Abstract
Objectives
To investigate whether liver fibrosis can be staged by deep learning techniques based on CT images.
Methods
This clinical retrospective study, approved by our institutional review board, included 496 CT examinations of 286 patients who underwent dynamic contrast-enhanced CT for evaluations of the liver and for whom histopathological information regarding liver fibrosis stage was available. The 396 portal phase images with age and sex data of patients (F0/F1/F2/F3/F4 = 113/36/56/66/125) were used for training a deep convolutional neural network (DCNN); the data for the other 100 (F0/F1/F2/F3/F4 = 29/9/14/16/32) were utilised for testing the trained network, with the histopathological fibrosis stage used as reference. To improve robustness, additional images for training data were generated by rotating or parallel shifting the images, or adding Gaussian noise. Supervised training was used to minimise the difference between the liver fibrosis stage and the fibrosis score obtained from deep learning based on CT images (FDLCT score) output by the model. Testing data were input into the trained DCNNs to evaluate their performance.
Results
The FDLCT scores showed a significant correlation with liver fibrosis stage (Spearman's correlation coefficient = 0.48, p < 0.001). The areas under the receiver operating characteristic curves (with 95% confidence intervals) for diagnosing significant fibrosis (≥ F2), advanced fibrosis (≥ F3) and cirrhosis (F4) by using FDLCT scores were 0.74 (0.64–0.85), 0.76 (0.66–0.85) and 0.73 (0.62–0.84), respectively.
Conclusions
Liver fibrosis can be staged by using a deep learning model based on CT images, with moderate performance.
Key Points
• Liver fibrosis can be staged by a deep learning model based on magnified CT images including the liver surface, with moderate performance.
• Scores from a trained deep learning model showed moderate correlation with histopathological liver fibrosis staging.
• Further improvement are necessary before utilisation in clinical settings.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Liver cirrhosis is an end stage of liver fibrosis, and is associated with hepatocellular carcinomas, oesophageal and/or gastric varices and hepatic failure [1, 2]. Diagnosis of liver fibrosis and liver cirrhosis is therefore clinically important. Liver fibrosis is staged from biopsied specimens by using METAVIR [3] or the new Inuyama classification system [4]. However, liver biopsy is invasive and associated with complications [5], and less invasive methods for staging liver fibrosis would be beneficial to patients.
Some non-invasive or minimally invasive modalities for staging liver fibrosis are available. Transient elastography (TE) is the most validated ultrasound elastography method for staging liver fibrosis [6, 7]; this is easy to perform, but its failure rate has been reported as 6–23% [6]. Magnetic resonance elastography (MRE) also allows the evaluation of liver elasticity [8], but it requires dedicated equipment that is not readily available.
Recently, deep learning has been gaining attention as a strategy for realising artificial intelligence [9]. Several types of deeply stacked artificial neural network have been used for deep learning, with deep convolutional neural networks (DCNNs) recognised as demonstrating high performance for image recognition tasks [10,11,12,13]. There has also been some initial success in applying deep learning to the assessment of radiological images [14,15,16,17,18,19,20,21], including liver imaging [22,23,24], suggesting that this approach may have the potential to stage liver fibrosis on the basis of radiological images. Given that CT is more readily available than magnetic resonance imaging, a deep learning model that enables the staging of liver fibrosis based on CT would benefit a large number of patients.
The aim of this study was to investigate whether liver fibrosis could be staged from dynamic contrast-enhanced CT images by using deep learning techniques.
Materials and methods
This clinical retrospective study was approved by our institutional review board, which waived the requirement for obtaining written informed consent from the patients.
Patients
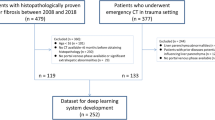
CT examinations of patients who underwent dynamic contrast-enhanced CT for evaluations of the liver at our institution between January 2014 and December 2015 were initially included in this study (4671 examinations). The following exclusion criteria were applied: lack of liver fibrosis staging evaluated histopathologically within 250 days from the CT (4144 examinations); surgery or transarterial chemoembolisation of the left lobe of the liver (29 examinations); massive tumours in the left lobe of the liver (1 examination) and an image that included a substantial metal artefact (1 examination).
