Abstract
Purpose
To evaluate the diagnostic relevance of T2-weighted (T2W) MRI-derived textural features relative to quantitative physiological parameters derived from diffusion-weighted (DW) and dynamic contrast-enhanced (DCE) MRI in Gleason score (GS) 3+4 and 4+3 prostate cancers.
Materials and Methods
3T multiparametric-MRI was performed on 23 prostate cancer patients prior to prostatectomy. Textural features [angular second moment (ASM), contrast, correlation, entropy], apparent diffusion coefficient (ADC), and DCE pharmacokinetic parameters (Ktrans and Ve) were calculated from index tumours delineated on the T2W, DW, and DCE images, respectively. The association between the textural features and prostatectomy GS and the MRI-derived parameters, and the utility of the parameters in differentiating between GS 3+4 and 4+3 prostate cancers were assessed statistically.
Results
ASM and entropy correlated significantly (p < 0.05) with both GS and median ADC. Contrast correlated moderately with median ADC. The textural features correlated insignificantly with Ktrans and Ve. GS 4+3 cancers had significantly lower ASM and higher entropy than 3+4 cancers, but insignificant differences in median ADC, Ktrans, and Ve. The combined texture-MRI parameters yielded higher classification accuracy (91%) than the individual parameter sets.
Conclusion
T2W MRI-derived textural features could serve as potential diagnostic markers, sensitive to the pathological differences in prostate cancers.
Key Points
• T2W MRI-derived textural features correlate significantly with Gleason score and ADC.
• T2W MRI-derived textural features differentiate Gleason score 3+4 from 4+3 cancers.
• T2W image textural features could augment tumour characterization.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Prostate cancer (PCa) diagnosis and assessment has improved significantly since the introduction of multiparametric-MRI (mp-MRI) into its management [1, 2]. Combining multiple MRI sequences such as T2-weighted imaging (T2WI), diffusion-weighted imaging (DWI), spectroscopy, and dynamic contrast-enhanced (DCE) imaging provides complementary information for detection, localization, staging, and grading of the disease, thereby improving accuracy, sensitivity, and specificity [1, 3, 4].
T2WI provides anatomical images of the prostate at high spatial resolution. DWI probes the restriction of free motion of water molecules in tissue to derive the apparent diffusion coefficient (ADC), which provides information about cell density and tissue microenvironment [5, 6]. DCE-MRI enables pharmacokinetic modelling of the dynamic uptake and extravasation of a contrast agent, and can be used to derive parameters such as the volume transfer constant (Ktrans), extravascular-extracellular volume fraction (Ve), and blood plasma volume fraction (Vp) [7–9], from which tissue pathophysiology (e.g. perfusion, vascular permeability) can be inferred [10].
The Gleason grading system [11, 12] is regarded as the gold standard for PCa diagnosis and aggressiveness assessment. However, PCa is a heterogeneous and multifocal disease; Gleason grading based on prostate biopsies, which only samples a small portion of the gland, may, therefore, not provide a complete representation of the disease and tends to have low specificity [13, 14], whereas prostatectomy Gleason grading is usually confined to intermediate- and high-risk patients. Furthermore, the Gleason grading is qualitative, subjective, and hence prone to intra and inter-observer variations. Despite the improvements offered by mp-MRI incorporation, the separation of Gleason score (GS) 3+4 cancers from GS 4+3 cancers using conventional MRI parameters tends to be less effective due to overlapping values [15, 16]. These two PCa patterns account for the largest group at biopsy or prostatectomy, and have significantly different prognoses irrespective of having the same GS of 7 [17–20]. Hence, misclassification could have serious clinical consequences. Accurate classification of these two GS patterns thus requires objective, sensitive, and reproducible quantitative image analysis methods.
Texture analysis is a promising technique known to have such attributes. It comprises the use of mathematical parameters, derived as functions of the spatial distribution of pixel intensities, to characterize the fundamental textures of objects in an image [21]. The applicability of texture analysis in medical image analysis has been demonstrated in several studies [22–27], predominantly for characterizing disease types or diseased from healthy tissues. Yet, unlike conventional quantitative MRI parameters such as ADC, Ktrans, and Ve [28, 29], little is known about the pathophysiological semantics of textural features. This knowledge gap poses a challenge in the development of texture-based computer-aided diagnosis tools and their integration into clinical practice. The aim of this study was to bridge this gap by evaluating how grey-level co-occurrence matrix (GLCM) based textural features [30] derived from T2W MRI relate to GS (3+4 vs. GS 4+3) cancer patterns, and MRI-based physiological parameters (ADC, Ktrans, and Ve). Furthermore, the study evaluated the utility of these features/parameters as potential markers for distinguishing between these two PCa patterns was evaluated.
