Abstract
Archaeal communities represent a significant fraction of the Antarctic marine microbial plankton and surely play a relevant role in the proper functioning of the ecosystem. We studied the archaeal community structure in surface water samples from Potter Cove, Antarctica. Temporal and spatial variability was investigated along a whole year cycle using DGGE and 16S rRNA gene sequencing from clone libraries. Additionally, photosynthetic pigments, suspended particulate matter (SPM), salinity and temperature were measured. The multivariate analysis performed using diversity, dominance and richness indexes, and environmental data evidenced a seasonal pattern in the archaeal community and revealed that spring–summer samples clustered separately from autumn to winter ones. High salinity and high values of diversity and richness were related to autumn–winter samples, whereas the spring–summer samples were associated mainly with higher values of temperature, SPM, Chl-a, carotenoids and archaeal dominance. The phylogenetic analysis of five independent clone libraries (467 sequences) showed that 448 sequences fell into a clade containing Nitrosopumilus maritimus and other sequences of ammonia-oxidizing archaea which belong to the Thaumarchaeota phylum. A high fraction of these sequences (62 %) constituted a single cluster containing only highly similar Potter Cove representatives, which probably belong to the same species. Fifteen sequences were affiliated to a group closely related to the order Thermoplasmatales (Euryarchaeota). This work represents a first step towards obtaining a deep understanding of the structure of archaeal communities from Antarctic coastal marine environments and contributes to cover the current gap in knowledge of the dynamics of the archaeoplankton in the Antarctic seas.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In the last decades, it has become clear that archaea thrive not only or exclusively in areas exhibiting extreme environmental conditions but also in those with moderate physicochemical parameters. This finding changed the old concept of archaea as being obligate extremophiles to a more realistic view as ecophysiologically relevant ubiquitous microorganisms (Schleper et al. 2005). Among their relevant ecological roles, there is the ability to grow chemoautotrophically using ammonium as electron donor (Hallam et al. 2006; Ingalls et al. 2006). Furthermore, their carbon assimilation pathways permit some archaea to live as strict autotrophs or as mixotrophs, utilizing both carbon dioxide and organic material as carbon sources (Hallam et al. 2006). Marine archaea are estimated to represent one-third of the prokaryotic plankton biomass (DeLong et al. 1994), but deeper knowledge of their ecological role is prevented by the impossibility to cultivate them. The division of the archaeal domain into phyla is now under debate, and what is known so far clearly represents a preliminary classification. Currently, the archaeal domain includes the most widely distributed and most often encountered phyla Crenarchaeota and Euryarchaeota, the “deep branching” Korarchaeota, the seemingly thermophilic group Nanoarchaeota and the recently proposed, diverse and abundant phylum Thaumarchaeota, formerly known as the Marine Group I Crenarchaeota (Brochier-Armanet et al. 2008).
Unlike other oceans, where archaea seem to be predominant only in deep waters, the Southern Ocean contains a significant fraction of archaea as a component of surface prokaryotic plankton (DeLong et al. 1994). Several authors also observed a significant seasonal variability of the archaeal communities from Antarctic marine surface waters (Murray et al. 1998; Massana et al. 1998). Cowie et al. (2010) found that Thaumarchaeota was the dominant archaeal phylotype in Antarctic sea ice. A similar finding was reported for central Arctic sea waters by Bano et al. (2004) and for Arctic sea ice (Collins et al. 2010). Although this group has also proved to be restricted to subsurface waters of temperate areas (Karner et al. 2001; Fuhrman and Hagström 2008), it has also been reported as abundant in surface waters of polar regions and the North Sea, mainly during winter or early spring (Church et al. 2003; Galand et al. 2008). Surface water Marine Group I Crenarchaeota populations seem to decline sharply during the Antarctic spring and summer (Massana et al. 1998; Murray et al. 1998). Some authors attributed such decline to competition with bacteria that bloom in response to increased phytoplankton production in the spring (Church et al. 2003; Wuchter et al. 2006; Herfort et al. 2007).
In this work, we studied the archaeal community structure from Potter Cove, a small fjord located in the south-west area of 25 de Mayo (King George) Island, South Shetland Islands, Antarctica. Potter Cove is strongly influenced by the seasonal input of the meltwater run-off. Potter Cove coastal land was designated Antarctic Specially Protected Area (ASPA) N° 132 by the Scientific Committee on Antarctic Research (SCAR). However, despite a long history of studies of different components of the Potter marine ecosystem including benthos, phytoplankton, zooplankton, micro- and macroalgae (Fuenets et al. 2008; Quartino and Boraso de Zaixso 2008; Schloss et al. 2012), no studies on the composition and structure of its microbial communities have been published. Yet microbial data for this ecological system is urgently needed for the completion of current models, aiming to describe the functioning of this Antarctic marine ecosystem (Schloss et al. 2002). Although Potter Cove seems a typical Antarctic coastal environment, it often lacks a spring phytoplankton bloom reported for many other Antarctic coastal environments and can be defined as a low phytoplankton biomass system (Schloss and Ferreyra 2002). We therefore hypothesize that this atypical characteristic will have a deep influence on the microbial community structure of the cove. In this work, we analysed, during an annual cycle, the presence, composition and spatial and temporal changes of the archaeal community from Potter Cove.