Because of the inevitable overfitting problem with deep learning techniques, we divided the patients into a training group and a testing group, ensuring that images of a patient obtained at different examinations were not included in both the training group and the testing group. The number of CT examinations for the testing group was chosen to be about 20% of the total number of CT examinations. The testing group patients were chosen randomly from those at each fibrosis stage among the patients who underwent a single CT examination, ensuring that the training and testing groups contained a similar proportion of patients at each fibrosis stage. The remaining patients were included in the training group.
Reference standard: histopathological liver fibrosis staging
A single radiologist (K.Y.) reviewed the patients’ clinical histopathological reports. Liver fibrosis stages were based on the new Inuyama classification (F0 = no fibrosis, F1 = fibrous portal expansion, F2 = bridging fibrosis, F3 = bridging fibrosis with architectural distortion and F4 = cirrhosis) [4]. When several stages were noted, the most advanced stage was recorded for this study; for example, if there were mixed findings of F3 and F2, the stage was recorded as F3.
For the training group, the median (interquartile range) time intervals between biopsy and CT examination and between surgery and CT examination were 101 days (31–154 days) and 58 days (21–144 days), respectively. For the testing group, these intervals were 132 days (46–175 days) and 60 days (23–153 days).
CT scanning
CT scanners from two vendors were used to obtain the images used in this study: from Canon Medical Systems (Aquilion One and Aquilion Prime) and from GE Healthcare (Discovery CT 750HD). The following scanning and image reconstruction parameters were used: tube potential, 120 kVp for both Canon and GE; tube current, SD of 13.0 for Canon and noise index of 11.36 for GE, with automatic tube current modulation used for both; helical pitch, 0.8125:1 for Canon and 0.984:1 for GE; gantry rotation speed, 0.5 s for both Canon and GE; detector configuration, 0.5 mm × 80 for Canon and 0.625 mm × 64 for GE; reconstruction algorithm, filtered back projection for both Canon and GE; kernel, FC03 for Canon and standard for GE; and slice thickness/interval, 0.3/0.3 mm for Canon and 0.25/0.25 mm for GE. The concentration of iodine contrast enhancement material was determined on the basis on the patient’s body weight: 350 mgI/ml for patients weighing less than 60 kg and 370 mgI/ml for patients weighing more than 60 kg. The volume was determined by multiplying the body weight (in kilograms) by 2, with an upper limit of 100 ml. The iodine contrast enhancement materials were injected within 30 s. A bolus-tracking technique was used to determine the timing of scan. A region of interest (ROI) was placed at the descending aorta at the level of the diaphragm, and portal phase images were scanned 55 s after the CT attenuation within the ROI reached 200 Hounsfield units.
Formatting the input images
A single radiologist (K.Y.) performed the input image formatting. Axial portal phase CT images in the Digital Imaging and Communications in Medicine (DICOM) format, acquired with the table position including the umbilical portion of the portal vein, were displayed with a commercial viewer (Centricity RA 1000, GE Healthcare). The images were magnified, referencing the scale bar displayed at the bottom of the window, so that the width of the image became about 10 cm. They were then shifted so that the ventral aspect of the liver was displayed horizontally across the centre of the window (Fig. 1). Then, using the viewer’s capture function, the image was captured in 8-bit Joint Photographic Experts Group (JPEG) format with 594 × 644 pixels. The JPEG images (the ‘original images’) were further processed with the Python 3.5 programming language (https://www.python.org) and the Python imaging library of Pillow 3.3.1 (https://pypi.python.org/pypi/Pillow/3.3.1). Regions with 350 × 350 pixels were cropped from the original images with the crop function and resized to 96 × 96 pixels with the resize function. These resized images were used as the input images for the DCNNs.
