, T2WI, or FLAIR images [46]. Therefore, our study results are in line with those previous studies which concluded that T1C conveys prognostic information. Furthermore, we extracted features not only from T1C but also from T2WI because the study cohort included lower-grade gliomas that may present with large proportion of nonenhancing tumor, and almost half of consistently selected texture features were derived from T2WI. A recent study suggested that progression-free survival in patients with grade II/III gliomas could be predicted accurately by radiomic features solely extracted from T2WI [19]. Thus, we believe that radiomics from conventional MRI, especially T2WI and T1C, may provide with useful information for predicting a patient’s prognosis. Fortunately, this is also more feasible in clinical practice with regard to its application and validation.
There was a significant difference in the extent of resection between the institutional cohort and the validation cohort. Gross total resection of the tumor was less frequently performed, but a biopsy was more frequently performed in our institution compared with the validation cohort. These differences may be due to the heterogeneous nature of the TCGA, which consisted of data from multiple institutions, which may have different standards for deciding on the extent of resection. In addition, the extent of resection was determined by comparing the pre- and postoperative T2WI and T1C in our institution by a neuroradiologist; however, it remains unclear how the extent of resection has been determined in the TCGA dataset.
There are several limitations in this study to be addressed. First, this is a retrospective study and the number of study population was small, especially in the validation cohort, because IDHwt tumors account for less than 30% of lower-grade gliomas. In addition, unfortunately, the postoperative treatment was not retrievable in all patients, while KPS was not available in 11 patients in the TCGA dataset. However, we managed to externally validate the prognostic value of RRS in patients with available clinical data and found that adding RRS to a model with clinical features increased the performance of model for survival prediction. The differences of performances between models with and without RRS were not statistically significant, which might be attributed to the small number of patients. Further studies using a larger cohort with more comprehensive clinical information may be needed to validate our results; however, our study results revealed a potential of RRS as an imaging biomarker for predicting survival. Second, patients were enrolled from relatively long time periods (from January 2007 to February 2018) in the institutional cohort; therefore, the specific MR parameters could be changed over time. In addition, in the external validation cohort, MRI protocols were heterogeneous as the TCGA dataset contains data from different institutions. Therefore, those different scan parameters and MRI protocols which lied in the institutional and validation cohort might affect the extracted radiomic features. We performed pre-processing such as resampling and intensity normalization to mitigate the effects which might occur due to those heterogeneities. We believe that our results showed the potential of radiomics for survival prediction even using MRI with heterogeneous protocols. Third, EGFR amplification or TERTp mutation has been reported to have prognostic implication in IDHwt lower-grade gliomas according to the cIMPACT-NOW recommendations. However, unfortunately, those genetic mutation results were available in only a small number of patients in the institutional cohort; therefore, we were not able to include genetic markers in the model. Future studies are required to validate the prognostic value of RRS in conjunction with EGFR amplification or TERTp mutation status.
Conclusion
In conclusion, radiomics enables noninvasive risk stratification and can improve survival prediction in patients with IDHwt lower-grade gliomas when integrated with clinical features.
Abbreviations
- iAUC:
-
Integrated area under the receiver operating characteristic curve
- IDH:
-
Isocitrate dehydrogenase
- IDHwt:
-
Isocitrate dehydrogenase wild-type
- KPS:
-
Karnofsky Performance Status
- OS:
-
Overall survival
- ROC:
-
Receiver operating characteristic
- RRS:
-
Radiomics risk score
- TCGA:
-
The Cancer Genome Atlas
- WHO:
-
World Health Organization
References
Brat DJ, Verhaak RG, Aldape KD et al (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–2498
Yan H, Parsons DW, Jin G et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820
Jiao Y, Killela PJ, Reitman ZJ et al (2012) Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 3:709–722
Metellus P, Coulibaly B, Colin C et al (2010) Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol 120:719–729
Eckel-Passow JE, Lachance DH, Molinaro AM et al (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372:2499–2508
Reuss DE, Kratz A, Sahm F et al (2015) Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol 130:407–417
Aibaidula A, Chan AK, Shi Z et al (2017) Adult IDH wild-type lower-grade gliomas should be further stratified. Neuro Oncol 19:1327–1337
Chan AK, Yao Y, Zhang Z et al (2015) TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Mod Pathol 28:177–186
Chan AK, Yao Y, Zhang Z et al (2015) Combination genetic signature stratifies lower-grade gliomas better than histological grade. Oncotarget 6:20885–20901
Brat DJ, Aldape K, Colman H et al (2018) cIMPACT-NOW update 3: recommended diagnostic criteria for “diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol 136:805–810