Materials And Methods
Patient Cohort
The cohort of 23 PCa patients used in this retrospective study was obtained from a previous (in 2011) prospective study of 30 patients approved by St. Olavs University Hospital, Trondheim, Norway, and the Regional Committee for Medical and Health Research Ethics, Central Norway. Each patient gave a written informed consent for data usage. Inclusion in the prospective study required that the patient was initially confirmed by means of core needle biopsy to have PCa, and was scheduled to undergo pre-operative mp-MRI involving T2W, DW and DCE-MRI at least 4 weeks after the last biopsy, followed by radical prostatectomy within 12 weeks after the mp-MRI.
MR Image Acquisition
All MRI were performed in feet first supine position, on a 3T MRI system (Magnetom Trio; Siemens Medical Solutions, Erlangen, Germany) using standard body and spine phased array matrix coils for signal detection. Glucagon (Novo Nordisk, Bagsværd, Denmark) was injected intramuscularly prior to imaging to suppress bowel movements.
The T2W images were acquired with a turbo spin-echo sequence (TR/TE 4000/101 ms; flip angle: 150°; FOV: 200×200; matrix: 320×320; slice thickness: 3 mm; interslice gap: 0.6 mm), and the DW images with a single-shot echo-planar imaging sequence, using four b-values: 0, 100, 400, and 800 s/mm2 (TR/TE: 3300/60 ms; flip angle: 90°; FOV: 260×211; matrix: 160×130; slice thickness: 3.6 mm). A 3D fast low-angle shot sequence (time-resolved imaging with stochastic trajectories) was used for the DCE-MRI (TR/TE: 3.85/1.42 ms; flip angle: 12°; FOV: 260×260; matrix: 160×160; slice thickness: 3.6 mm). It included the acquisition of pre-contrast T1W image scans using four variable flip angles (2°, 5°, 10°, and 20°), followed by 70 dynamic scans at 4.22 s temporal resolution. After the third dynamic scan, the gadolinium-based contrast agent gadoterate meglumine (Dotarem®; Amersham Health, Oslo, Norway) was administrated intravenously by bolus injection (0.1 mmol/kg; rate: 2 mL/s) using a Spectris™ MR injection system (Medrad Inc. PA, USA), followed by a 20-mL normal saline flush. The images were oriented along the longest axis of the prostate, perpendicular to the urethra to best match routine histologic sectioning of the prostate and covered the entire prostate volume.
Histopathology and Regions of Interest Delineation
After radical prostatectomy, the excised gland was fixed in formalin and serially sectioned from the apex to the base into 4-mm axial slices, from which 3.5-μm sections were stained with haematoxylin-erythrosine-saffron (HES). The HES-stained slides were examined by an experienced pathologist who outlined cancer foci, described cancer location, and graded all foci in accordance with the Gleason scoring system [11, 12].
For each patient, the histopathology slide with the index tumour—the largest histopathologically defined, clinically significant (volume ≥ 0.5 cm3) cancer focus with the highest GS—was selected. The best corresponding axial T2W image slice was then identified with the guidance of anatomical landmarks (Fig. 1a). To support region of interest (ROI) delineation in the T2W image, both images (histopathology and T2W) were overlaid with a radial grid using the same anatomical landmarks as reference, while taking into account the offsets in orientation and distance between the urethra and the base for the two images. Subsequently, a ROI replicating the index tumour was manually outlined in the T2W image based on the location in the histology section and its appearance on the T2W image.
(a) Histopathology slide spatially matched to T2-weighted (T2W) image slice based on anatomical landmarks (e.g. urethra, ejaculatory ducts, size/shape of the peripheral zone, apex/base proximity), (b) cancer region of interest delineated in T2W image based on histology and rigidly registered to diffusion-weighted (DW) and dynamic contrast-enhanced (DCE) images, and (c) textural features, apparent diffusion coefficient (ADC), volume transfer constant (Ktrans), and extravascular-extracellular volume fraction (Ve) computation.