Adjustment of the input images for training
For the training data, the radiologist (K.Y.) adjusted the images in various ways so that the model would robustly handle differences in the location of the liver within an image, the angle of rotation of the liver (especially for small angles) and the amount of image noise. Axial images were captured in three slightly different table positions (all including the umbilical portion of the portal vein) for the training group. These original images were processed with Python 3.5 and Pillow 3.3.1. Fifteen cropped images (with regions of 350 × 350 pixels) were generated from each original image, changing the location of the cropping region. Further new images were created by rotating these 15 new images through 5, 90, 180, 270 and 355°, and by adding Gaussian noise (with mean of 0 and sigma of 15) to the image. Thus, 105 (= 15 × (1 + 5 + 1)) images were generated from each original image.
Implementation of the deep convolutional neural network
Deep learning was implemented on a computer with 64 GB of random access memory, a Core i7-6700K 4.00-GHz central processing unit (Intel) and a GeForce GTX 1080 graphics processing unit (NVIDIA), using the Python 3.5 programming language and the Chainer 1.24.0 framework for neural networks (http://chainer.org/). The structure of the DCNN is shown schematically in Fig. 2. In summary, the input images were fed into the DCNN, where they were down-sampled with the ‘max pooling’ functions, ultimately to single-pixel images, before being processed with fully connected layers [13]. The patient’s age and sex (0 = male; 1 = female) were concatenated to the data at the second fully connected layers. The DCNN returned a single value for each image as the output. The rectified linear unit (Relu) function [25] was used as an activation function for the DCNN. This function returns the input value when the value is greater than 0; otherwise, it returns 0. The DCNN used batch normalisation, which normalises the values of the input data for each minibatch; this is known to reduce the risk of the overfitting problem [26, 27].
The structure of the deep convolutional neural network (DCNN). The input images were processed with four convolutional layers with filters with 3 × 3 pixels. The data were then processed with three fully connected layers. This DCNN was structured to output a single continuous value. BN batch normalisation, C channel, Conv convolutional layer, FC fully connected layer, Relu rectified linear unit function, S stride, U unit
Training and testing the deep convolutional neural network
The DCNN was trained from scratch by feeding in the training images and data. The training was supervised to minimise the difference between the actual liver fibrosis stages and the fibrosis scores obtained from deep learning based on CT images (FDLCT scores) output by the DCNN. This was achieved by using the error function (in this study, the mean squared error function) to calculate the error between the FDLCT scores and liver fibrosis stage data; this error was then backpropagated to the DCNN and the parameters within the DCNN were updated by using the optimiser of AdaGrad [28]. Minibatch learning was performed using the data for batches of 15 patients. To obtain the DCNN model, five epochs were performed (i.e. each set of patient data was utilised five times). For each epoch, the sets of patient data were shuffled before they were assigned to the minibatches. Because there was randomness in the initial weight and patient selection for the minibatches, the training was performed 15 times (i.e. resulting in 15 DCNN models).
After the training was completed, the testing data were used as input into the trained DCNN models, and the resulting FDLCT scores were used to evaluate each model’s performance.
Statistical analysis
Statistical analyses were performed with EZR version 1.33 (http://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmedEN.html) [29], which is a graphical user interface of R version 3.3.1 (https://www.r-project.org/). All the statistical analyses were performed on the basis of the testing data.
The relationship between liver fibrosis stages and FDLCT scores was analysed with Spearman’s correlation analysis for each model. And for each model, receiver operating characteristic (ROC) analyses were used to assess the models’ efficacy in using FDLCT scores for diagnosing significant fibrosis (≥ F2), advanced fibrosis (≥ F3) and cirrhosis (F4), by calculating the areas under the ROC curve (AUCs). The results are expressed as the median values for the 15 models with ranges. For the model which showed median Spearman’s correlation coefficient among 15 models, p value for Spearman’s correlation coefficient and 95% confidence interval (CI) for AUC were calculated by using test data (100 examinations).
