Abstract
Objectives
The conceptus dose during diagnostic imaging procedures for pregnant patients raises health concerns owing to the high radiosensitivity of the developing embryo/fetus. The aim of this work is to develop a methodology for automated construction of patient-specific computational phantoms based on actual patient CT images to enable accurate estimation of conceptus dose.
Methods
We developed a 3D deep convolutional network algorithm for automated segmentation of CT images to build realistic computational phantoms. The neural network architecture consists of analysis and synthesis paths with four resolution levels each, trained on manually labeled CT scans of six identified anatomical structures. Thirty-two CT exams were augmented to 128 datasets and randomly split into 80%/20% for training/testing. The absorbed doses for six segmented organs/tissues from abdominal CT scans were estimated using Monte Carlo calculations. The resulting radiation doses were then compared between the computational models generated using automated segmentation and manual segmentation, serving as reference.
Results
The Dice similarity coefficient for identified internal organs between manual segmentation and automated segmentation results varies from 0.92 to 0.98 while the mean Hausdorff distance for the uterus is 16.1 mm. The mean absorbed dose for the uterus is 2.9 mGy whereas the mean organ dose differences between manual and automated segmentation techniques are 0.07%, − 0.45%, − 1.55%, − 0.48%, − 0.12%, and 0.28% for the kidney, liver, lung, skeleton, uterus, and total body, respectively.
Conclusion
The proposed methodology allows automated construction of realistic computational models that can be exploited to estimate patient-specific organ radiation doses from radiological imaging procedures.
Key Points
• The conceptus dose during diagnostic radiology and nuclear medicine imaging procedures for pregnant patients raises health concerns owing to the high radiosensitivity of the developing embryo/fetus.
• The proposed methodology allows automated construction of realistic computational models that can be exploited to estimate patient-specific organ radiation doses from radiological imaging procedures.
• The dosimetric results can be used for the risk-benefit analysis of radiation hazards to conceptus from diagnostic imaging procedures, thus guiding the decision-making process.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
One of the most frequently asked questions associated with the increasing clinical use of radiologic imaging is the management of radiation dose for patients presenting with high radiosensitivity, including pregnant female and pediatric patients [1,2,3]. Radiologic imaging should be avoided on pregnant females; however, in some very specific situations, computed tomography (CT) or even positron emission tomography (PET)/CT becomes unavoidable [4]. Under these circumstances, the radiation risks to the fetus are a significant concern. At fetal doses greater than 50 mGy, the potential hazard effects include embryonic death, intra-uterine growth limitation, average intelligence quotient (IQ) loss, mental retardation, organ malformation, and small head size [5,6,7]. Stochastic effects might also occur at fetal doses below 50 mGy [8,9,10]. According to ICRP publications [6, 11], the termination of pregnancy at fetal doses below 100 mGy cannot be justified based on radiation risks to the fetus while for fetal doses between 100 and 500 mGy, the decision of pregnancy termination should be considered based on individual circumstances. In this context, the accurate estimation of conceptus dose plays a key role in managing safety and quality in CT imaging procedures for pregnant patients, keeping in mind the current international recommendations in terms of good practice and diagnostic reference levels [12].
Different techniques have been adopted for estimating the conceptus dose in pregnant patients, including Monte Carlo calculations using dedicated computational anthropomorphic models [2, 13,14,15,16,17,18,19,20,21,22,23,24] and experimental measurements using physical phantoms [25,26,27,28]. However, these techniques naturally bear many limitations, including the difficulty of accurate modeling of individual patient’s anatomy considering the location, shape and size of internal organs, and the uterus/fetus within the target region of the pregnant patient’s body [29]. The inherent assumptions in the modeling and simulation setups might result in substantial overestimation or underestimation of the conceptus dose. Monte Carlo calculations are commonly integrated with computational phantoms to simulate radiation transport and account for all aspects of particle interactions within the human body. The technique is deemed to be the most accurate method for radiation absorbed dose calculation [30]. The relevance and reliability of Monte Carlo–based calculations are closely related to the adopted computational model, which reflects the physical characteristics and anatomical features of the human body. In this regard, the accurate delineation of the anatomical structures of the human body is required to estimate organ doses and radiation risks [31]. This task is referred to as “image segmentation” and is commonly part of the process of constructing computational phantoms. However, the manual delineation of contours and internal organs is usually time-consuming and suffers from intra- and inter-observer variability.