Materials and methods
Site description and sampling
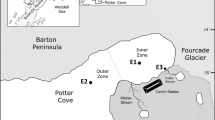
Sampling was carried out in Potter Cove (62°14′S, 58°40′W), 25 de Mayo (King George) Island, Antarctica. The cove waters exhibit marked seasonal changes, with cyclic processes of freezing and thawing of the surface, and a huge input of meltwater during the spring and summer that significantly alter the physicochemical characteristics of the environment. Potter Cove is divided into an outer and an inner basin separated by a 30-m-deep transversal sill. These topological features determine that the dominant streams as well as the major creek draining freshwater into the marine basin mainly affect the inner part of the cove (Curtosi et al. 2007). The outer part of the cove is less affected by these factors and exhibits similar characteristics to those of the open waters of the Admiralty Bay.
Three sampling sites were selected: station 1 (E1; 62°13.935′S, 58°39.990′W) located at the inner zone, station 2 (E2, 62°14.011′S, 58°41.443′W) at the outer zone and station 3 (E3; 62°14.064′S, 58° 39.402′W) at the mouth of the Potter Creek (Fig. 1). From spring to summer, the Potter Creek is the main source of run-off waters which are the product of precipitations and glacier-ice melting. Our sampling campaign covered a 15-month period from December 2007 to February 2009. During the spring and summer, when the cove was ice free, samples were taken every 15 days at each sampling station. During autumn and winter, sampling was performed monthly at E1 and E2. E3 station was not sampled in this period because the flow of freshwater from Potter Creek ceased and the shallow marine waters close to the mouth were completely frozen. Surface sea water samples (10 L) were obtained using Niskin bottles manipulated from Zodiac boats. Niskin bottles were sterilized by washing them with 1 N HCl and further rinsed with sterile distilled water. The water samples were successively filtered onto 3 and 0.22 μm cellulose acetate membranes (Millipore, USA). Filters were kept at −80 °C for further processing in the laboratory. Further molecular studies were performed using 0.22-μm membranes, focusing attention on the planktonic cell fraction.
Measurement of environmental parameters
Vertical profiles of temperature and salinity were obtained using a MicroCTD Falmouth FSI, MCTD3 model. The total suspended particulate matter (SPM) was measured by gravimetry. Briefly, we weighed pre-combusted (500 °C, 5 h) Whatman GF/F filters (Whatman International Ltd., Maidstone, UK), which were then used to filter 1 L of water samples. After filtration, filters were rinsed twice with distilled water to remove salts, then dried 24 h at 60 °C and weighed (W SPM). The total SPM was obtained by subtracting the weight of the pre-combusted filter from the weight of the dried SPM filter. Finally, the proportion of organic matter (OM) was obtained by combusting the SPM filters for 5 h at 500 °C and reweighing the filter. The difference between weight of the SPM filter before and after combustion provided the OM value.
To measure the photosynthetic pigments concentration, 1 L of surface water was filtered onto Whatman GF/F filters. Photosynthetic pigments were extracted from filters in 90 % acetone for 24 h at 4 °C in the dark. Chlorophyll-a (Chl-a) concentration was spectrophotometrically determined according to Strickland and Parsons (1972) using a UV–VIS 139 spectrophotometer (Hitachi, Japan). The following equation was applied:
The phaeopigment concentration was calculated as follows (Lorenzen 1967):
where o.d.664a is absorbance at 664 nm, after acidification with 1 N HCl, o.d.664b is absorbance at 664 nm, before acidification with 1 N HCl, v is volume of acetone (mL), l is cuvette length (cm), V is volume of filtered water (L).
Carotenoid concentrations were estimated using equation of Strickland and Parsons (1968):
where o.d.480 is absorbance at 480 nm, v is volume of acetone (ml), l is cuvette length (cm), V is volume of filtered water (L).
DNA extraction, PCR and DGGE
Genomic DNA (gDNA) was extracted from the 0.22-μm membrane filters using the UltraClean soil DNA isolation kit (# 12800-100, MoBIO, USA). A fragment of the 16S rRNA gene was amplified using the archaeal primer set Arc21F (5′-TCCGGTTGATCCYGCCGG-3′) and Arc958R (5′-YCCGGCGTTGAMTCCAATT-3′) where Y = C or T (DeLong 1992). These primers were analysed using BLASTn in order to test the correct annealing with all groups of archaeal 16S rRNA available gene sequences. The reaction mixture (50 μL total volume) consisted of 1× PCR buffer, 2.5 Mm MgCl2, 2 % DMSO, 0.2 mM dNTPs, 0.2 μM of each primer and 2 U of Taq DNA polymerase (Gold Taq, Eurogentec, Belgium). For DGGE purposes, a PCR was performed from the genomic DNA (20–30 ng per reaction) to amplify a fragment of approximately 190 bp from the V3 region of the archaeal 16S rRNA gene using the 344f-GC (modified with a GC clamp at the 5′-end) and 517R primer set as described in Raskin et al. (1994). Reaction and cycling conditions described in Ferrari and Hollibaugh (1999) were used. The quality and the concentration of the PCR products were checked by electrophoresis on 1 % agarose gels in 1× TAE buffer and subsequent staining with ethidium bromide. Size and yield of PCR products were estimated by comparison with a DNA smart ladder (Eurogentec, Belgium).
