Abstract
Objectives
To build a reliable radiomics model from multiregional and multiparametric magnetic resonance imaging (MRI) for pretreatment prediction of O6-methylguanine-DNA methyltransferase (MGMT) promotor methylation status in glioblastoma multiforme (GBM).
Methods
In this retrospective multicentre study, 1,705 multiregional radiomics features were automatically extracted from multiparametric MRI. A radiomics model with a minimal set of all-relevant features and a radiomics model with univariately-predictive and non-redundant features were built for MGMT methylation prediction from a primary cohort (133 patients) and tested on an independent validation cohort (60 patients). Predictive models combing clinical factors were built and evaluated. Both radiomics models were assessed on subgroups stratified by clinical factors.
Results
The radiomics model with six all-relevant features allowed pretreatment prediction of MGMT methylation (AUC=0.88, accuracy=80 %), which significantly outperformed the model with eight univariately-predictive and non-redundant features (AUC=0.76, accuracy=70 %). Combing clinical factors with radiomics features did not benefit the prediction performance. The all-relevant model achieved significantly better performance in stratified analysis.
Conclusions
Radiomics model built from multiregional and multiparameter MRI may serve as a potential imaging biomarker for pretreatment prediction of MGMT methylation in GBM. The all-relevant features have the potential of offering better predictive power than the univariately-predictive and non-redundant features.
Key Points
• Multiregional and multiparametric MRI features reliably predicted MGMT methylation in multicentre cohorts.
• All-relevant imaging features predicted MGMT methylation better than univariately-predictive and non-redundant features.
• Combing clinical factors with radiomics features did not benefit the prediction performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Glioblastoma multiforme (GBM) is the most common malignant brain tumour [1]. The poor prognosis (median survival less than 15 months) is mainly due to its intratumour genetic heterogeneity [2]. Among all identified genetic alterations, MGMT promotor methylation has been used as an important predictive molecular marker in clinical settings [3]. This alteration is common in GBM, and less common in lower grade glioma [1]. GBM patients with methylated MGMT are more sensitive to temozolomide and radiotherapy, hence have improved prognosis [4, 5]. Currently, the detection of MGMT methylation status relies on genetic profiling approaches, requiring tissue obtained via biopsy or surgical resection. Although tissue is accessible in most patients undergoing gross total resection, tumour heterogeneity poses clear barriers to biopsy-based method [2, 6, 7]. Substantial methylation assessment requires sampling multiple regions of the heterogeneous tumour to capture its full clonal history. In several multiregional-biopsy studies, intratumour heterogeneity in MGMT methylation status had been identified in 14 % of cases [8, 9]. Medical image provides three-dimensional characteristics of the entire tumour in a non-invasive and repeatable way. Recent advances in imaging-genomics permit correlating imaging phenotypes with molecular data. Identification of a possible association between imaging phenotypes and MGMT methylation status has undoubted clinical benefits.
Early evidence has shown the correlation between MRI features and MGMT methylation [10,11,12]. Korfiatis et al. [13] made use of texture features from 2D multiregional MR slices. A recent study [14] investigated VASARI features [15] and volumetric variables of 3D tumour subregions. These features may not fully characterize image phenotypes, thus limiting the potential of the models. Recent advances in an emerging technique, radiomics, permit comprehensive quantification of the imaging phenotypes in heterogeneous tumours. Radiomics converts medical images into minable data through extracting high-throughput quantitative imaging features [16,17,18]. For GBM, several radiomics signatures associated with survival, molecular characteristics and treatment response have been reported [19,20,21,22,23,24,25].
Recent studies have revealed the multiregional and microenvironmental heterogeneity in GBM [2, 7, 26]. They highlight the value of multiregional image analysis in spatially distinct habitats, some of which harbour heterogeneous tumour populations [27, 28]. To our knowledge, although it is recognized that a radiomics model from multiparametric and multiregional MRI holds prognostic and predictive value [19,20,21,22], little work has been done on such an imaging-genomics model associated with MGMT methylation status in GBM. Currently, radiomics models built with a minimal set of all-relevant multiregional imaging features for predicting MGMT methylation status are still required. Particularly, identifying a minimal set of features that are relevant to classification of MGMT methylation status, rather than a larger set of features that are merely useful for predicting outcome without interpretable relevance, might help researchers to explore the mechanism underlying an imaging-genomics model. Moreover, most previous studies use a relatively small cohort from a single-centre, where bias may occur especially when machine learning methods are used.