Results
In this study, 496 CT examinations (286 patients) were included. Of these, 396 CT examinations (186 patients) (mean age, 66.2 ± 11.6; 281 men and 115 women) and 100 CT examinations (100 patients) (mean age, 66.1 ± 11.6; 73 men and 27 women) were assigned to the training and testing groups, respectively. The distribution of fibrosis stages (F0/F1/F2/F3/F4) in the training and testing groups were 113/36/56/66/125 and 29/9/14/16/32, respectively. These were based on biopsy specimens (n = 112 and 34 for the training and testing groups, respectively) or surgical specimens (n = 284 and 66). In the training group, 261 and 135 examinations were performed with the Canon and GE scanners, respectively; in the testing group, these numbers were 67 and 33.
The median (range) Spearman's correlation coefficients between FDLCT score and liver fibrosis stage calculated in each model was 0.48 (0.41–0.53). There were significant correlations for all 15 models (all p < 0.001). The median (range) AUCs for diagnosing significant fibrosis, advanced fibrosis and cirrhosis by using FDLCT scores were 0.73 (0.69–0.76), 0.76 (0.71–0.81) and 0.74 (0.69–0.77), respectively.
Table 1 summarises the diagnostic performance of the model with the median (r = 0.48, p < 0.001) Spearman's correlation coefficient. Figure 3 shows the relationship between FDLCT scores and fibrosis stages for the model with the median correlation coefficient. For this model, the AUCs (with 95% CIs) for diagnosing significant fibrosis, advanced fibrosis and cirrhosis by using FDLCT scores calculated by using a test group data (100 examinations) were 0.74 (0.64–0.85), 0.76 (0.66–0.85) and 0.73 (0.62–0.84), respectively (Fig. 4).
Box and whisker plot for the relationship between pathological liver fibrosis stage and the model’s output fibrosis score obtained from deep learning based on CT images (FDLCT score). These data are for the model that showed the median Spearman's correlation coefficient. The thick black lines, boxes, whiskers and circles denote the median, interquartile range, 10 and 90 percentiles and outliers, respectively
Receiver operating characteristic curves (grey background colour denotes 95% confidence intervals [CI]) for predicting a significant fibrosis (≥ F2) (AUC [with 95% CI] = 0.74 [0.64–0.85]), b advanced fibrosis (≥ F3) (AUC = 0.76 [0.66–0.85]) and c cirrhosis (F4) (AUC = 0.73 [0.62–0.84]). These results were for the model that showed the median Spearman's correlation coefficient between FDLCT score and liver fibrosis stage
Discussion
In this study, we showed that liver fibrosis can be staged, with moderate performance, by using a deep learning model based on dynamic contrast-enhanced portal phase CT images.
There are several less-invasive modalities available for the diagnosis of liver fibrosis. TE and MRE are known to perform well in liver fibrosis staging, with meta-analyses reporting their performance (assessed as AUCs) in diagnosing significant fibrosis, advanced fibrosis and cirrhosis as 0.84, 0.89 and 0.94, respectively, for TE [30] and 0.88–0.95, 0.93–0.95 and 0.92–0.93 for MRE [31, 32]. Although the performance of our model was not as high as these values, our model can be applied retrospectively to CT images, providing the potential to estimate a patient’s past course of liver fibrosis. Deep learning has previously been applied to liver fibrosis staging based on gadoxetic acid-enhanced hepatobiliary phase MR images. The AUCs assessing the performance of that model in diagnosing significant fibrosis, advanced fibrosis and cirrhosis were reported to be 0.85, 0.84 and 0.84, respectively [23]. The performance of our model, based on portal phase CT images, was not as high; but, because CT is more readily available than MRI, our model would offer more opportunities for use in clinical settings. CT can be acquired for patients who are not eligible for MRI, such as those with claustrophobia or the presence of mechanical devices or metal. Our model could be used for such patients.