The delineated T2W image ROI was then transformed (using Elastix toolbox [31]) to the corresponding DW and DCE images (Fig. 1b) and their parametric maps (Fig. 1c) via intensity-based rigid registration using the Mattes mutual information similarity metric. To optimize the registration, possible intensity-based artefacts due to artificial global intensity inhomogeneity were corrected for according to Cohen et al [32] prior to registration. The resulting registration transformations were then applied to the original/unfiltered images or ROIs. The quality of the inter-protocol ROI registration was assessed visually in the three images together with the ADC and Ktrans maps.
Quantitative Analysis
Haralick textural features [30] and MRI-based physiological parameters consisting of ADC, Ktrans, and Ve were computed from the respective delineated ROIs as follows:
Two-dimensional GLCM-based texture analysis [30] was performed on the T2W image ROIs (mean area: 212 pixels, range: 116–416 pixels) by first rescaling (histogram equalization) the intensities within each ROI to a range of 0-255 grey levels. The GLCM—a square matrix in which each element GLCM(i,j) represents the number of times that a pair of pixels with grey levels i and j co-occur at a specified distance and direction in relation to each other [30]— was computed using 64 bins and at one pixel distance for each direction of 0°, 45°, 90°, and 135°. Haralick textural features were then calculated from each GLCM, and the average of each parameter over the four directions was obtained. A single value was computed for each textural feature for a defined ROI. Haralick et al [30] originally proposed fourteen GLCM textural features, most of which are highly correlated and reflect similar information. We therefore selected four distinct textural features—angular second moment (ASM), contrast, correlation, and entropy—based on the literature [26, 30, 33] for analysis. ASM measures homogeneity of an image; contrast measures local variations; correlation measures linear dependency of the grey-levels; and entropy measures randomness or complexity [30]. The mathematical expressions for these features are given in the appendix.
ADC maps were calculated voxel-wise from the DW images using b-values of 100, 400, and 800 s/mm2 by fitting the DWI signal as a function of b-value to the monoexponential decay model. The median ADC value for each ROI was calculated.
The extended Tofts model [9] (two-compartment with intravascular tracer contribution) was applied to the DCE-MRI time series images for pharmacokinetic modelling. Prior to parameter estimation, the DCE and the pre-contrast T1W images were rigidly co-registered [31] in time as described above to correct for possible patient motion during imaging. Pre-contrast T1-maps were calculated (from the pre-contrast images) for the conversion of signal intensity into contrast agent concentration. Voxels with T1 values outside [500–3000 ms] were excluded [34]. Fitting of image data to the model was done on a voxel-by-voxel basis from which maps of Ktrans, Ve, and Vp were computed. The population-averaged arterial input function (AIF) reported by Parker et al [35] was used, and the time delay of the tissue enhancement curve relative to the AIF was accounted for. The contrast agent relaxivity was 3.5 s-1mM-1. Voxels for which Ktrans < 0, Ktrans > 10, Ve < 0, or Ve > 1 were also discarded, and the median values computed for each ROI. Vp was not used for further analysis.
Median T2W signal intensity (T2WSI) was also computed for each ROI. First, the image intensities (original/non-scaled) were corrected for nonstandardness by aligning the image intensity histogram of each patient image to match the mean intensity histogram across the different patients [36], thereby ensuring that the image intensities are comparable and have consistent tissue-specific meaning.
All the analyses were performed using MATLAB R2014a (Mathworks, Natick, MA, USA).
Statistical Analysis
Point-biserial correlation coefficients (rpb) and Spearman correlation coefficients (ρ) were calculated to quantify the relationships between the textural features and GS and between the textural features and MRI-based physiological parameters (ADC, Ktrans, and Ve), respectively.
Each computed parameter was tested for significant differences between GS 3+4 and GS 4+3 cancers using two-tailed Mann-Whitney U tests. A generalized linear model with a logistic regression model component was used to perform receiver operating characteristic (ROC) analysis to classify the two cancer patterns. The nonparametric approach by DeLong et al [37] for comparing the areas under two or more correlated ROC curves was further used to evaluate the classification capabilities of the parameters. p-values < 0.05 were considered to be statistically significant. The p-values were adjusted for multiple testing using Benjamini and Hochberg’s approach [38]. The statistical analyses were performed in MATLAB.