Molecular fingerprints of the archaeoplanktonic community were generated by denaturing gel gradient electrophoresis (DGGE) using the DGGE PhorU system (Ingeny, Goes, The Netherlands). Optimal band separation was obtained on a 6 % polyacrylamide gel using a 40 to 60 % urea-formamide DNA denaturing gradient. A 100 % urea-formamide was defined as 7 M urea (Bio-Rad, Veenendaal, The Netherlands) and 40 % deionized formamide (Sigma-Aldrich Chemie B.V.) by Muyzer et al. (1993, 2004). For each sample, we loaded 200 ng PCR product supplemented with 1× loading buffer (0.05 % w/v bromophenol blue, 40 % sucrose, 0.1 M EDTA pH 8.0, 0.5 % sodium lauryl sulphate). The DGGE was run for 16 h at 60 °C and 100 V in 0.5× TAE buffer. (TAE buffer is 0.04 M Tris base, 0.02 M sodium acetate and 10 mM EDTA [pH 7.4]). A marker sample was added to each gel as a reference for subsequent band pattern analysis.
Analysis of DGGE profiles
The DGGE band profiles were digitalized and normalized using flanking marker bands with BioNumerics version 3.5 (Applied Maths NV, Belgium) as described in Piquet et al. (2008). Band patterns were translated into both a presence/absence matrix and a relative abundance matrix also using BioNumerics. The relative abundance matrix was used to estimate the Shannon diversity (H), richness (R) and dominance (D). We used the above-mentioned values and environmental variables data (Chl-a, phaeopigments, SPM, OM, IM, salinity and temperature) for multivariate analysis (PCA, Discriminant analysis [DA], MANOVA). MANOVA was tested using Wilks, Pillai, Lawley-Hotelling and Roy statistics. Samples for different seasons were compared using Hotteling test with Bonferoni’s corrected level (InfoStat program, Di Rienzo et al. 2001).
Construction of 16S rDNA clone libraries
Five 16S rDNA clone libraries were constructed as described previously (Piquet et al. 2010). The PCR primers used to amplify the 16S rDNA of archaea were 21F and 958R. Based on the results obtained from the cluster analysis of the DGGE pattern, we decided to pool samples as follows: E1 spring–summer, E1 autumn–winter, E2 spring–summer, E2 autumn–winter and E3 spring–summer. First, we amplified each sample individually, and then equal amounts of all the amplicons corresponding to each library were mixed. Pooled PCR products were cloned in the pGEM-T easy vector system (Promega Benelux B.V.) and transformed in Escherichia coli strain TOP10 according to the manufacturer’s protocol. One hundred colonies with positive insert were randomly selected and amplified by growth of colonies on LB broth. The plasmid was isolated using a HP GenElute Plasmid Miniprep kit (Sigma-Aldrich). Rarefaction curves indicating archaeal 16S rRNA richness were made by using Analytic Rarefaction (Holland 2003).
Sequencing of 16S rDNA clone library and phylogenetic analysis
For each pooled sample, 100 white colonies were selected and amplified from the vector’s T7-SP6 sites; 30 ng of PCR product was cleaned using ExoSAP-IT (Affymetrix, Cleveland, USA) and used as template in the sequencing reaction performed with BigDye® 3.1 Terminator buffers (Applied Biosystems) and 0.2 mM T7 primer. Sequence products were cleaned by standard isopropanol precipitation and analysed on an automated Applied Biosystems 3730xl Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequences were aligned using the online Clustal Omega Multiple Sequence Alignment (http://www.ebi.ac.uk/Tools/services/web_clustalo/). Alignment was manually edited using BioEdit. Identity matrix was calculated from this alignment by using the accessory to BioEdit. The 16S rDNA phylogenetic trees were constructed from 881-nucleotide-length sequences. Estimation of the reliability of phylogenetic reconstructions was made by bootstrap analysis (1,000 replicates) by means of the Mega 5 package using evolutionary distances (Kimura 2 plus G: 0.29) and the neighbour joining and maximum likelihood methods. That is, fewer than 5 % alignment gaps, missing data and ambiguous bases were allowed at any position. There were a total of 620 positions in the final dataset. Operational taxonomic units (OTUs) were ascribed to a taxonomic group based on the closest match obtained with GenBank database using BLAST (version 3.0), which is available at http://www.ncbi.nlm.nih.gov/blast/. OTUs were defined as group of sequences with at least 97 % identity.
Nucleotide sequence accession numbers
The sequences obtained in this study are available in the GenBank database under accession numbers: KF558400 to KF558493 (ArcE2spr–sum), KF558494 to KF558588 (ArcE3spr–sum), KF558589 to KF558684 (ArcE1aut–win), KF558685 to KF558777 (ArcE1spr–sum) and KF558778 to KF558865 (AcrE2aut–win).
Results
Environmental parameters
The average values for the environmental parameters grouped by season for each station are shown in Table 1. Changes in water temperature over time showed similar trends at the three sampling locations. Minimum values were −1.69, −1.40 and −0.29 °C for E1, E2 and E3, respectively. E3 temperatures tended to be higher than the other two stations during the periods when this station could be monitored. The highest temperatures were recorded at sampling stations within the cove. The highest temperature recorded at E1 and E3 were 2.89 and 3.94 °C, respectively, while a highest temperature at E2 was 1.98 °C.