In this retrospective multicentre study, we investigated the wealth of radiomics features from multiple tumour subregions in multiparametric MR images. The aim was to build a reliable multiregional and multiparametric MRI radiomics model for pretreatment prediction of MGMT promotor methylation status in GBM.
Material and methods
Patient enrolment
In this retrospective multicentre study, a cohort of 193 patients was recruited from The Cancer Imaging Archive (TCIA) and three local institutions between 2011 and 2016. TCIA is a publicly available data set that de-identifies and hosts a large archive of medical images of cancer (www.cancerimagingarchive.net). Institutional Review Board approval for TCIA data was not required. Institutional Review Board approvals from the three local institutions were obtained, and informed patient consent was waived. The primary cohort of 133 patients comprised 57 from TCIA and 76 from Guangzhou General Hospital of Guangzhou Military Command. Another cohort of 60 patients comprising 39 from the Third Affiliated Hospital of Sun Yat-Sen University and 21 from Sun Yat-Sen University Cancer Center were used for independent validation. The inclusion criteria were that patients with (1) newly diagnosed histologically-confirmed GBM (grade IV, WHO classification) and (2) pretreatment MR imaging including T1-weighted, T1-weighted gadolinium contrast-enhanced, T2-weighted and T2-weighted FLAIR (T1w, T1c, T2w and FLAIR, respectively) and (3) available MGMT methylation status. The MGMT methylation data of the TCIA patients obtained from The Cancer Genome Atlas (TCGA), which includes genomics data corresponding to TCIA patients. The clinical variables consisted of age, sex and Karnofsky performance score (KPS). Patient and tumour characteristics of the primary and validation cohorts are summarized in Table 1.
MR imaging
All local MR images were acquired with 3.0-T MR imaging systems (Magnetom Verio or Trio TIM, Siemens Healthcare, or Discovery MR 750, GE). T1w images were acquired at repetition time 380–586 ms; echo time 4.3–20 ms; section thickness 2.0–5.0 mm. T1c were acquired at repetition time 460–720 ms; echo time 4.3–20 ms; section thickness 2.0–5.0 mm. T2w were obtained with repetition time 2,137–5,360 ms; echo time 80–119 ms; section thickness 4.0–5.0 mm. FLAIR were obtained with a repetition time msec, 8002-11000; echo time msec, 91-155; section thickness, 4.0-5.0mm.
MGMT promoter methylation status testing
MGMT promoter methylation status of local patients was assessed by the pyrosequencing technology with standard protocol described in [29]. The DNA was isolated from paraffin sections of tumuor tissue with the QIAamp DNA FFPE Tissue Kit (Qiagen). The prepared DNA was modified with a sodium bisulphite treatment using the EpiTect Bisulfite Kit (Qiagen). Pyrosequencing analysis was performed using the PyroMark Q96 system (Qiagen) in the CpG island region of MGMT.
Image preprocessing and tumour subregion segmentation
A preprocessing pipeline was applied on T1w, T1c, T2w and FLAIR for image standardization. First, N4ITK was applied to correct the bias field distortion [30]. After skull stripping and isotropic voxel resampling, rigid registration was performed with the mutual information similarity metric. Due to the intensity variation between different MRI acquisitions especially for multicentre studies, an efficient landmark-based approach with piecewise intensity mapping was used for intensity standardization across multicentre patients [31]. Then, a convolutional neural network (CNN)-based method was used to automatically segment the tumour into four subregions: necrosis, oedema, non-enhancement area and enhancement area [32]. The four subregions were defined in detail by the Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) [33]. To train the CNN model, real patient MR data from the BRATS 2015 was used. The segmentation procedure was accomplished using the deep learning software TensorFlow [34].