Results
Patient Characteristics
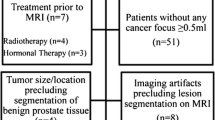
Seven of the 30 patients were excluded from this retrospective study due to unavailable histopathology (n = 5), poor MR image quality, or histopathology-MR matching (n = 2). A total of 32 clinically significant (volume ≥ 0.5 cm3) cancer foci according to the histopathology were present in the remaining 23 patients (details of patient and tumour exclusion are given in Fig. 2). Twenty-three respective index tumours were analysed (Table 1). Because of incomplete DCE data acquisition, data sets for 20 patients were included in the statistical analysis of the DCE pharmacokinetic (Ktrans and Ve) parameters.
Flowchart of patient and tumour inclusion and exclusion. PZ = peripheral zone; TZ = transition zone. One cancer focus per patient, i.e. the index tumour, the largest histopathologically defined, clinically significant (volume ≥ 0.5 cm3) cancer focus with the highest Gleason score was considered for analysis. Excluded multifocal cancers: two 3+3, three 3+4 and four 4+3 (two from the same patient) Gleason score cancers.
T2W MRI-derived Textural Features Correlate Significantly with GS and ADC
T2W image textural features ASM and entropy correlated significantly (p < 0.05) with GS after multiple test correction. ASM correlated moderately negatively (rpb = -0.52), and entropy moderately positively (rpb = 0.49). ASM, entropy, and contrast also correlated significantly (p < 0.05) with median ADC. The correlations were highly positive (ρ = 0.82), highly negative (ρ = -0.80) and moderately negative (ρ = -0.44), respectively. None of the textural features correlated significantly with median Ktrans or Ve (Table 2).
T2W MRI-Derived Textural Features Distinguish GS 3+4 from 4+3 Cancers
Patients with GS 4+3 cancers had significantly lower T2W image textural ASM and higher entropy than those with GS 3+4 cancers (Fig. 3), but no significant difference was found for the conventional MRI parameters (Fig. 4). Table 3 depicts the areas under the curves (AUCs) and 95% confidence intervals (CIs) for the ROC analysis classification of the two cancer patterns using the individual parameters. ASM and entropy were the best performing individual parameters, whereas correlation and contrast performed worst. The combined textural features resulted in a higher classification accuracy (AUC = 82%, CI = 0.61–0.99, p < 0.05) than the MRI-based physiological parameters combined (AUC = 75%, CI = 0.52–0.98, p < 0.05). The highest classification accuracy (AUC = 91%, CI = 0.75–0.99, p < 0.05) was however achieved when all computed parameters were combined (Fig. 5). The differences in AUCs were not significant.
Box plots comparing the distributions of median T2W signal intensity (T2WSI), apparent diffusion coefficient (ADC), volume transfer constant (Ktrans), and extravascular-extracellular volume fraction (Ve) between Gleason score 3+4 and 4+3 cancers. Black diamonds indicate mean values. p-values corrected for multiple testing.
ROC curves for T2W MRI-derived textural features (angular second moment, contrast, correlation, and entropy), MRI-based physiological parameters (apparent diffusion coefficient, volume transfer constant, and extravascular-extracellular volume fraction), and combined texture-MRI parameters for distinguishing Gleason score 3+4 versus Gleason score 4+3 cancers.
Discussion
The clinical applicability of texture analysis remains limited despite having been shown to have high accuracy and sensitivity in classifying tumour patterns and discriminating healthy from diseased tissues [22–26, 39]. The latest prostate imaging reporting and data system (PI-RADS) [40] describes the texture of prostate tissue as an important feature for identifying prostate cancer, especially in the transition zone (TZ), yet the evaluation procedure is qualitative. Successful integration of texture-based computer-aided diagnosis tools (i.e. texture analysis) into clinical practice could make this process quantitative and less subjective. However, texture analysis being a relatively new field in medical image analysis, the pathophysiological semantics and relevance of the textural features are currently lacking. In addition, the existence of numerous texture analysis approaches [21, 41] and subsequent large number of possible textural features serve as challenges in selecting the most relevant features for prostate cancer. This preliminary study therefore investigated the diagnostic relevance of T2W MRI-derived textural features (ASM, contrast, correlation, entropy) in relation to parameters with known pathophysiological significance (i.e. GS, ADC, Ktrans, and Ve).