Surface salinities did not show similar trends at the three sampling stations over time. Salinities measured at the inner cove (E1) and near the meltwater stream (E3) were variable and showed a clear decrease during summer. E3 values accompanied the changes observed in E1, reaching values lower than 29.62 psu during summer (2008/2009). In contrast, salinities from E2 showed more stability over time and stayed relatively high throughout the summer (E2 summer average = 33.58 ± 0.63 psu). Chl-a and carotenoids showed similar patterns at E1 and E2 stations, reaching the maximum average values during spring. The maximal Chl-a value measured was 2.3 μg L−1, showing no major phytoplankton biomass build-up. For phaeopigments, maximum values were detected in different sampling times in the three stations.
Finally, SPM showed higher values during summer, when freshwater run-off was maximal, indicating that SPM is mainly of terrestrial origin. The analysis of the SPM composition showed that the SPM was mostly composed of inorganic matter. The organic fraction of SPM was lower (annual average for E1: 18.6 %, E2: 23.6 % and E3: 8.8 %) and constant throughout (results not shown). The inorganic fraction of the SPM was responsible for the cyclic changes. Additional figures showing the values of the environmental parameters all along the sampling period are given as Online Resource 1 to 6.
DGGE analysis of archaeal communities
The DGGE analysis revealed a heterogeneous community with spatial and temporal variability. The richness (total number of DGGE bands per sample) was low (5.04 ± 2.55). Differences in the number and position of the bands were observed even in water samples from the same station taken consecutively. These results demonstrate continuous and fast temporal changes of the archaeal community. Differences between sampling stations at the same time were also evident (Fig. 2). A clear decrease in the number of DNA bands (4.48 ± 2.67) was observed in the spring–summer samples compared with those observed in the autumn–winter samples (6.5 ± 1.50). The features observed from the DGGE gel were confirmed by the analysis of diversity (Table 2). Diversity was low for all stations, with the lowest values observed in E3. In this station, some of the samples showed no archaeal bands at all (Fig. 2). Maximum diversity values were observed for E2 and E1 samples from autumn. At stations E1 and E2, the archaeal diversity showed a diversity increase in autumn–winter and a decrease towards summer. The drop in diversity is explained by the decrease in species richness and the increase in dominance during summer.
Multivariate analysis: PCA, DA and MANOVA
Environmental and biological data were used to conduct multivariate analyses. The PCA showed a separation of the samples into two groups, with the main two components explaining 84.3 % of the variance (Fig. 3). The first component explained 70.9 % of the variance and separated the autumn–winter samples from spring to summer ones. The seasonal grouping of samples was also confirmed by the DA results (Fig. 4). In the PCA, autumn and winter samples were explained by the following variables: salinity, diversity and richness, whereas spring and summer samples were related to temperature, SPM, OM, Chl-a, carotenoids and archaeal dominance. The variable “phaeopigments” explained most of the variability of component 2 and showed the maximum concentration in summer and autumn. It is interesting to note that during autumn and winter, E1 and E2 (the only two stations sampled in these seasons) were closely grouped, while E1 and E3 grouped during summer and spring. These results indicate that E1 is strongly influenced in spring and summer by the freshwater run-off and more by oceanic conditions in winter–autumn. E2 exhibited a different pattern; during summer, it shared more similarity with E1 and E3, while in spring it maintained an intermediate position on the CP1 axis.
First two principal components (PC) from PCA analysis, using archaeal data: richness, diversity (H) and dominance, and environmental data: Salinity, Temperature, Chlorophyll-a (Chl-a), Carotenoides (Carotenoid.), Phaeopigments (Phaeopigm.), suspended particulated matter (SPM), organic matter (OM) and inorganic matter (IM)
Two principal canonical axes from discriminant analysis (DA), using archaeal data: richness, diversity (H) and dominance, and environmental data: Salinity, Temperature, Chlorophyll-a (Chl-a), Carotenoides (Carotenoid.), Feopigments (Feopigm.), suspended particulated matter (SPM), organic matter (OM) and inorganic matter (IM). White diamond summer, grey circle spring, grey square autumn, black circle winter
The same variables considered for PCA were used in a DA that consider only seasons as grouping criteria. Results based on the confidence ellipses obtained confirmed the grouping of the samples in an autumn–winter and a summer–spring clusters (Fig. 4).
Multivariate analysis of variance (MANOVA) used to compare samples obtained in the different seasons gave a high level of significance (p < 0.001). MANOVA showed no significant differences between winter and autumn samples, nor between summer and spring samples. However, samples from autumn–winter differed significantly from the spring–summer (p < 0.05), confirming the statistical significance of the clustering observed with the PCA and DA. The robustness of the previous results that grouped autumn–winter and spring–summer samples led us to consider the pooling of samples based on this temporal-based criterion to perform the clone libraries.
Phylogenetic analysis of Potter Cove environmental sequences
A total of 478 clones from the five independent 16S rRNA gene libraries were obtained and sequenced. Eleven sequences were identified as likely chimeras using the Bellerophon program (Huber et al. 2004) and excluded from further analyses. The phylogenetic analysis of the 467 remaining sequences led to the identification of 92 OTUs (at 97 % of identity level). Rarefaction analyses of collector’s curves, calculated either from the combination of the all clones or from the individual clone libraries at this identity level were near saturation (Online Resource 7). These curves indicated that our sampling effort covered most of the archaeal diversity from Potter Cove.