Multiregional and multiparametric MRI radiomic feature extraction
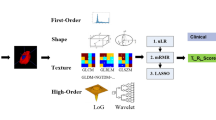
Based on the segmented subregions, we extracted four groups of features: (1) location features, (2) geometry features, (3) intensity features and (4) texture features. The features (2–4) were extracted from four modalities within six extraction subregions, including necrosis, enhancement area, non-enhancement area, oedema, solid core (the whole tumour except oedema) and whole tumour.
The location features were defined as the locations of the tumour geographic epicentre according to the VASARI guidelines [15], and determined by three neurologists (H.B. with 12 years’ experience in neuroradiology, and Y.C. and C.L., each with 5 years’ experience in neuroradiology) and a radiologist (Y.Z., with 8 years’ experience in neuroradiology). There were seven locations in each part of the right, left and bilateral areas, resulting in 21 location features. Twenty-eight geometry features were extracted to describe the 3D characteristics of the tumour shape. 288 intensity features were extracted from six extraction subregions and four MR modalities. These intensity features described the first-order distribution of the multiregional intensities. The texture features were extracted using four methods, including the grey-level co-occurrence matrix (GLCM), grey-level run length matrix (GLRLM), grey-level size zone matrix (GLSZM) and neighbourhood grey-tone difference matrix (NGTDM) methods. The calculation of the texture features can be found in Aerts et al. [18]. 1,248 texture features were computed from six subregions and four modalities, describing the patterns or the high-order distributions of the intensities. The features extracted are summarized in Table 1. Finally, for each patient 1,705 quantitative features were extracted.
Feature selection and classification
Having high-dimensional multiregional features, we aimed to develop a reliable model with a minimal set of relevant features. Selection of relevant features was different from selection of univariately-predictive and non-redundant ones, as we cannot rely on classification accuracy and data redundancy as the selection criteria. Thus a machine learning-based algorithm, Boruta, was used to select all-relevant features and finally build a reliable classification model [35]. Boruta was a wrapper algorithm for all-relevant feature selection, where relevant features were searched in a top-down manner by comparing the importance of the original features with the importance achieved by artificially added random features. A random forest algorithm [36] was performed in each iteration to evaluate the classification and measure the feature importance, where irrelevant features were eliminated progressively. To obtain statistically significant results, the algorithm repeatedly calculated all possible subsets of the features and finally selected the minimal set of the most relevant features for an optimal classification. The R package Boruta was used to build the model [35].
A radiomics model with univariately-predictive and non-redundant features was also built. For each feature, the Mann-Whitney U test was used to assess its univariate predictive power. The Benjamini-Hochberg method was used to correct for multiple testing p-values. From all 1,705 features, features with p<0.05 were selected for further analysis. Then, Spearman’s correlation coefficient was used for redundancy evaluation. For feature pairs with a correlation coefficient ≥ 0.80, the more predictive feature was retained. Based on the selected features, a random forest algorithm was used for classification. Furthermore, two combined models based on radiomics features and clinical factors (sex, age and KPS) were built using both feature selection methods followed by a random forest. A predictive model based on clinical factors alone was also built using random forest.
Statistical analysis
All statistical analysis was done with R software, version 3.4.0 (https://www.r-project.org/). The statistical significance levels were set at .05. The differences in sex, age, KPS and MGMT methylation status between the primary and validation cohorts were assessed.
Model validation
All predictive models were trained on the primary cohort and tested on the independent validation cohorts. The performance was assessed using accuracy, sensitivity, specificity and area under the receiver operating characteristic (ROC) curve (AUC). The DeLong method was used for statistical comparison of the ROC curves [37].
Stratified analyses
Stratified analyses were performed to assess the potential association of both radiomics models with MGMT methylation status in patient subgroups stratified by clinical factors. Specifically, the patients from the validation cohort were stratified by age (≤ 65 or > 65 years), sex (female or male) and KPS (≤ 70 or > 70).