Several limitations of this study should be acknowledged. First, because of the limitation of computer resources, we used a single resized JPEG format image per patient. Resizing and capturing of the image (as JPEG format) require human interaction; however, we believe that they can be relatively easily performed on commercial viewers. Use of multiple non-resized images as volume data could potentially have improved the performance of the model. The overall morphological features of the liver (such as atrophic right lobe, hypertrophy of the caudate and lateral left lobes, expanded gallbladder fossa, etc.) and total splenic volume are known to be useful information in predicting the liver fibrosis [6, 33], and such information might be able to be included in the model if the volume data of the liver and/or the spleen are utilised. However, the use of such a large volume of input data would have resulted in a large DCNN. Training of a large DCNN generally requires much more input data, much greater computation and time, and a more powerful computer with a large amount of random access memory. Second, we did not include information regarding hepatitis B or C and alcohol consumption because the retrospective nature of this study meant these were not available for all the patients. Future prospective studies with large numbers of patients are expected. Third, unlike TE and MRE, our model did not use information about the elasticity of the liver for estimating the liver fibrosis stage. Instead, our model was developed to directly associate CT images with liver fibrosis stage. Fourth, the performance of the models was moderate. In the previous similar study, deep learning models based on gadoxetic acid-enhanced hepatobiliary phase MR images were developed by using 534 examinations and showed high performance for staging of liver fibrosis [23]. Considering that the current study was performed in a similar way to that study, the number of CT examinations utilised for building models would not be considerably small. The difference in imaging modality’s ability to capture the features of liver parenchyma might be a main reason for the difference in performance between this study and the previous study. Because deep learning technologies are evolving, performance improvement in models is expected by applying new technologies or by using high-performance computers (so that volume data of the liver can be utilised as input data) in the future. Finally, the histopathological liver fibrosis stages were evaluated from specimens obtained by biopsy or surgery and so they may not have reflected the degree of fibrosis across the whole liver or the ventral aspect of the liver. However, it is difficult to obtain whole liver specimens for a large number of patients.
In conclusion, liver fibrosis can be staged, with moderate performance, by using deep learning models based on dynamic contrast-enhanced portal phase CT images. This model allowed liver fibrosis staging with minimal human interaction; however, further improvement in the performance of the model would be required before being integrated into clinical strategy.
Abbreviations
- AUC:
-
Area under the receiver operating characteristic curve
- DCNN:
-
Deep convolutional neural network
- DICOM:
-
Digital Imaging and Communications in Medicine
- FDLCT :
-
Fibrosis score obtained from deep learning based on CT images
- IQR:
-
Interquartile range
- JPEG:
-
Joint Photographic Experts Group
- MRE:
-
Magnetic resonance elastography
- ROC:
-
Receiver operating characteristic
- ROI:
-
Region of interest
- TE:
-
Transient elastography
References
Forner A, Llovet JM, Bruix J (2012) Hepatocellular carcinoma. Lancet 379:1245–1255
Schuppan D, Afdhal NH (2008) Liver cirrhosis. Lancet 371:838–851
Bedossa P, Poynard T (1996) An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 24:289–293
Ichida F, Tsuji T, Omata M et al (1996) New Inuyama classification; new criteria for histological assessment of chronic hepatitis. Int Hepatol Commun 6:112–119
Rockey DC, Caldwell SH, Goodman ZD, Nelson RC, Smith AD (2009) Liver biopsy. Hepatology 49:1017–1044
Horowitz JM, Venkatesh SK, Ehman RL et al (2017) Evaluation of hepatic fibrosis: a review from the society of abdominal radiology disease focus panel. Abdom Radiol (NY) 42:2037–2053
Foucher J, Chanteloup E, Vergniol J et al (2006) Diagnosis of cirrhosis by transient elastography (FibroScan): a prospective study. Gut 55:403–408
Huwart L, Sempoux C, Salameh N et al (2007) Liver fibrosis: noninvasive assessment with MR elastography versus aspartate aminotransferase-to-platelet ratio index. Radiology 245:458–466