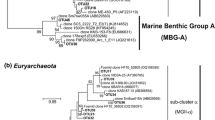
The NJ phylogenetic analysis of our sequence data showed a prevalence of thaumarchaeotal 16S rRNA gene sequences (Fig. 5): Overall, 449 out of 467 sequences fell into a thaumarchaeotal clade, which also contained Nitrosopumilus maritimus and other environmental sequences of ammonia-oxidizing archaea (AOA). For an in-depth analysis of the thaumarchaeotal clade, we generated a phylogenetic tree (Fig. 6) obtained with sequences exclusively belonging to the phylum Thaumarchaeota, whose ribotypes dominated all the Potter Cove libraries constructed and represented 96 % of the total sequences obtained. As can be observed in Fig. 6, the thaumarchaeotal clade contained several clusters, some of them (mixed clusters M1–M7) containing both, Potter Cove sequences and environmental sequences from databank. One of the mixed clusters (M4) contained Nitrosopumilus maritimus sequence. The other thaumarchaeotal clusters only contained Potter Cove thaumarchaeotal sequences and are referred to as nonmixed (NM) clusters. One of those consisted of Potter Cove sequences collected from different sites and on different sampling days, while the NM2 cluster consisted exclusively of sequences isolated from E3 (26 sequences).
The remaining 18 archaeal 16S rRNA gene sequences from this study were not affiliated to the thaumarchaeotal clade and were distributed into five different branches/groups (Fig. 5). The NJ method revealed that sequence ArcE3-48spr–sum grouped with members of the Crenarchaeota AK31 clade. Sequence ArcE3-90spr–sum appeared to be a distant relative of an unclassified environmental sequence obtained from a subsurface microbial community at the saturated zone of the Hanford Site (Lin et al. 2012). ArcE3-13spr–sum clone shared most of its identity with a euryarchaeotal sequence distantly related to Methanocella paludicola. A complementary ML analysis indicated that this sequence was related to Methanospirillum lacunae (AB517986.1), a methane producing archaeon isolated from Japanese soil (Iino et al. 2010).
The remaining 15 sequences seem to represent a novel lineage of Archaea domain that grouped into a not well-supported (NJ method, bootstrap value <50) clade related to the order Thermoplasmatales (Fig. 5). We decided to name this clade “Putative Cold Marine Water Euryarchaeota” (PCMWE). This group of sequences (supported by a bootstrap value of 85) consists of two well-supported clusters (bootstrap values 99 for both clades), PCMWE1 and PCMWE2. Sequences from these groups shared a high identity with several environmental sequences from different cold marine areas. An additional analysis by ML method (Fig. 7), also revealed a very well-supported PCMWE group (bootstrap value 100), which includes two well-supported clades (bootstrap values 100 and 98). Both clades comprise marine water sequences from different cold regions, and a subcluster is composed exclusively by Potter Cove sequences. Using this method, the PCMWE cluster was affiliated as a brother group (bootstrap value of 80) to the order Thermoplasmatales, which contains Ferroplasma cupricumulans, Ferroplasma acidiphilum (a nonthermophilic acidophile genus), Picrophilus oshimae, Thermoplasma acidophilum and Thermogymnomonas acidicola (a moderately thermophilic genus). This result supports the euryarchaeotal affiliation of these 15 Potter Cove archaeal sequences. However, among the type strains, the closest relatives of these organisms from BLASTn analysis had only 78 % identity (data not shown).
Discussion
Twenty years ago, the pioneer work of DeLong (1992) demonstrated the presence and abundance of archaeoplankton in Antarctic marine environments, even in surface waters. Yet, to this day, knowledge about the identification of these archaea and their role in polar ecosystems, where they thrive, remains scarce. In the present study, we try to contribute to the advance on this issue by applying a strong sampling effort over a 15-month period on three stations from Potter Cove, a shallow water coastal marine area in the northern Antarctic Peninsula. In summer, Potter Cove waters are strongly influenced by the input of freshwater from melting glaciers. We therefore included measurements for several physicochemical parameters (salinity, temperature, SPM), in order to follow the strong seasonal oscillations. Multivariate analyses of the DGGE band patterns and the environmental variables significantly separated all samples taken in autumn–winter from the spring–summer samples. Spring and summer represent the period when solar radiation and temperature increase in Antarctica, enabling the onset of primary production. Also in Potter Cove, spring and summer Chl-a values increased, resulting from phytoplankton biomass build-up and greater primary production rates. Yet, during this period, Potter Cove also undergoes a significant increase in the SPM, in particular of inorganic matter originating from freshwater run-off. The freshwater input led to a decrease in salinity in the inner part of the cove, which mainly affected sampling sites E1 and E3. In the same period, we observed a decrease in archaeal diversity, caused by a decrease in richness and an increase in dominance of some archaeal groups that probably grow faster by taking advantage of the higher productivity of the Potter ecosystem in these seasons. The increased dominance took place to the detriment of other archaeal groups with a lower interspecific competition success.
Contrary to this, during the autumn–winter period, the decrease in the incident solar radiation (that reached minimum values in June and July) leads to a decrease in water temperature, resulting in the freezing of the surface waters and stopping the freshwater input. Under these conditions, salinity values rose and SPM dropped, accompanying an increase in archaeal diversity.