A number of studies have used texture analysis of histopathology images for automated Gleason grading with high accuracy [42–44]. Other studies found GLCM-based textural features derived from ADC maps to be capable of differentiating between benign and malignant tumours in the peripheral zone (PZ) [26] and GS 3+3 from GS ≥ 7 cancers [45]. In this study, however, we focused the texture analysis on T2W images because they are easy to acquire, less prone to artefacts, non-invasive, and suitable for every patient. Importantly, T2WI offers high signal-to-noise ratio, spatial resolution and soft tissue contrast images of the prostate structures [46], an important feature in PI-RADS [40]. Hence, texture analysis on these high-resolution images could reveal subtle alterations in tissue architecture due to cancer. Moreover, this study further improves our understanding of textural features in that it explored additional quantitative physiological MRI parameters, Ktrans and Ve, which the previous studies did not touch on.
We found the T2W image-derived textural features (ASM and entropy) to correlate significantly with GS. Our results support the findings of Vignati et al [45], who also found significant correlations between T2W image textural features [homogeneity (ASM) and contrast] and GS. Although Wibmer et al [26] did not find T2W image textural features to correlate significantly with GS, their study and that of Rosenkrantz et al [15] found that ADC map textural features correlated significantly with GS and the percentage of Gleason grade 4 in GS 7 cancers, respectively. The differences in data acquisition (resolution) and analysis (e.g. pooling together PZ and TZ cancers) between these studies possibly contributed to the discordance.
More interestingly, we found these two textural features (ASM and entropy) to be significantly different between GS of 3+4 and 4+3 cancers. A similar observation was also reported in [26] for ASM. These findings further underscore the need to consider each cancer group separately in clinical practice during staging or therapeutic intervention, irrespective of having the same GS of 7, as indicated in previous studies [19, 20]. Median T2WSI did not differ significantly between the two cancer groups, an indication that T2W image textural features could be more sensitive in revealing underlying tissue morphology than ordinary summary statistics of T2W image intensities.
Cancer growth requires formation of new blood vessels to sustain the proliferating cells. The abnormal nature of such neo-vasculature causes fast extravasation of injected contrast agent and thus an increase in Ktrans when compared to healthy tissues [10, 47]. DCE pharmacokinetic analysis in particular probes blood vessel function and not the underlying morphology of the tissue per se, a possible explanation for the lack of association with the textural features. Moreover, GS 7 cancers could grow relatively slowly, without heavy demand for new blood vessels, as opposed to more aggressive cancers.
Generally, PCa is characterized by high cellularity and decreased extracellular space as a result of reduced luminal space and stromal matrix [46, 48]. Movement of extracellular and intraductal water molecules is, therefore, restricted, resulting in low ADC. These tissue microenvironment characteristics, which serve as determinants of ADC, also define the underlying textural properties of the tissue and could explain the significant correlation observed between the textural features (ASM, contrast, entropy) and ADC (Table 2).
Textural features ASM (homogeneity) and entropy (randomness/complexity) were particularly found to correlate significantly with both GS and ADC. Increased PCa aggressiveness is characterized by decreased ADC, as well as deterioration of the architectural patterns depicting cellular integrity of the prostate gland due to poor differentiation and glandular structure deformation (i.e. high Gleason grade). All of these could result in less homogeneity and increased complexity of the tissue, which is reflected by the decreased ASM and increased textural entropy, possibly explaining the observed correlations with GS and ADC. Arguably, these two features could be considered the most intuitive textural descriptors of the Gleason grading system.
Vignati et al [45] also found significant correlations between T2W image textural features (ASM and contrast) and ADC values. Similar to Fehr et al [33], our results also indicate that combining traditional MRI parameters and T2W MRI-derived textural features could achieve a higher classification accuracy (91%) than the traditional MRI parameters alone. ADC calculations are dependent on the range of b-values used [40]. Our ADC calculation excluded the b0 image, which minimized the contribution of perfusion to some extent. Though relatively high, the majority of the ADC values are within a range ([816–1891 μmm2/s] for GS 3+4 and [753–1405 μmm2/s] for GS 4+3 cancers) comparable to other studies [15, 26, 28]. Contrary to other studies [28, 49], we did not find median ADC to differ significantly between GS 3+4 and 4+3 cancers, as also reported in [15]. The most plausible reason for this observation is the relatively low number of patients in our cohort; nonetheless, the textural features could significantly differentiate the two cancer patterns with higher accuracy.