Deep learning algorithms have recently been applied in the field of image analysis and radiation therapy [32]. These techniques proved superior compared with previous state-of-the-art segmentation methods. Instead of describing the tissue patterns manually, deep learning methods discover multiple levels of representation and abstraction and use hierarchical layers of learned abstraction to understand image characteristics. Different categories of deep learning algorithms were used, like recurrent neural networks [33], gated recurrent units [34], deep belief networks [35], Hopfield neural networks [36], generative adversarial networks [37], Boltzmann machines [38], or convolutional neural networks (CNNs) [39]. The latter have many interesting features, such as simple structure, less training parameters, and adaptability. Their network structure resembles biological neural networks, which rendered them suitable for pattern recognition and medical image analysis. Zhou et al [40] trained a CNN for mapping each voxel automatically on 3D CT images to an anatomical label whereas Liu et al [41] adopted super-pixel and CNNs for automatic segmentation of the liver and lung from CT images. Likewise, Weston et al [42] performed segmentation of abdominal CT images for body composition analysis using CNNs based on the U-Net architecture.
Image segmentation of organ/tissues from medical images is the first step for the construction of computational models, commonly manual and time-consuming. In this work, we report on an automated methodology for the construction of computational models using CNN-based image segmentation for estimating the conceptus doses of pregnant patients from CT examinations.
Materials and methods
Data acquisition
This study included thirty-two pregnant patients referred to the emergency unit of Geneva University Hospital (HUG) for abdominal and pelvic CT scans. The institutional ethics committee approved this retrospective study and written informed consent was waived. The patients were scanned using a volumetric CT data acquisition protocol on the Discovery CT 750 HD CT scanner (GE Healthcare) (120-kVp tube voltage, 22.9 effective mAs tube current without current modulation, a pitch of 1.375, slice thickness from 0.625 to 3 mm, and table speed of 55 mm/rotation). CT images were reconstructed using a matrix size of 512 × 512. All pregnant patients receiving low-dose CT scans between March 2011 and September 2018 were included. The 32 pregnant patients (age range 19–45 years) had gestational ages ranging from 8 to 35 weeks. The body contour, skeleton, liver, kidney, lung, and uterus were manually contoured by an experienced medical physicist under the supervision of qualified radiologists. Thirty-two voxelized computational models resulting from the manual segmentation of pregnant patients’ CT images were created to derive reference dosimetric estimates. The right and left kidneys and lungs were both segmented together as one organ. The maternal perimeter determined by the maximum outer perimeter of the patient on the image containing the uterus was measured automatically from the manually constructed model of each patient. Likewise, the average distance from the skin to the closest surface of the uterus was used as an indicator of the conceptus depth. These two measurements were used to estimate the size of the maternal body and the location of the uterus within the mother’s abdomen, respectively.
The segmented volumetric images and original CT data of 32 patients were resampled to a unified matrix size of 128 × 128 × 64 with a slice thickness of 2 mm. Data augmentation was performed to increase the size of samples to 128 datasets by randomly cropping the edges of the original volumetric CT images. The original CT and corresponding segmented images were similarly cropped for consistency. Eighty percent (102) of the 128 datasets were randomly selected for training whereas the remaining (26) datasets were used for testing. The training dataset was used to adjust the parameters of the proposed CNNs, whereas the test set was used to assess their performance. The manual segmentation served as reference for evaluation.
Convolutional neural network–based segmentation
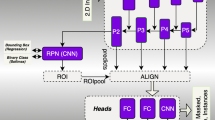
CNNs have become the most popular deep learning algorithms for medical image analysis. The U-Net architecture of CNNs integrates spatial and contextual information in a network architecture comprising an analysis path and a synthesis path for pixel-wise prediction of the label probability of classified tissues and organs. In this work, we adopted a modified 3D U-Net [43, 44] model to segment the internal organs from CT images for Monte Carlo–based radiation dose calculations. Figure 1 shows the geometry of the proposed CNNs, which consists of an encoder module and a decoder module, each with three resolution steps of 32, 64, and 128 feature maps. In the encoder module, each layer contains two 3D convolutions followed by a rectified linear unit (ReLu), and then a 3D max pooling with stride size of 2 × 2 × 2. In the decoder module, each layer consists of 3D upsampling with 2 × 2 × 2 strides followed by two 3D convolutions and a ReLu. For each convolutional layer, the size of all convolutional kernels is 3 × 3 × 3.