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Kickingereder P, Gotz M, Muschelli J et al (2016) Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin Cancer Res 22:5765–5771
Bae S, Choi YS, Ahn SS et al (2018) Radiomic MRI phenotyping of glioblastoma: improving survival prediction. Radiology 289:797–806
Hu LS, Ning S, Eschbacher JM et al (2017) Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol 19:128–137
Kickingereder P, Burth S, Wick A et al (2016) Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 280:880–889
Zhou H, Vallieres M, Bai HX et al (2017) MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 19:862–870
Liu X, Li Y, Qian Z et al (2018) A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. Neuroimage Clin 20:1070–1077
Zhang X, Tian Q, Wang L et al (2018) Radiomics strategy for molecular subtype stratification of lower-grade glioma: detecting IDH and TP53 mutations based on multimodal MRI. J Magn Reson Imaging 48:916–926
Ren Y, Zhang X, Rui W et al (2019) Noninvasive prediction of IDH1 mutation and ATRX expression loss in low-grade gliomas using multiparametric MR radiomic features. J Magn Reson Imaging 49:808–817
Li Y, Qian Z, Xu K et al (2018) MRI features predict p53 status in lower-grade gliomas via a machine-learning approach. Neuroimage Clin 17:306–311
Liu X, Li Y, Li S et al (2019) IDH mutation-specific radiomic signature in lower-grade gliomas. Aging (Albany NY) 11:673–696
Pedano N, Flanders A, Scarpace L et al (2016) Radiology data from the cancer genome atlas low grade glioma [TCGA-LGG] collection. Cancer Imaging Archive
Tustison NJ, Avants BB, Cook PA et al (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320
Sled JG, Zijdenbos AP, Evans AC (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17:87–97
Shinohara RT, Sweeney EM, Goldsmith J et al (2014) Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin 6:9–19
Fedorov A, Beichel R, Kalpathy-Cramer J et al (2012) 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30:1323–1341
Taha AA, Hanbury A (2015) Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 15:29
Lee M, Woo B, Kuo MD, Jamshidi N, Kim JH (2017) Quality of radiomic features in glioblastoma multiforme: impact of semi-automated tumor segmentation software. Korean J Radiol 18:498–509
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Takano S, Tian W, Matsuda M et al (2011) Detection of IDH1 mutation in human gliomas: comparison of immunohistochemistry and sequencing. Brain Tumor Pathol 28:115–123
Choi J, Lee EY, Shin KJ, Minn YK, Kim J, Kim SH (2013) IDH1 mutation analysis in low cellularity specimen: a limitation of diagnostic accuracy and a proposal for the diagnostic procedure. Pathol Res Pract 209:284–290
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33:1–22
Microsoft HO (2017) glmnetUtils: utilities for ‘Glmnet’. R package version 1.1. Available via https://CRAN.R-project.org/package=glmnetUtils
Simon N, Friedman J, Hastie T, Tibshirani R (2011) Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw 39:1–13
Heagerty PJ, Zheng Y (2005) Survival model predictive accuracy and ROC curves. Biometrics 61:92–105
Akaike H (1974) A new look at the statistical model identification. IEEE Transactions on Automatic Control 19:716–723
Contal C, O’Quigley J (1999) An application of changepoint methods in studying the effect of age on survival in breast cancer. Comput Stat Data Anal 30:253–270
Kickingereder P, Neuberger U, Bonekamp D et al (2018) Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol 20:848–857
Ingrisch M, Schneider MJ, Norenberg D et al (2017) Radiomic analysis reveals prognostic information in T1-weighted baseline magnetic resonance imaging in patients with glioblastoma. Invest Radiol 52:360–366
Soni N, Priya S, Bathla G (2019) Texture Analysis in Cerebral Gliomas: A Review of the Literature. AJNR Am J Neuroradiol 40:928–934
Suárez-García JG, Hernández-López JM, Moreno-Barbosa E, de Celis-Alonso B (2020) A simple model for glioma grading based on texture analysis applied to conventional brain MRI. bioRxiv. https://doi.org/10.1101/2020.01.29.924712
Meyer M, Reimand J, Lan X et al (2015) Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A 112:851–856
Liu Y, Zhang X, Feng N et al (2018) The effect of glioblastoma heterogeneity on survival stratification: a multimodal MR imaging texture analysis. Acta Radiol 59:1239–1246
Yang D, Rao G, Martinez J, Veeraraghavan A, Rao A (2015) Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med Phys 42:6725–6735
Acknowledgments
The authors acknowledge M.S. Haesol Shin from the Department of Biostatistics and Computing, Yonsei University College of Graduate, for her support in statistical analysis. We also acknowledge Sang Wook Kim from the Department of Biomedical Engineering, Korea University, and Dongmin Choi from the Department of Computer Science, Yonsei University, for their support in radiomic feature extraction.
Funding
This research received funding from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, Information and Communication Technologies & Future Planning (2017R1D1A1B03030440).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Sung Soo Ahn.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise. Kyunghwa Han, Ph.D., from the Department of Radiology and Research Institute of Radiological Science, Yonsei University College of Medicine, has significant statistical expertise and one of the authors.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 280 kb)
Rights and permissions
About this article
Cite this article
Park, C.J., Han, K., Kim, H. et al. Radiomics risk score may be a potential imaging biomarker for predicting survival in isocitrate dehydrogenase wild-type lower-grade gliomas. Eur Radiol 30, 6464–6474 (2020). https://doi.org/10.1007/s00330-020-07089-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07089-w