This study has some limitations. The sample size is relatively small (23 patients), and our focus on separating only GS 3+4 and GS 4+3 PCa patterns could be regarded as limited in terms of tumour diversity. The sample size did not allow for separate analyses of PZ and TZ cancers as in other studies [25, 26, 33]. As all the analyzed data represent index tumours of GS 3+4 or 4+3, this small tumour diversity could also be explained as sample homogeneity, which contributes to the quality of our data. Including multiple cancers from the same patient (multifocal cancers) as separate data points could be a way to increase the sample size, but would have hampered sample homogeneity. Furthermore, in clinical practice, treatment decision mostly relies on the Gleason score of the index tumour (i.e. the largest histopathologically defined, clinically significant (volume ≥ 0.5 cm3) cancer focus with the highest GS). Hence, including multifocal cancers could generate confounding results that do not accurately represent clinical practice.
Secondly, standardized T2WSI were used for cancer characterization, rather than directly quantified T2 values through T2-mapping. Further, we employed 2D texture analysis instead of the reported improved 3D texture analysis [50, 51] since the latter is not considered optimal for acquisitions with interslice gaps. Finally, the lack of spatial co-registration of the histopathology slides and the MR images, which caused us to delineate the ROIs directly on the T2W MR images is also worth noting.
Conclusions
Our study shows that T2W MRI-derived textural features correlate with the underlying pathophysiology of prostate cancer tissue, and thus could augment existing prostate cancer classification techniques. However, for texture analysis to gain a stronger place in prostate cancer management, more efforts need to be directed towards understanding the biological and/or pathological semantics of the textural features, determining the most relevant features, and standardizing texture analysis methods. To this effect, a validation of this study in a larger, preferably multi-centre cohort with higher tumour diversity should be pursued.
References
Scheenen TW, Rosenkrantz AB, Haider MA, Futterer JJ (2015) Multiparametric Magnetic Resonance Imaging in Prostate Cancer Management: Current Status and Future Perspectives. Invest Radiol 50:594–600
Vos EK, Kobus T, Litjens GJ et al (2015) Multiparametric Magnetic Resonance Imaging for Discriminating Low-Grade From High-Grade Prostate Cancer. Invest Radiol 50:490–497
Kitajima K, Kaji Y, Fukabori Y, Yoshida K, Suganuma N, Sugimura K (2010) Prostate cancer detection with 3 T MRI: comparison of diffusion-weighted imaging and dynamic contrast-enhanced MRI in combination with T2-weighted imaging. J Magn Reson Imaging 31:625–631
Delongchamps NB, Rouanne M, Flam T et al (2011) Multiparametric magnetic resonance imaging for the detection and localization of prostate cancer: combination of T2-weighted, dynamic contrast-enhanced and diffusion-weighted imaging. BJU Int 107:1411–1418
Koh DM, Collins DJ (2007) Diffusion-weighted MRI in the body: applications and challenges in oncology. AJR Am J Roentgenol 188:1622–1635
Malayeri AA, El Khouli RH, Zaheer A et al (2011) Principles and applications of diffusion-weighted imaging in cancer detection, staging, and treatment follow-up. Radiographics 31:1773–1791
Brix G, Griebel J, Kiessling F, Wenz F (2010) Tracer kinetic modelling of tumour angiogenesis based on dynamic contrast-enhanced CT and MRI measurements. Eur J Nucl Med Mol Imaging 37 Suppl 1:S30–S51
Parker GJM, Buckley DL (2005) Tracer Kinetic Modelling for T1-Weighted DCE-MRI. In: Jackson A, Buckley DL, Parker GJM (eds) Dynamic Contrast-Enhanced Magnetic Resonance Imaging in Oncology. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 81–92
Tofts PS, Brix G, Buckley DL et al (1999) Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging 10:223–232
Vos EK, Litjens GJ, Kobus T et al (2013) Assessment of prostate cancer aggressiveness using dynamic contrast-enhanced magnetic resonance imaging at 3 T. Eur Urol 64:448–455
Gleason DF, Mellinger GT (1974) Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol 111:58–64