Results
There was no significant difference in patient and tumour characteristics between the primary and validation cohorts (p=0.56 to 0.85). Figure 1 shows one example of the segmentation results. Subregions are shown in red (necrosis), yellow (enhancement region), green (non-enhancement region) and blue (oedema).
Feature selection
For both radiomics models, six all-relevant features were selected by the Boruta algorithm while eight features remained after univariately-predictive and non-redundant feature selection, as shown in Table 3 and Table 4. For both combined models, the same six all-relevant features were selected after Boruta selection while age and the same eight features remained after univariately-predictive and non-redundant feature selection.
Model validation
The radiomics model with six relevant features achieved an AUC of 0.95 and an accuracy of 87 % in the primary cohort. The predictive performance was further confirmed in the validation cohort with an AUC of 0.88 and an accuracy of 80 %. To further reveal the relevance of the selected six features with the MGMT methylation, the feature maps are presented in Fig. 2 for an unmethylated patient and a methylated patient. Meanwhile, the radiomics model with eight univariately-predictive and non-redundant features reached an AUC of 0.94 and an accuracy of 88 % in the primary cohort. In the validation cohort its performance declined to an AUC of 0.76 and an accuracy of 70 %. The ROC curves of both six-feature and eight-feature radiomics models are shown in Fig. 3. The DeLong analysis found a significant difference (p=.012) between the AUCs of both models in the validation cohort. The performance of the radiomics models, the clinical model and the combined models (radiomics features + clinical factors) are summarized in Table 5.
Stratified analyses
The performance of both radiomics models for patients in the validation cohort stratified by age, sex and KPS is shown in Fig. 4. Significant differences (p<0.05) were found between the AUCs of both radiomics models in all subgroups by using the DeLong analysis.
Performance comparison between all-relevant radiomics model and univariately-predictive non-redundant radiomics model for patients in the validation cohort stratified by age, sex and Karnofsky performance score (KPS). AR and PN indicate, respectively, all-relevant and univariately-predictive non-redundant. (a) Age > 60 years. (b) Age≤ 60 years. (c) Female. (d) Male. (e) KPS > 70. (f) KPS ≤ 70
Discussion
According to the radiomics hypothesis, imaging phenotypes could be the expression of underlying biological or genetic heterogeneity [16, 17]. Previous studies have investigated the visually-assessed features [10,11,12], texture features from 2D MRI slices [13] and VASARI features [14] for MGMT methylation prediction. In these studies, the AUCs ranged from 0.75 to 0.85 while the accuracies ranged from 58 % to 73.6 %. Note that all these studies used cross-validation rather than an independent validation cohort for performance evaluation. To the best of our knowledge, our study was the first multicentre study with an independent validation cohort, and our all-relevant radiomics model achieved higher accuracy (80 %) and AUC (0.88) compared with previous models. Our study was based on 1,705 quantitative features derived from multiple 3D tumour subregions in multiparametric MR images (Table 2), allowing for a more comprehensive characterization of the intratumour heterogeneity. This may offer potential to improve the prediction performance. The inherent intensity variability across MRI acquisitions may severely reduce the stability of quantitative features, especially in a multicentre study. To overcome this, we normalized the images via an effective landmark-based mapping method [31] that allowed for stable feature extraction from multiparametric MRI.
Feature selection was one key step in the discovery of predictors from high-throughput features. Many existing imaging-genomics studies built their classification models by selecting a set of non-redundant features with the best univariate predictive power [18, 20, 21]. This study built two radiomics models, one with six all-relevant features and the other with eight univariately-predictive and non-redundant ones. Despite the comparable performance in the training cohort, the six-feature model outperformed the eight-feature one in the validation cohort with a significantly better AUC (Fig. 2). This demonstrated the reliability of the employed all-relevant feature selection method.