The multivariate analyses show the clear grouping of the spring and summer samples from E1 and E3, which was mostly explained by the high SPM and temperature data, as well as low salinities. This strongly indicates that archaeal communities from stations E3 (located at the mouth of the creek) and E1 (located at the inner part of the cove) are strongly influenced by the freshwater input. In autumn–winter, samples from E1 grouped with E2 samples. In this period, stations E1 and E2 exhibit more oceanic characteristics, such as higher salinity and lower SPM. Remarkably, the spring and summer samples from E2 stand aside from the previous groups, which indicates that E2 is less influenced by freshwater input and potentially is a more stable site throughout the year. From these data, we can conclude that Potter Cove is mostly influenced by meltwater effects in spring–summer, while autumn–wintertime conditions are mostly oceanic, and E2 remains “oceanic” throughout most of the year.
Analysis of the sequences from clone libraries revealed a high dominance of members of the phylum Thaumarchaeota, formerly known as Marine Group I Crenarchaeota. A possible bias in the primer annealing as cause of the observed dominance of thaumarchaeotal sequences was discarded based on the BLASTn primer analysis in which all type of archaeal sequences were retrieved. This primer set has also been successfully used by other authors (DeLong et al. 1994; Bano et al. 2004) with no clear dominance of the Thaumarchaeota group. Furthermore, a recent metagenomic dataset obtained from surface sediments of Potter Cove collected close to station 1 sampling site (unpublished data) also revealed a great dominance of Thaumarchaeota among the members of the Archaea domain (JGI 2013). Overall, these data support our finding for Potter Cove archaeoplankton composition. The Thaumarchaeota phylum seems to occupy a broad variety of ecosystems and habitats. Members of the Thaumarchaeota have been recovered from the very warm to the extremely cold environments and also from soils, marine and freshwaters, ice and the body of other organisms. It is considered to be a very diversified and old group of archaea (Pester et al. 2011; Brochier-Armanet et al. 2012).
This group acquired even more ecological relevance when it was reported that its members have the ability to oxidize ammonia (Ingalls et al. 2006). The isolation, characterization and complete genome sequencing of Nitrosopumilus maritimus (Köneke et al. 2005) more clearly allowed us to infer the relevance of the ammonia-oxidation-based chemoautotrophic metabolism. These findings lead to the further proposal of classifying these archaeal microorganisms into a new phylum named Thaumarchaeota (Brochier-Armanet et al. 2008). Based on the characteristics of this group, the dominance of Thaumarchaeota in our clone libraries suggests that it could be playing a relevant role in the nitrogen cycle of Potter Cove. Given the key role played by ammonia-oxidizing bacteria (AOB) in the dynamics of the coastal marine environments (Zehr and Ward 2002), future deep work should be carried out in order to prove whether thaumarchaeotal AOAs inhabiting Potter Cove are playing the role that AOB play in other marine ecosystems. The dominance of AOA among the archaeal microorganisms in Potter Cove water samples was observed at the temporal and spatial scale. Dominance of Thaumarchaeota among archaeal populations from polar marine waters was reported in both Antarctic and Arctic surface waters (Bano et al. 2004; Amano-Sato et al. 2013). However, other authors have reported a significant seasonal oscillation of the archaeal abundance in coastal Antarctic waters, with near absence of archaea during summer season (Kalanetra et al. 2009). Our findings differ from those data. Although we observed a clear decrease in the marine archaeal richness and diversity during summer, we detected the presence of archaea throughout our entire sampling period. Only a few samples collected from the creek station E3 lacked any archaeal fingerprint, suggesting some deleterious effect of freshwater inflow on the community.
The analysis of the phylogenetic tree obtained for the thaumarchaeotal sequences offers several points to highlight. Four of the thaumarchaeotal clusters were exclusively composed of Potter Cove sequences, which were not affiliated with any sequence previously reported from the GeneBank database. Despite the growing number of archaeal and thaumarchaeotal sequences available in the NCBI databases, it appears that several thaumarchaeotal groups inhabiting are Potter Cove waters specific, as they have no known close relatives from other marine environments.
The analysis of the phylogenetic tree revealed that the NM4 cluster (a Potter Cove cluster) not only exhibited a high dominance (62 % of the whole thaumarchaeotal sequences) but also showed that a great proportion of its sequences have 99 % of similarity and probably are the same species. In brief, our work suggests that approximately 59 % of the archaeal sequences recovered from Potter Cove surface waters belong to so far unknown thaumarchaeotal strains. Among the other three Potter-exclusive clusters: NM1, NM2 and NM3, M2 was an E3-exclusive cluster, as it only comprised sequences from the spring–summer E3 library. We speculate that the freshwater flowing from the creek might have been a source for this M2 group, since E3 is strongly influenced by the freshwater input originating from the glacier melting.
A closer inspection of sequences obtained from the database and included within our mixed thaumarchaeotal clusters showed that all had been obtained from Arctic or Antarctic coastal areas (0–50 m depth). Surprisingly, when these sequences originated in temperate or tropical areas, the samples always were deep sea samples. Low temperatures might be the factor conditioning this observation, because temperature represents one of the factors shared by these two, in other ways very dissimilar, marine environments (the surface polar waters and the deep waters from temperate and tropical regions). A similar observation can be made in relation to the euryarchaeal sequences. The cluster that we called “putative cold marine water Euryarchaeota” (Fig. 5) comprised 16 sequences grouped in two mixed sub-clusters. Moreover, phylogenetic analysis showed that both subclusters were composed of two strongly supported groups of sequences: a Potter-exclusive group and a databank-exclusive one (see Fig. 7). Again, the databank sequences closely related to both Potter-exclusive groups of sequences were obtained from cold sea waters. In addition, this “putative cold marine water Euryarchaeota” group, which represents a deep-branching lineage of Archaea domain and that lack any cultivated representatives, showed to be related to the order Thermoplasmatales. The presence of marine Euryarchaeota related to the Thermoplasmatales was reported near 20 years ago for Antarctic waters (DeLong et al. 1994). However, the absence of culturable members of this group prevents any solid inference about their physiological or ecological role in the environment where they come from.