Epstein JI, Allsbrook WC Jr, Amin MB, Egevad LL (2005) The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol 29:1228–1242
Stav K, Judith S, Merald H, Leibovici D, Lindner A, Zisman A (2007) Does prostate biopsy Gleason score accurately express the biologic features of prostate cancer? Urol Oncol 25:383–386
Divrik RT, Eroglu A, Sahin A, Zorlu F, Ozen H (2007) Increasing the number of biopsies increases the concordance of Gleason scores of needle biopsies and prostatectomy specimens. Urol Oncol 25:376–382
Rosenkrantz AB, Triolo MJ, Melamed J, Rusinek H, Taneja SS, Deng FM (2015) Whole-lesion apparent diffusion coefficient metrics as a marker of percentage Gleason 4 component within Gleason 7 prostate cancer at radical prostatectomy. J Magn Reson Imaging 41:708–714
Verma S, Rajesh A, Morales H et al (2011) Assessment of Aggressiveness of Prostate Cancer: Correlation of Apparent Diffusion Coefficient With Histologic Grade After Radical Prostatectomy. Am J Roentgenol 196:374–381
Chan TY, Partin AW, Walsh PC, Epstein JI (2000) Prognostic significance of Gleason score 3+4 versus Gleason score 4+3 tumor at radical prostatectomy. Urology 56:823–827
Khoddami SM, Shariat SF, Lotan Y et al (2004) Predictive value of primary Gleason pattern 4 in patients with Gleason score 7 tumours treated with radical prostatectomy. BJU Int 94:42–46
Stark JR, Perner S, Stampfer MJ et al (2009) Gleason score and lethal prostate cancer: does 3 + 4 = 4 + 3? J Clin Oncol 27:3459–3464
Epstein JI, Zelefsky MJ, Sjoberg DD et al (2016) A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur Urol 69:428–435
Castellano G, Bonilha L, Li LM, Cendes F (2004) Texture analysis of medical images. Clin Radiol 59:1061–1069
Freeborough PA, Fox NC (1998) MR image texture analysis applied to the diagnosis and tracking of Alzheimer's disease. IEEE Trans Med Imaging 17:475–479
Ganeshan B, Burnand K, Young R, Chatwin C, Miles K (2011) Dynamic contrast-enhanced texture analysis of the liver: initial assessment in colorectal cancer. Invest Radiol 46:160–168
Teruel JR, Heldahl MG, Goa PE et al (2014) Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed 27:887–896
Viswanath SE, Bloch NB, Chappelow JC et al (2012) Central gland and peripheral zone prostate tumors have significantly different quantitative imaging signatures on 3 Tesla endorectal, in vivo T2-weighted MR imagery. J Magn Reson Imaging 36:213–224
Wibmer A, Hricak H, Gondo T et al (2015) Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol 25:2840–2850
Nketiah G, Savio S, Dastidar P, Nikander R, Eskola H, Sievanen H (2015) Detection of exercise load-associated differences in hip muscles by texture analysis. Scand J Med Sci Sports 25:428–434
Boesen L, Chabanova E, Logager V, Balslev I, Thomsen HS (2015) Apparent diffusion coefficient ratio correlates significantly with prostate cancer gleason score at final pathology. J Magn Reson Imaging 42:446–453
Oto A, Yang C, Kayhan A et al (2011) Diffusion-weighted and dynamic contrast-enhanced MRI of prostate cancer: correlation of quantitative MR parameters with Gleason score and tumor angiogenesis. AJR Am J Roentgenol 197:1382–1390
Haralick RM, Shanmugam K, Dinstein I (1973) Textural Features for Image Classification. IEEE Trans Syst Man Cybernet SMC-3:610–621
Klein S, Staring M, Murphy K, Viergever MA, Pluim JP (2010) elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29:196–205
Cohen MS, DuBois RM, Zeineh MM (2000) Rapid and effective correction of RF inhomogeneity for high field magnetic resonance imaging. Hum Brain Mapp 10:204–211
Fehr D, Veeraraghavan H, Wibmer A et al (2015) Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A 112:E6265–E6273
Fennessy FM, Fedorov A, Gupta SN, Schmidt EJ, Tempany CM, Mulkern RV (2012) Practical considerations in T1 mapping of prostate for dynamic contrast enhancement pharmacokinetic analyses. Magn Reson Imaging 30:1224–1233
Parker GJ, Roberts C, Macdonald A et al (2006) Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med 56:993–1000
Nyul LG, Udupa JK, Zhang X (2000) New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 19:143–150
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845
Benjamini Y, Hochberg Y (2000) On the Adaptive Control of the False Discovery Rate in Multiple Testing With Independent Statistics. J Educ Behav Stat 25:60–83