In imaging-genomics models a relevant feature selection might be a more interesting problem than dimension reduction. It is related to the so-called all-relevant problem – the identification of all features that are in certain circumstances relevant to the classification [38]. The all-relevant principle may be less well known, although it has received much interest in genomics analysis that identifies genes related to cancer [39, 40]. Here we built the six-feature model by using a Boruta algorithm [35], which has been successfully used for all-relevant feature selection in radiomic [41] and radiogenomic [42] studies. Interestingly, the features selected in the two models were totally different (Tables 3 and 4). This implies that a relevant feature does not mean a strong predictive feature, and a relevant classifier might contain weakly predictive features. Similarly, in the polygenic inheritance situation, the identified relevant genes are often great in quantity but small in individual effect [43]. In our study, univariately-predictive feature selection ignored the potential correlation or interaction between features, which may be important to the MGMT methylation. This may cause the performance degradation of the eight-feature model. Previous studies found that locations, volumetric variables and diffusion tensor imaging (DTI) parameters were correlated with MGMT methylation [10,11,12,13,14]. In the eight-feature model, one geometry feature was selected because of its higher univariately-predictive power, while in the six-feature model, the selected all-relevant features consisted of two intensity features and four texture features. This indicates that multiregional intensity and texture features may be more relevant to MGMT methylation than location and geometric features.
Although our selected features were relevant to classification, interpretation of the relevance remains challenging. Here we tried to understand the relevance in a fundamental way. Radiological intratumour heterogeneity usually reflects multiregional variations in blood flow, oedema and necrosis, whereas genetic heterogeneity is typically due to random mutations. From tumour evolutionary and ecological perspectives, the imaging-genomics association might be ascribed to the link between imageable microenvironmental variations and cellular adaptive strategies [44], which is governed by interactions between Darwin selection forces and cell phenotype in a predictable and reliable manner [45, 46]. This link may enable clinical imaging to make reliable predictions about cellular or even molecular properties of cancer [46]. Based on this hypothesis, features from multiregional and multiparametric MR images could identify microenvironmental variations and radiologically characterize intratumour Darwin dynamics. The feature maps in Fig. 3 give an example of how the selected features radiologically quantified the multiregional variations. Specifically, the feature f1 measured the intensity asymmetry within the tumour core; f2 was the quadratic intensity in oedema; f3 measured the amount of local variations present in necrosis; f4 quantified the intensity variance between homogeneous subregions within the enhancement area; f5 characterized the texture homogeneity of the oedema area; f6 described the spatial rate of intensity change within the tumour core. Several similar attributes have been reported previously, such as a certain image uniformity in tumour core and a certain image pattern in the enhancement area for methylated tumours [13, 14]. Lemée et al. [28] revealed that GBM genetic heterogeneity also involves the oedema area. Our method offers an automatic way to identify methylation-relevant multiregional features that in the radiological level characterize the intratumour microenvironmental variations.
A previous study showed that GMB MGMT methylation may be more prevalent in the elderly [47]. Our results show that although patient age was selected as a predictive univariable, it was abandoned by the all-relevant selection algorithm. From Table 5 we can observe that in the validation cohort combing age with univariately-predictive image features resulted in a slight performance degradation (AUC=0.75, accuracy=68 %) compared with using univariately-predictive image features alone (AUC=0.76, accuracy=70 %). All the radiomics and combined models outperformed the model built with clinical factors alone. Our results highlight the value of the radiomics feature rather than the clinical factor in methylation prediction. When the patients were stratified by age, sex and KPS, the six-feature model outperformed the eight-feature model in all subgroups with significantly better AUCs.
The clinical relevance of this multicentre study lies in the advancement of the radiomics-based non-invasive pretreatment prediction of GBM MGMT methylation, and in the extension of existing knowledge by the all-relevant multiregional and multiparametric MRI features that may serve as putative imaging-genomics biomarker [48]. However, this study still had several limitations. First, only four standard-of-care MR modalities were used. Previous studies have suggested that DTI and dynamic contrast-enhanced (DCE) images are promising in predicting MGMT methylation. Our model may be potentially improved by incorporating these new images. Second, although reasonable accuracy of 80 % and AUC of 0.88 had been achieved on an independent validation cohort, we need to be cautious about direct use to aid clinical decision making. Notwithstanding the virtue of pretreatment non-invasive detection, currently the biopsy-based assessment may still outperform the prediction accuracy of our model. More efforts should be made to improve the prediction performance. Thirdly, although this study was based on multicentre cohorts, larger data sets from more institutes should be involved to demonstrate the potential clinical utility of our model. A larger training data set also has great potential to improve the prediction performance of our machine learning-based model.