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Krizhevsky A, Sutskever I, Hinton G (2012) ImageNet classification with deep convolutional neural networks. Advances in Neural Information Processing System 25 (NIPS 2012). https://papersnipscc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks. Accessed 20 Jan 2018
Szegedy C, Liu W, Jia Y et al (2014) Going deeper with convolutions. Cornell University Library. https://arxivorg/abs/14094842. Accessed 20 Jan 2018
He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition. Cornell University Library. https://arxivorg/abs/151203385. Accessed 20 Jan 2018
Andrearczyk V, Whelan PF (2016) Using filter banks in convolutional neural networks for texture classification. Cornell University Library. https://arxiv.org/abs/1601.02919. Accessed 20 Jan 2018
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284:574–582
Prevedello LM, Erdal BS, Ryu JL et al (2017) Automated critical test findings identification and online notification system using artificial intelligence in imaging. Radiology 285:923–931
Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB (2018) Deep learning MR imaging-based attenuation correction for PET/MR imaging. Radiology 286:676–684
Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP (2018) Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology 287:313–322
Leynes AP, Yang J, Wiesinger F et al (2017) Direct pseudoCT generation for pelvis PET/MRI attenuation correction using deep convolutional neural networks with multi-parametric MRI: zero echo-time and Dixon deep pseudoCT (ZeDD-CT). J Nucl Med. https://doi.org/10.2967/jnumed.117.198051
Gonzalez G, Ash SY, Vegas Sanchez-Ferrero G et al (2018) Disease staging and prognosis in smokers using deep learning in chest computed tomography. Am J Respir Crit Care Med 197:193–203
Chang K, Bai HX, Zhou H et al (2018) Residual convolutional neural network for determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res 24:1073–1081
Nakao T, Hanaoka S, Nomura Y et al (2018) Deep neural network-based computer-assisted detection of cerebral aneurysms in MR angiography. J Magn Reson Imaging 47:948–953
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286:887–896
Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S (2018) Liver fibrosis: deep convolutional neural network for staging by using gadoxetic acid-enhanced hepatobiliary phase MR images. Radiology 287:146–155
Ben-Cohen A, Klang E, Diamant I et al (2017) CT image-based decision support system for categorization of liver metastases into primary cancer sites: initial results. Acad Radiol 24:1501–1509
Nair V, Hinton G (2010) Rectified linear units improve restricted Boltzmann machines. International conference on machine learning. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.165.6419&rank=1. Accessed 20 Jan 2018
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. Cornell University Library. http://arxiv.org/abs/1502.03167. Accessed 20 Jan 2018
Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O (2018) Deep learning with convolutional neural network in radiology. Jpn J Radiol. https://doi.org/10.1007/s11604-018-0726-3
Duchi J, Hazan E, Singer Y (2011) Adaptive subgradient methods for online learning and stochastic optimization. J Mach Learn Res 12:2121–2159
Kanda Y (2013) Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant 48:452–458
Friedrich-Rust M, Ong MF, Martens S et al (2008) Performance of transient elastography for the staging of liver fibrosis: a meta-analysis. Gastroenterology 134:960–974
Singh S, Venkatesh SK, Wang Z et al (2015) Diagnostic performance of magnetic resonance elastography in staging liver fibrosis: a systematic review and meta-analysis of individual participant data. Clin Gastroenterol Hepatol 13(440-451):e446
Kim YS, Jang YN, Song JS (2018) Comparison of gradient-recalled echo and spin-echo echo-planar imaging MR elastography in staging liver fibrosis: a meta-analysis. Eur Radiol 28:1709–1718
Pickhardt PJ, Malecki K, Hunt OF et al (2017) Hepatosplenic volumetric assessment at MDCT for staging liver fibrosis. Eur Radiol 27:3060–3068
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Koichiro Yasaka.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the institutional review board.
Ethical approval
Institutional review board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Rights and permissions
About this article
Cite this article
Yasaka, K., Akai, H., Kunimatsu, A. et al. Deep learning for staging liver fibrosis on CT: a pilot study. Eur Radiol 28, 4578–4585 (2018). https://doi.org/10.1007/s00330-018-5499-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-018-5499-7