Langenberger H, Shimizu Y, Windischberger C et al (2003) Bone homogeneity factor: an advanced tool for the assessment of osteoporotic bone structure in high-resolution magnetic resonance images. Invest Radiol 38:467–472
Weinreb JC, Barentsz JO, Choyke PL et al (2016) PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol 69:16–40
Haralick RM (1979) Statistical and structural approaches to texture. Proc IEEE 67:786–804
Almuntashri A, Agaian S, Thompson I, Rabah D, Al-Abdin OZ, Nicolas M (2011) Gleason grade-based automatic classification of prostate cancer pathological imagesSystems, Man, and Cybernetics (SMC), 2011 I.E. International Conference on, pp 2696-2701
Jafari-Khouzani K, Soltanian-Zadeh H (2003) Multiwavelet grading of pathological images of prostate. IEEE Trans Biomed Eng 50:697–704
Diamond J, Anderson NH, Bartels PH, Montironi R, Hamilton PW (2004) The use of morphological characteristics and texture analysis in the identification of tissue composition in prostatic neoplasia. Hum Pathol 35:1121–1131
Vignati A, Mazzetti S, Giannini V et al (2015) Texture features on T2-weighted magnetic resonance imaging: new potential biomarkers for prostate cancer aggressiveness. Phys Med Biol 60:2685–2701
Hegde JV, Mulkern RV, Panych LP et al (2013) Multiparametric MRI of prostate cancer: an update on state-of-the-art techniques and their performance in detecting and localizing prostate cancer. J Magn Reson Imaging 37:1035–1054
Buckley DL, Roberts C, Parker GJ, Logue JP, Hutchinson CE (2004) Prostate cancer: evaluation of vascular characteristics with dynamic contrast-enhanced T1-weighted MR imaging--initial experience. Radiology 233:709–715
Tamada T, Sone T, Jo Y, Yamamoto A, Ito K (2014) Diffusion-weighted MRI and its role in prostate cancer. NMR Biomed 27:25–38
Nagarajan R, Margolis D, Raman S et al (2012) Correlation of Gleason scores with diffusion-weighted imaging findings of prostate cancer. Adv Urol 2012:374805
Georgiadis P, Cavouras D, Kalatzis I et al (2009) Enhancing the discrimination accuracy between metastases, gliomas and meningiomas on brain MRI by volumetric textural features and ensemble pattern recognition methods. Magn Reson Imaging 27:120–130
Mahmoud-Ghoneim D, Toussaint G, Constans JM, de Certaines JD (2003) Three dimensional texture analysis in MRI: a preliminary evaluation in gliomas. Magn Reson Imaging 21:983–987
Acknowledgments
The research on which this study was based is funded by The Norwegian Cancer Society.
Gleason grading of the histopathology specimen was performed by Trond Viset, a senior pathologist at St. Olavs University Hospital, Trondheim. The scientific guarantor of this publication is Tone F. Bathen (tone.f.bathen@ntnu.no). The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article. No complex statistical methods were necessary for this paper. Institutional Review Board approval was obtained from St. Olavs University Hospital, Trondheim, and the Regional Committee for Medical and Health Research Ethics, Central Norway. Written informed consent was obtained from all subjects (patients) in this study. Approval from the institutional animal care committee was not required because this study is on humans. The study subjects or cohorts have not been previously reported. Methodology: Retrospective, diagnostic or prognostic study, performed at one institution.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Notation
G is the number of distinct levels in the histogram equalized image; \( N\left(i,j\right) \) is the \( \left(i,j\right) \) th entry in a normalized spatial GLCM; and \( {p}_x(i) \) is the i th entry in the marginal-probability matrix obtained by summing the rows of \( p\left(i,j\right)={\sum}_jp\left(i,j\right) \).
Textural Features
where \( {\mu}_x \) and \( {\sigma}_x \) are the mean and standard deviation of \( {p}_x \), respectively.
Rights and permissions
About this article
Cite this article
Nketiah, G., Elschot, M., Kim, E. et al. T2-weighted MRI-derived textural features reflect prostate cancer aggressiveness: preliminary results. Eur Radiol 27, 3050–3059 (2017). https://doi.org/10.1007/s00330-016-4663-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-016-4663-1