Shortcut connections between layers of equal resolution transfer the high-resolution features from the analysis path to the synthesis path. The numbers of channels were doubled before max pooling (sample-based discretization process) to avoid bottlenecks. Batch normalization was introduced before each ReLu to normalize the batches with mean and standard deviation, thus updating the global statistics during training. The input to the network is a 128 × 128 × 64 voxel matrix of the image with 7 channels. Our output in the final layer is 120 × 120 × 128 voxels in x, y, and z directions, respectively. The architecture has 3,192,871 parameters in total and comprises a dilated convolutional layer, 16 convolutional layers, and a series of pooling options.
The input images and their corresponding segmentation maps were used to train the network with the adaptive moment estimation (Adam) implementation of Keras [45], which computes adaptive learning rates for each parameter and keeps an exponentially decaying average of past gradients:
where mt and vt are estimates of the mean and the uncentered variance of the gradients, respectively. gt is the gradient at subsequent time step t and a1 and a2 are exponential decay rates with a1, a2∈ [0, 1).
The categorical cross-entropy loss function was used in the network training to evaluate the price paid for inaccuracy of predictions in classification. It is defined as:
where LCE refers to the cross-entropy loss evaluation, f(p)i is the softmax function, pi are the scores inferred by the net for each class in C, ti indicates the ground truth label for voxel i.
The CNN model was implemented using the open-source Keras package [45] and ran on NVIDIA Quadro K5000 GPU with 32 GB of memory and ubuntu operating system. To minimize the overhead and make maximum use of the GPU memory, the network was trained with a mini-batch size of 2 and for 100 epochs. The learning rate was initialized as 10−3, and the network weight was initialized as normal distribution with zero mean and a standard deviation of 0.01.
Computational phantoms
Overall, the manual segmentation of the original CT images for 32 patients took approximately 1 month while the automated segmentation of one CT dataset, following training that took 22 days, takes 20 s. A total of 6 organs/tissues were automatically or manually segmented from 3200 transaxial slices followed by assignment of a label to each one. Organ masses were calculated by multiplying the number of segmented voxels by the voxel size and the corresponding tissue density reported in the ICRP publication 89 [46]. Figure 2 shows the representative transverse, sagittal, and coronal slices of the original CT images and those generated by manual and automated segmentation techniques. Figure 3 shows the 3D coronal and sagittal views of the computational models generated using manual and automated segmentation techniques where the body is made transparent for enhanced viewing of internal organs and the skeleton. A selected set of metrics were used to measure the agreement between manual and automated segmentations, including the Jaccard similarity coefficient, Dice similarity coefficient (DSC), sensitivity, positive predictive value (PPV), volume difference, and Hausdorff distance (HD). The Jaccard coefficient measures the similarity between two images and is defined as the volume of the intersection divided by the volume of the union of manual and automatic segmentations. DSC is similar to Jaccard and is calculated as DSC = 2 × Jaccard/(1 + Jaccard). The sensitivity describes the ability of the CNN to correctly classify an individual voxel as belonging to the right target organ and equals the true positive divided by the sum of true positive and false negative rates. The PPV is the percentage of the correct identification in the automated segmentation results and can be defined as follows: PPV = true positive/(true positive + false positive). The measurement of HD reflects the translations and shape discrepancies between the computational phantoms produced using manual and automated segmentation techniques. The voxel-to-voxel distance can be calculated as:
where ||ai − bj|| is the Euclidean distance of point a in the manually segmented phantom to point b in the automatically segmented phantom.
Monte Carlo simulations and radiation dose calculations
Monte Carlo modeling was performed using a previously validated GE 750HD CT source where the CT gantry geometry model includes a Performix Pro VCT 100 x-ray tube with 7° target angle and 56° fan-beam angle, allowing for a beam collimation of 40 mm [47]. At 120 kVp, the half-value layer was measured as 7.8 mm Al, whereas the quality equivalent filtration of the x-ray tube is 4.3 mm of Al. The source-to-isocenter and the source-to-detector distances for this CT scanner are 54 cm and 95 cm, respectively. The computational phantoms and CT source and gantry models were integrated in the MCNPX Monte Carlo code [48] for simulation of low-dose CT examinations with a helical source path and total collimation width of 64 × 0.625 mm. The energies deposited in the computational phantoms produced through manual and automated segmentation techniques were recorded and used to calculate organ absorbed doses. The estimated organ-level absorbed doses (in mGy) were used to compare the dosimetric characteristics of the computational models generated using manual and automated segmentation techniques.