In conclusion, the presented multiregional and multiparametric MRI radiomics model has the potential to non-invasively detect MGMT promoter methylation status in GBM. The all-relevant features predict MGMG methylation with significantly better performance than the univariately-predictive and non-redundant features. Despite the limitations, the proposed model may serve as a potential imaging biomarker to guide preoperative patient care and make a step forward in radiomics-based precision medicine for GBM patients.
Abbreviations
- AUC:
-
Area under the ROC curve
- FLAIR:
-
Fluid-attenuated inversion recovery
- GBM:
-
Glioblastoma multiforme
- GLCM:
-
Grey-level co-occurrence matrix
- GLRLM:
-
Grey-level run length matrix
- GLSZM:
-
Grey level size zone matrix
- KPS:
-
Karnofsky performance score
- MGMT:
-
O6-methylguanine-DNA methyltransferase
- MRI:
-
Magnetic resonance imaging
- NGTDM:
-
Neighbourhood grey-tone difference matrix
- ROC:
-
Receiver operating characteristic curve
- TCGA:
-
The Cancer Genome Atlas
- TCIA:
-
The Cancer Imaging Archive
- VASARI:
-
Visually Accusable Rembrandt Images
References
Ostrom QT, Gittleman H, Xu J, et al (2016) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol 18:v1–v75
Reardon DA, Wen PY (2015) Glioma in 2014: unravelling tumour heterogeneity-implications for therapy. Nat Rev Clin Oncol 12:69–70
Weller M, Stupp R, Reifenberger G et al (2010) MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol 6:39–51
Hegi ME, Diserens AC, Gorlia T et al (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352:997–1003
Stupp R, Hegi ME, Mason WP et al (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10:459–466
Sottoriva A, Spiteri I, Piccirillo SG et al (2013) Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci 110:4009–4014
Patel AP, Tirosh I, Trombetta JJ et al (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401
Dunn J, Baborie A, Alam F et al (2009) Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Brit J Cancer 101:124–131
Parker NR, Hudson AL, Khong P et al (2016) Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci Rep 6:22477
Drabycz S, Roldán G, De Robles P et al (2010) An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage 49:1398–1405
Carrillo JA, Lai A, Nghiemphu PL et al (2012) Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. Am J Neuroradiol 33:1349–1355
Moon WJ, Choi JW, Roh HG, Lim SD, Koh YC (2012) Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology 54:555–563
Korfiatis P, Kline TL, Coufalova L et al (2016) MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys 43:2835–2844
Kanas VG, Zacharaki EI, Thomas GA, Zinn PO, Megalooikonomou V, Colen RR (2017) Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput Meth Prog Bio 140:249–257
VASARI Research Project, Available via https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project. Accessed 2 June 2017
Lambin P, Leijenaar RT, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Gillies RJ, Kinahan PE, Hricak H (2015) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by non-invasive imaging using a quantitative radiomics approach. Nat Comm 5
Itakura H, Achrol AS, Mitchell LA et al (2015) Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med 7:303ra138–303ra138
Cui Y, Tha KK, Terasaka S et al (2015) Prognostic imaging biomarkers in glioblastoma: development and independent validation on the basis of multiregion and quantitative analysis of MR images. Radiology 278:546–553
Li Q, Bai H, Chen Y et al (2017) A Fully-Automatic Multiparametric Radiomics Model: Towards Reproducible and Prognostic Imaging Signature for Prediction of Overall Survival in Glioblastoma Multiforme. Sci Rep 7:14331
Kickingereder P, Götz M, Muschelli J et al (2016) Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin Cancer Res 22:5765–5771
Cui Y, Ren S, Tha KK, Wu J, Shirato H, Li R (2017) Volume of high-risk intratumoral subregions at multi-parametric MR imaging predicts overall survival and complements molecular analysis of glioblastoma. Eur Radiol 27:3583–3592