In conclusion, the present work permitted us to confirm that archaeal communities from Potter Cove suffer seasonal oscillations, with higher diversity during winter. However, different from previous reports, our results showed that archaea in Potter Cove stayed detectable, all throughout the year. This fact could be related to the lack of a spring algal biomass build-up, preventing a significant growth of the bacterial cells and then giving archaea the chance to continue growing in spring–summer.
We observed the existence of a clear temporal and spatial dynamics in this community but also a great dominance of a unique thaumarchaeotal species. These two characteristics could be conditioned by the strong variation in the environmental factors present in Potter Cove, determining clear changes between seasons, but also the presence of only a few groups successfully adapted to this changing environment. We cannot discard, though, the influence of some more indirect effects, such as competition with eubacterial species, especially during summer and spring.
The absence of a strong springtime organic matter input in Potter Cove could favour the presence of archaeal species fulfilling the role of AOA. We expect that the oscillations in archaeal community structure as well as the dominance of Thaumarchaeota be shared by other Antarctic coastal marine environments of the West Antarctic Peninsula (WAP) exhibiting low productivity. However, as this study represents, as far as we know, one of the first studies dealing with this subject in the mentioned region, further studies in different shallow water areas of the WAP will be needed in order to corroborate this hypothesis.
References
Amano-Sato C, Akiyama S, Uchida M, Shimada K, Utsumi M (2013) Archaeal distribution and abundance in water masses of the Arctic Ocean, Pacific sector. Aquat Microb Ecol 69:101–112
Bano N, Ruffin S, Ransom B, Hollibaugh JT (2004) Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl Environ Microbiol 70:781–789
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P (2008) Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6:245–252
Brochier-Armanet C, Gribaldo S, Forterre P (2012) Spotlight on the Thaumarchaeota. ISME J 6:227–230
Church MJ, DeLong EF, Ducklow HW, Karner MB, Preston CM, Karl DM (2003) Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol Oceanogr 48:1893–1902
Collins RE, Rocap G, Deming JW (2010) Persistence of bacterial and archaeal communities in sea ice through an Arctic winter. Environ Microbiol 12:1828–1841
Cowie ROM, Masa EW, Ryan KG (2010) Archaeal diversity revealed in Antarctic sea ice. Antarct Sci 23:531–536
Curtosi A, Pelletier E, Vodopivez CL, Mac Cormack WP (2007) Distribution pattern of PAHs in soil and surface marine sediments near Jubany Station (Antarctica). Possible role of permafrost as a low-permeability barrier. Sci Total Environ 383:193–204
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689
DeLong EF, Wu KY, Prezelin BB, Jovine RVM (1994) High abundance of Archaea in Antarctic marine picoplankton. Nature 371:695–697
Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW (2001) InfoStat versión 2011. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. http://www.infostat.com.ar
Ferrari VC, Hollibaugh T (1999) Distribution of microbial assemblages in the central Arctic Ocean basin studied by PCR/DGGE: analysis of a large data set. Hydrobiologia 401:55–68
Fuenets VL, Schnack-Schiel SB, Schloss IR, Esnal GG (2008) Mesozooplankton of Potter Cove: community composition and seasonal distribution in 2002–2003. Rep Pol Mar Res 571:75–84
Fuhrman J, Hagström Å (2008) Bacterial and archaeal community structure and its patterns. In: Kirchman DL (ed) Microbial ecology of the oceans. Wiley, Hoboken, pp 45–90
Galand PE, Lovejoy C, Pouliot J, Vincent WF (2008) Heterogeneous archaeal communities in the particle-rich environment of an arctic shelf ecosystem. J Mar Syst 74:774–782
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF (2006) Pathways of carbon assim-ilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4:520–536, E95. doi:10.1371/journal.pbio.0040095
Herfort L, Schouten S, Abbas B, Veldhui MJW, Coolen MJL, Wuchter C (2007) Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. FEMS Microbiol Ecol 62:242–257
Holland SM (2003) Analytic rarefaction. See http://www.uga.edu/strata/software/
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon, a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Iino T, Mori K, Suzuki K (2010) Methanospirillum lacunae sp. Nov., a methane-producing archaeon isolated from a puddly soil, and emended descriptions of the genus Methanospirillum and Methanospirillum hungatei. Int J Syst Evol Microbiol 60:2563–2566
Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ER, Pearson A (2006) Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Nat Acad Sci USA 103:6442–6447
JGI (Joint Genome Institute) (2013) https://img.jgi.doe.gov/cgi-bin/mer/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300000136
Kalanetra KM, Bano N, Hollibaugh JT (2009) Ammonia-oxidizing Archaea in the Arctic Ocean and Antarctic coastal waters. Environ Microbiol 11:2434–2445
Karner MB, DeLong EF, Karl DM (2001) Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510