Results
The results of automated segmentation are compared with those of manual segmentation of all patients with respect to the total body, skeleton, liver, lung, kidney, and uterus using different metrics (Table 1). The Jaccard similarity coefficient for the different segmented organs varies between 0.85 and 0.96 with an average of 0.90 ± 0.04; while the DSC varies between 0.92 and 0.98 with an average of 0.94 ± 0.02. Conversely, the sensitivity varies between 0.9 and 0.97 with an average of 0.95 ± 0.03, while the PPV varies between 0.92 and 0.98 with an average of 0.94 ± 0.02. The size and location accuracy of the segmented organs between the phantoms constructed by the two methods were evaluated by calculating the volume difference and HD. The former ranges from − 4.9 to 2.96% with an average of 0.26% ± 2.67%, while the latter varies between 10.71 and 50.07 mm with an average of 23.62 mm ± 12.86 mm among organs. It should be emphasized that the uterus dose served as surrogate for the conceptus dose in this work. For segmented uterus among patients, the Jaccard similarity coefficient, DSC, sensitivity, PPV, volume differences, and HD are 0.88 ± 0.06, 0.94 ± 0.04, 0.94 ± 0.05, 0.93 ± 0.03, 1.49% ± 4.19%, and 16.1 mm ± 8.16 mm, respectively.
Table 2 summarizes the calculated conceptus dose of pregnant patients according to gestation age, which varies between 8 and 35 weeks. The conceptus doses for the thirty-two pregnant patients estimated using computational models developed based on manual segmentation and automated segmentation are also given. The measured maternal perimeter varies from 65.2 to 127.7 cm with a mean value of 93.5 cm. The mean conceptus dose is 2.91 ± 0.7 mGy. Figure 4 compares the absorbed doses to the kidney, liver, lung, uterus, skeleton, and total body between the manual and automated segmentation models. Figure 5 shows the relative differences of absorbed doses in target organs between the two segmentation methods. The relative absorbed dose differences for the total body among the thirty-two patients range from − 0.21 to 1.48% with an average of 0.28% ± 0.39% whereas the dose differences for the skeleton vary between − 2.26 and 1.33% with an average of − 0.48% ± 0.91%. The dose difference for the uterus ranges from − 5.98 to 6.31% with an average of − 0.12% ± 2.62%. The absorbed dose in target regions with low density and small volume is more easily affected by organ’s shape and position. Therefore, the dose difference between manual and automated segmentations observed for the lungs is higher than that for the liver and kidneys while the skeleton and total body have the least dose differences. Figure 6 compares the relative differences of organ and conceptus absorbed doses between manual and automated segmentation techniques at different gestational periods, respectively. The mean conceptus dose differences between the two methods are 0.93% ± 2.5%, − 1.19% ± 2.97%, 0.053% ± 2.51%, and − 0.38% ± 1.28% at early pregnancy, the first trimester, the second trimester, and the third trimester, respectively. The error bars in Figs. 5 and 6 represent the minimum and maximum dose differences between manual and automatic segmentations among patients. The sources of uncertainty reflect the anatomic divergences of the segmented target regions. The dependence of the conceptus dose upon the maternal perimeter estimated using the technique described by Angel et al [13] is also compared in Table 2 and Fig. 6b.
Comparison between absorbed doses from abdominal CT scans to the kidney, liver, lung, uterus, skeleton, and total body estimated using the pregnant computational models produced by manual vs. automated segmentation. The error bars represent ± STD (standard deviations) of organ doses among the patients
Relative differences between absorbed doses (in %) from abdominal CT scans estimated in target organs identified in Fig. 4 when using computational models produced by manual vs. automated segmentation
a Relative dose differences to the uterus (in %) from abdominal CT scans when using computational models produced using manual segmentation and automated segmentation at different gestational periods. b The same as a between the computational models produced by manual segmentation and dose estimates using the model by Angel et al [13]
Discussion
A deep learning–based automated algorithm was optimized for successful automated segmentation of CT images of pregnant patients who presented with a range of anatomical variability. Produced patient-specific computational models were suitable for incorporation in dedicated Monte Carlo codes for radiation dose calculations. The evaluation of the proposed CNN algorithms using thirty-two clinical studies demonstrated a good agreement between automated and manual segmentation results (mean values of DSC, PPV, volume difference, and HD are equal to 0.95, 0.95, 0.26%, and 23.62 mm, respectively).