Yoon RG, Kim HS, Paik W, Shim WH, Kim SJ, Kim JH (2017) Different diagnostic values of imaging parameters to predict pseudoprogression in glioblastoma subgroups stratified by MGMT promoter methylation. Eur Radiol 27:255–266
Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P (2017) Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur Radiol 27:4188–4197
Vartanian A, Singh SK, Agnihotri S et al (2014) GBM's multifaceted landscape: highlighting regional and microenvironmental heterogeneity. Neuro-oncology 16:1167–1175
Gatenby RA, Grove O, Gillies RJ (2013) Quantitative imaging in cancer evolution and ecology. Radiology 269:8–14
Lemée JM, Clavreul A, Menei P (2015) Intratumoral heterogeneity in glioblastoma: don't forget the peritumoral brain zone. Neuro-oncology 17:1322–1332
Havik AB, Brandal P, Honne H et al (2012) MGMT promoter methylation in gliomas-assessment by pyrosequencing and quantitative methylation-specific PCR. J Transl Med 10:36
Tustison NJ, Avants BB, Cook PA et al (2010) N4ITK: improved N3 bias correction. IEEE T Med Imaging 29:1310–1320
Lötjönen JM, Wolz R, Koikkalainen JR et al (2010) Fast and robust multi-atlas segmentation of brain magnetic resonance images. NeuroImage 49:2352–2365
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE T Med Imaging 35:1240–1251
Menze BH, Jakab A, Bauer S et al (2015) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE T Med Imaging 34:1993–2024
Abadi M, Agarwal A, Barham P et al (2016) Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint:1603.04467
Kursa MB, Rudnicki WR (2010) Feature selection with the Boruta package. J Stat Softw 36:1–13
Breiman L (2001) Random forests. Mach Learn 45:5–32
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics:837-845
Nilsson R, Peña JM, Björkegren J, Tegnér J (2007) Consistent feature selection for pattern recognition in polynomial time. J Mach Learn Res 8:589–612
Kursa MB (2014) Robustness of Random Forest-based gene selection methods. BMC bioinformatics 15:8
Guo P, Luo Y, Mai G et al (2014) Gene expression profile based classification models of psoriasis. Genomics 103:48–55
Baessler B, Mannil M, Oebel S, Maintz D, Alkadhi H, Manka R (2017) Subacute and Chronic Left Ventricular Myocardial Scar: Accuracy of Texture Analysis on Nonenhanced Cine MR Images. Radiology https://doi.org/10.1148/radiol.2017170213
Yamamoto S, Korn RL, Oklu R et al (2014) ALK molecular phenotype in non–small cell lung cancer: CT radiogenomic characterization. Radiology 272:568–576
Falconer DS, Mackay TFC (1996) Introduction to Genetics, Fourth edn. Addison Wesley Longman, Harlow, Essex, UK
Burrell RA, McGranahan N, Bartek J, Swanton C (2013) The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501:338–345
McCarthy N (2012) Tumour heterogeneity: Darwin's finches. Nat Rev Cancer 12:317–317
Polyak K (2014) Tumor heterogeneity confounds and illuminates: a case for Darwinian tumor evolution. Nat Med 20:344–346
Reifenberger G, Hentschel B, Felsberg J et al (2012) Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer 131:1342–1350
O'connor JP, Aboagye EO, Adams JE et al (2016) Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 14:169–186
Funding
This study received funding by National Natural Science Foundation of China (No. 61571432), National Basic Research Program of China (973 Program, No. 2015CB755500), and Shenzhen Basic Research Program (JCYJ20170413162354654).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Hairong Zheng.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors (Zhi-Cheng Li) has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained from the three local institutions. Institutional Review Board approval for the TCIA data was not required.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Multicentre study
Rights and permissions
About this article
Cite this article
Li, ZC., Bai, H., Sun, Q. et al. Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: A multicentre study. Eur Radiol 28, 3640–3650 (2018). https://doi.org/10.1007/s00330-017-5302-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-017-5302-1