Lin X, Kennedy D, Fredrickson J, Bjornstad B, Konopka A (2012) Vertical stratification of subsurface microbial community composition across geological formations at the Hanford Site. Environ Microbiol 14:414–425
Lorenzen CJ (1967) Determination of chlorophyll and phaeopigments: spectrophotometric equations. Limnol Oceanogr 12:343–346
Massana R, Taylor LT, Murray AE, Wu KY, Jeffrey WH, DeLong EF (1998) Vertical distribution and temporal variation of marine planktonic archaea in the Gerlache Strait, Antarctica, during early spring. Limnol Oceanogr 43:607–617
Murray AE, Preston CM, Massana R, Taylor LT, Blakis A, Wu K, DeLong EF (1998) Seasonal and spatial variability of Bacterial and Archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol 64:2585–2595
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rDNA. Appl Environ Microb 59:695–700
Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C (2004) Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Kowalchuk GA, de Bruin FJ, Head IM, Akkermans DL, Van Elsas JD (eds) Molecular microbial ecology manual. Kluwer, Dordrecht, pp 743–770
Pester M, Schleper C, Wagner M (2011) The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14:300–306
Piquet AM-T, Bolhuis H, Davidson AT, Thomson PG, Buma AGJ (2008) Diversity and dynamics of Antarctic marine microbial eukaryotes under manipulated environmental UV radiation. FEMS Microbiol Ecol 66:352–366
Piquet AM-T, Bolhuis H, Davidson AT, Buma AG (2010) Seasonal succession and UV sensitivity of marine bacterioplankton at an Antarctic coastal site. FEMS Microbiol Ecol 73:68–82
Quartino ML, Boraso de Zaixso AL (2008) Summer macroalgal biomass in Potter Cove, South Shetland Islands, Antarctica: its production and flux to the ecosystem. Pol Biol 31:281–294
Raskin L, Stromley JM, Rittmann BE, Stahl DA (1994) Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol 60:1232–1240
Schleper C, Jurgens G, Jonuscheit M (2005) Genomic studies of uncultivated Archaea. Nat Rev Microbiol 3:479–488
Schloss IR, Ferreyra GA (2002) Primary production, Light and vertical mixing in Potter Cove, a shallow bay in the maritime Antractica. Pol Biol 25:41–48
Schloss IR, Ferreyra GA, Ruiz-Pino D (2002) Phytoplankton biomass in Antarctic shelf zones: a conceptual model based on Potter Cove, King George Island. J Mar Syst 36:129–143
Schloss IR, Abele D, Demers S, Bers V, González O, Ferreyra GA (2012) Response of phytoplankton dynamics to 19-year (1991–2009) climate trends in PotterCove (Antarctica). J Mar Syst 92:53–66
Strickland JDH, Parsons TR (1968) A practical handbook of seawater analysis. Pigment analysis, Bull Fish Res Bd, Otawa, Canada, 167, 311 pp
Strickland JDH, Parsons DR (1972) A practical handbook of seawater analysis. J Fish Res Board Can Bull 167:1–310
Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P (2006) Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103:12317–12322
Zehr J, Ward BB (2002) Nitrogen Cycling in the Ocean: new perspectives on processes and paradigms. Appl Environ Microbiol 79:1015–1024
Acknowledgments
This research was carried out under an agreement between the Instituto Antártico Argentino and the Facultad de Farmacia y Bioquímica of the Universidad de Buenos Aires. This work was supported by grants PICTO 2010-0124 from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) and UBA 20020100100378 from Universidad de Buenos Aires. Also we had the financial support from the European Commission through the Marie Curie Action IRSES, project no 318718, IMCONet (Interdisciplinary Modelling of climate change in coastal Western Antarctica—Network for staff Exchange and Training). We thank Gustavo Latorre, Gastón Aguirre and Oscar Gonzalez for their technical assistance and Cecilia Ferreiro for the correction of the English manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
300_2014_1569_MOESM1_ESM.tif
Annual variation of chlorophyll a in Potter Cove. E1 (inner cove), E2 (outer cove), E3 (mouth of Potter Creek) (TIFF 44 kb)
300_2014_1569_MOESM2_ESM.tif
Annual variation of carotenoids in Potter Cove. E1 (inner cove), E2 (outer cove), E3 (mouth of Potter Creek) (TIFF 42 kb)
300_2014_1569_MOESM3_ESM.tif
Annual variation of phaeopigments in Potter Cove. E1 (inner cove), E2 (outer cove), E3 (mouth of Potter Creek) (TIFF 42 kb)
300_2014_1569_MOESM4_ESM.tif
Annual variation of suspended particulate matter (SPM) in Potter Cove. E1 (inner cove), E2 (outer cove), E3 (mouth of Potter Creek) (TIFF 38 kb)
300_2014_1569_MOESM5_ESM.tif
Annual variation of temperature in Potter Cove. E1 (inner cove), E2 (outer cove), E3 (mouth of Potter Creek) (TIFF 35 kb)
300_2014_1569_MOESM7_ESM.tif
Rarefaction curves of the archaeal 16S rRNA sequences from Potter Cove sea waters at 97 % identity. a) Rarefaction curve from 5 combined libraries, b) rarefaction curves from each library. The total number of sequenced clones is plotted against the number of OTUs observed in the same library (TIFF 194 kb)
Rights and permissions
About this article
Cite this article
Hernández, E.A., Piquet, A.MT., Lopez, J.L. et al. Marine archaeal community structure from Potter Cove, Antarctica: high temporal and spatial dominance of the phylum Thaumarchaeota. Polar Biol 38, 117–130 (2015). https://doi.org/10.1007/s00300-014-1569-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-014-1569-8