Accurate delineation of organs at risk for radiation dose estimation in radiation protection or treatment planning is critical, yet efforts to automate the process of constructing patient-specific computational models remain elusive. Our CNN-based approach provides a fully automated solution for radiation dosimetry in CT imaging. The approach does not require user interaction and enables efficient and automated segmentations for fast computational model construction and accurate radiation dose calculation, thus facilitating eventual use in the clinic. The goal of our framework is to perform automated delineation of the contours of internal organs at risk from abdominal CT scans with the aim to build patient-specific computational models for radiation dosimetry. Taking advantage of convolutional neural network architecture, automated feature extraction is conducted on patient CT images, demonstrating capability in identifying target organs.
The absorbed dose for the uterus is commonly used as a surrogate for the absorbed dose to the embryo/fetus in medical radiation dosimetry [5] while different methods consider the larger uterine volume compared with the non-pregnant female [5, 49]. The dose calculation process reported in this work has the advantage of taking the individual characteristics of the patient’s anatomy into the dose calculation algorithm. The radiation dose in diagnostic imaging procedures is normally lower than the dose from radiation therapy but still carries a risk that cannot be eliminated entirely.
The conceptus dose for pregnant patients from CT examinations varies within the range 1.8–4.7 mGy with a mean value of 2.9 mGy. The differences of mean conceptus absorbed doses between the two segmentation techniques are 1.66%, 2.55%, 1.84%, and 1.18% at early pregnancy, first trimester, second trimester, and third trimester, respectively. It is not clear why the highest difference was observed at the first trimester. The differences of mean conceptus absorbed doses between manual segmentation and the technique proposed by Angel et al [13] are 19.33%, 19.39%, 10.97%, and 16.37% at early pregnancy, the first trimester, the second trimester, and the third trimester, respectively. Overall, the automated segmentation algorithm generated computational phantoms that resulted in almost similar estimates of individual conceptus doses as manual segmentation.
This study demonstrates the feasibility of an automated construction of patient-specific computational models for organ-level dose estimation, using deep learning approaches. The quality of typical low-dose CT images made it difficult to segment all organs, and as such, we had to limit the number of segmented organs. Another limitation of this study is that the uterus dose is used as a surrogate for radiation dose estimation to the fetus in the computational model while the fetal position may vary among patients and at different gestation periods. This assumption adds uncertainties to the dose estimation process. Another limitation is that only a limited number of organs have been segmented from CT images, which prevented the estimation of all organ doses for pregnant patients. Overall, the computational time required for construction of patient-specific models using automatic segmentation for one dataset is approximately 20 s. With advances in computer technology, particularly grid and cloud computing, and advanced variance reduction techniques, the proposed approach could be clinically implemented for real-time calculation of individual radiation doses to patients.
Conclusion
Since the individual anatomic characteristics have a noticeable impact on conceptus dose estimates, this study shows that patient-specific computational models can be created using an automated deep learning–based segmentation algorithm. Therefore, radiologists could perform accurate patient-specific dose estimation for a variety of radiation exposure situations and specifically for the evaluation of conceptus dose. In most situations encountered in emergency units, the benefit of performing CT outweighs the radiation risk. However, the proposed approach for automated computational modeling and dose calculation can be useful for retrospective evaluation of radiation dose, for instance if the pregnancy was unknown when emergency CT was performed, for the decision-making process for high-dose procedures in clinical setting and also in research studies involving retrospective data analysis.
Abbreviations
- Adam:
-
Adaptive moment estimation
- CNNs:
-
Convolutional neural networks
- CT:
-
Computed tomography
- DSC:
-
Dice similarity coefficient
- HD:
-
Hausdorff distance
- HUG:
-
Geneva University Hospital
- IQ:
-
Intelligence quotient
- PET:
-
Positron emission tomography
- PET/CT:
-
Positron emission tomography/computed tomography
- PPV:
-
Positive predictive value
- ReLu:
-
Rectified linear unit
References
Lazarus E, Debenedectis C, North D, Spencer PK, Mayo-Smith WW (2009) Utilization of imaging in pregnant patients: 10-year review of 5270 examinations in 3285 patients—1997–2006 Radiology 251:517–524
Xie T, Zaidi H (2014) Fetal and maternal absorbed dose estimates for positron-emitting molecular imaging probes. J Nucl Med 55:1459–1166
Zanotti-Fregonara P, Laforest R, Wallis JW (2015) Fetal radiation dose from [18F]-FDG in pregnant patients imaged with PET, PET/CT and PET/MRI. J Nucl Med 56:1218–1222
Poletti PA, Botsikas D, Becker M et al (2019) Suspicion of appendicitis in pregnant women: emergency evaluation by sonography and low-dose CT with oral contrast. Eur Radiol 29:345–352
American College of Radiology (2013) ACR-SPR practice parameter for imaging pregnant or potentially pregnant adolescents and women with ionizing radiation. Revised 2013 (Resolution 48)
ICRP (2000) ICRP publication 84: pregnancy and medical radiation. Ann ICRP 30:iii–viii, 1-43
ICRP (2003) ICRP publication 90: biological effects after prenatal irradiation (embryo and fetus). Ann ICRP 33:5–206
Altman KI, Lett JT (2013) Relative radiation sensitivities of human organ systems. Elsevier
McCollough CH, Schueler BA, Atwell TD et al (2007) Radiation exposure and pregnancy: when should we be concerned? Radiographics 27:909–917 discussion 917-908
Pearce MS, Salotti JA, Little MP et al (2012) Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet 380:499–505
(2007) ICRP publication 105. Radiological protection in medicine. Ann ICRP 37:1–63
Vañó E, Miller DL, Martin CJ et al(2017) ICRP publication 135: diagnostic reference levels in medical imaging. Ann ICRP 46:1–144
Angel E, Wellnitz CV, Goodsitt MM et al (2008) Radiation dose to the fetus for pregnant patients undergoing multidetector CT imaging: Monte Carlo simulations estimating fetal dose for a range of gestational age and patient size. Radiology 249:220–227
Damilakis J, Perisinakis K, Tzedakis A, Papadakis AE, Karantanas A (2010) Radiation dose to the conceptus from multidetector CT during early gestation: a method that allows for variations in maternal body size and conceptus position. Radiology 257:483–489
Damilakis J, Tzedakis A, Perisinakis K, Papadakis AE (2010) A method of estimating conceptus doses resulting from multidetector CT examinations during all stages of gestation. Med Phys 37:6411–6420
Helmrot E, Pettersson H, Sandborg M, Altén JN (2007) Estimation of dose to the unborn child at diagnostic X-ray examinations based on data registered in RIS/PACS. Eur Radiol 17:205–209
Gu J, Xu XG, Caracappa PF, Liu B (2013) Fetal doses to pregnant patients from CT with tube current modulation calculated using Monte Carlo simulations and realistic phantoms. Radiat Prot Dosimetry 155:64–72
Winer-Muram HT, Boone JM, Brown HL, Jennings SG, Mabie WC, Lombardo GT (2002) Pulmonary embolism in pregnant patients: fetal radiation dose with helical CT. Radiology 224:487–492
Hoseinian-Azghadi E, Rafat-Motavalli L, Miri-Hakimabad H (2014) Development of a 9-months pregnant hybrid phantom and its internal dosimetry for thyroid agents. J Radiat Res 55:730–747
Lopez-Rendon X, Walgraeve MS, Woussen S et al (2017) Comparing different methods for estimating radiation dose to the conceptus. Eur Radiol 27:851–858
Maynard MR, Long NS, Moawad NS et al (2014) The UF family of hybrid phantoms of the pregnant female for computational radiation dosimetry. Phys Med Biol 59:4325–4343
Xie T, Zaidi H (2016) Development of computational pregnant female and fetus models and assessment of radiation dose from positron-emitting tracers. Eur J Nucl Med Mol Imaging 43:2290–2300
Xie T, Zanotti-Fregonara P, Edet-Sanson A, Zaidi H (2018) Patient-specific computational model and dosimetry calculations for a patient pregnant with twins undergoing a PET/CT examination. J Nucl Med 59:1451–1458
Gilet AG, Dunkin JM, Fernandez TJ, Button TM, Budorick NE (2011) Fetal radiation dose during gestation estimated on an anthropomorphic phantom for three generations of CT scanners. AJR Am J Roentgenol 196:1133–1137
Chatterson LC, Leswick DA, Fladeland DA, Hunt MM, Webster S, Lim H (2014) Fetal shielding combined with state of the art CT dose reduction strategies during maternal chest CT. Eur J Radiol 83:1199–1204
Jaffe TA, Neville AM, Anderson-Evans C et al (2009) Early first trimester fetal dose estimation method in a multivendor study of 16- and 64-MDCT scanners and low-dose imaging protocols. AJR Am J Roentgenol 193:1019–1024
Solomou G, Papadakis AE, Damilakis J (2015) Abdominal CT during pregnancy: a phantom study on the effect of patient centering on conceptus radiation dose and image quality. Eur Radiol 25:911–921
Kelaranta A, Kaasalainen T, Seuri R, Toroi P, Kortesniemi M (2015) Fetal radiation dose in computed tomography. Radiat Prot Dosimetry 165:226–230
Zaidi H, Xu XG (2007) Computational anthropomorphic models of the human anatomy: the path to realistic Monte Carlo modeling in medical imaging. Annu Rev Biomed Eng 9:471–500
Zaidi H, Ay MR (2007) Current status and new horizons in Monte Carlo simulation of X-ray CT scanners. Med Biol Eng Comput 45:809–817
Xie T, Poletti PA, Platon A, Becker CD, Zaidi H (2018) Assessment of CT dose to the fetus and pregnant female patient using patient-specific computational models. Eur Radiol 28:1054–1065
Litjens G, Kooi T, Bejnordi BE et al (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Mikolov T, Karafiát M, Burget L, Černocký J, Khudanpur S (2010) Recurrent neural network based language model. 11th annual conference of the International Speech Communication Association (INTERSPEECH-2010), Makuhari, Chiba, Japan, September 26–30. 2010, pp 1045–1048
Chung J, Gulcehre C, Cho K, Bengio Y (2015) Gated feedback recurrent neural networks. Proceedings of the 32nd International Conference on International Conference on Machine Learning. J PMLR 37:2067–2075
Mohamed A, Dahl GE, Hinton G (2012) Acoustic modeling using deep belief networks. IEEE Trans Audio Speech Lang Process 20:14–22
Nasrabadi NM, Choo CY (1992) Hopfield network for stereo vision correspondence. IEEE Trans Neural Netw 3:5–13
Emami H, Dong M, Nejad-Davarani SP, Glide-Hurst CK (2018) Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med Phys 45:3627–3636
Sum J, Leung CS (2019) Learning algorithm for Boltzmann machines with additive weight and bias noise. IEEE Trans Neural Netw Learn Syst. https://doi.org/10.1109/TNNLS.2018.2889072
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60:84–90
Zhou X, Takayama R, Wang S, Zhou X, Hara T, Fujita H (2017) Automated segmentation of 3D anatomical structures on CT images by using a deep convolutional network based on end-to-end learning approach. Medical imaging: image processing
Liu X, Guo S, Yang B et al (2018) Automatic organ segmentation for CT scans based on super-pixel and convolutional neural networks. J Digit Imaging 31:748–760
Weston AD, Korfiatis P, Kline TL et al (2018) Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology 290:669–679
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham Springer, pp 234–241
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science, vol 9901. Springer, Cham, pp 424–432
Chollet F (2018) Keras: the Python deep learning library. Astrophysics Source Code Library. Available at https://keras.io/
(2002) Basic anatomical and physiological data for use in radiological protection: reference values. A report of age- and gender-related differences in the anatomical and physiological characteristics of reference individuals. ICRP Publication 89. Ann ICRP 32:5–265
Akbarzadeh A, Ay MR, Ghadiri H, Sarkar S, Zaidi H (2010) Measurement of scattered radiation in a volumetric 64-slice CT scanner using three experimental techniques. Phys Med Biol 55:2269–2280
Pelowitz DB (2005) MCNPX user’s manual Version 2.5.0. Los Alamos National Laboratory, Los Alamos
Stovall M, Blackwell CR, Cundiff J et al (1995) Fetal dose from radiotherapy with photon beams: report of AAPM Radiation Therapy Committee Task Group No. 36. Med Phys 22:63–82
Acknowledgements
This work was supported by the Swiss National Science Foundation under grant SNSF 320030_176052 and Qatar National Research Fund under grant NPRP10-0126-170263. No other potential conflicts of interest relevant to this article exist.
Funding
This study has received funding by the Swiss National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Habib Zaidi.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived in this study.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Prospective
• Experimental
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xie, T., Zaidi, H. Estimation of the radiation dose in pregnancy: an automated patient-specific model using convolutional neural networks. Eur Radiol 29, 6805–6815 (2019). https://doi.org/10.1007/s00330-019-06296-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06296-4