Abstract
The bivalve Lissarca notorcadensis is one of the most abundant species in Antarctic waters and has colonised the entire Antarctic shelf and Scotia Sea Islands. Its brooding reproduction, low dispersal capabilities and epizoic lifestyle predict limited gene flow between geographically isolated populations. Relationships between specimens from seven regions in the Southern Ocean and outgroups were assessed with nuclear 28S rDNA and mitochondrial cytochrome oxidase subunit I (COI) genes. The 28S dataset indicate that while Lissarca appears to be a monophyletic genus, there is polyphyly between the Limopsidae and Philobryidae. Thirteen CO1 haplotypes were found, mostly unique to the sample regions, and two distinct lineages were distinguished. Specimens from the Weddell and Ross Sea form one lineage while individuals from the banks and islands of the Scotia Sea form the other. Within each lineage, further vicariance was observed forming six regionally isolated groups. Our results provide initial evidence for reproductively isolated populations of L. notorcadensis. The islands of the Scotia Sea appear to act as centres of speciation in the Southern Ocean.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antarctica represents an unrivalled laboratory for undertaking evolutionary studies of the origins and maintenance of biodiversity (Clarke 2000). The Antarctic continent and islands are separated from neighbouring landmasses by sheer distance and in the marine realm the Polar Front acts as a barrier to dispersal. Within Antarctica, marine and terrestrial habitats can be isolated by geological (e.g. distance, fragmented landscapes), physical (e.g. oceanic gyres, ice sheets/shelves, temperature) or ecological (e.g. niche variety, food availability) barriers. Habitat fragmentation can hinder dispersal among populations, limit gene flow and lead to allopatric speciation (e.g. Avise 2004; Frankham et al. 2004; Wagner and Liebherr 1992). The Antarctic fauna provides an excellent opportunity to study colonisation and gene flow between separated habitats, along environmental gradients and over evolutionary time-scales. Previously, the marine and terrestrial Antarctic invertebrate fauna had been considered to be low in species richness but having a high level of endemism (e.g. Boenigk et al. 2006; Clarke and Johnston 2003; Gutt et al. 2000; Maslen and Convey 2006; Stary and Block 1998). Recent studies using molecular genetics have revealed that the species richness in Antarctica is higher than previously suggested (Allcock et al. 1997; Freckman and Virginia 1997; Held and Waegele 2005; Lörz et al. 2006; Pawlowski et al. 2002; Stevens et al. 2006). On land, glacial events are important in structuring the genetic diversity of terrestrial arthropods (Allegrucci et al. 2006; Frati et al. 2001; Stevens and Hogg, 2003, 2006). In the sea, ice-sheet cycles, covering the continental shelf, and separation of water masses are of great consequence in the make-up of marine biodiversity and the radiation of species (Held 2000; Held and Wägele 2005; Page and Linse 2002; Raupach et al. 2004; Raupach and Wägele 2006). Few studies have analysed the intraspecific genetic variability in Antarctic marine invertebrates (Held and Leese 2006; Held and Wägele 2005; Raupach and Wägele 2006) and none so far have focussed on molluscan species.
Since the early days of exploration of the Southern Ocean, molluscs were a major subject of scientific investigations (Dell 1990; Hain 1990; Powell 1951). Overall the bivalve and gastropod fauna of Antarctic waters is probably described more completely than any other group of benthic marine invertebrates (Clarke and Johnston 2003). Current biogeographic results using SOMBASE, the Southern Ocean Mollusc Database (Griffiths et al. 2003), showed that the Scotia-Arc and Peninsula region appears to be a hot spot of species richness (Linse et al. 2006). In order to address fundamental evolutionary questions pertaining to the origins and diversification of Antarctic marine fauna (such as why the Scotia Arc region is so species-rich, where the origin of Antarctic species is, how the isolation of water masses supports radiation and what are the roles of islands and shallow shelves as possible stepping stones of colonisation and radiation in Antarctica?), we queried SOMBASE for a suitable taxon/species-complex. The ideal target species or group had to have a circum-Antarctic and sub-Antarctic distribution. This might give the possibility to link the evolutionary history of the species with plate tectonics (Barker and Burrell 1977; Lawver and Gahagan 2003) and oceanographic current systems such as the north-bound currents along the western Antarctic Peninsula towards the Scotia Arc islands (Hofmann et al. 1998), its counter current in the Bransfield Strait (Stein and Heywood 1994; Whitworth et al. 1994), and water mass circulation in the Weddell Sea (Fahrbach et al. 1992, 1994; Orsi et al. 1993).
Among the species found to be suitable for this study the small philobryid bivalve Lissarca notorcadensis Melvill and Standen 1907 stood out because of its wide distributional range (Dell 1990; Hain 1990), reproductive mode (Prezant et al. 1992), ecological success (Brey and Hain 1992; Brey et al. 1993) and regional morphological differences (Cope and Linse 2006). The species is attached by byssus threads to the spines of cidaroid sea urchins reaching densities of 350 individuals/cidaroid (Brey and Clarke 1993; Brey et al. 1993; Hain 1990). Morphometric population studies on L. notorcadensis indicated that large-shelled adults live on the top end of the spines while the juveniles live on the more protected bases of the spines and lead to the hypothesis that parents and offspring live on the same cidaroid (Brey et al. 1993). Following this hypothesis gene flow within and between populations would be limited and genetic differences should be detectable with molecular techniques.
The aim of this study is to test three hypotheses using L. notorcadensis as the model species: (1) The current species richness of bivalves in the Antarctic and sub-Antarctic regions is higher than suggested because of existence of cryptic species, (2) The isolated Scotia Arc Islands have acted as centres for speciation in the Scotia Sea region, and (3) the Weddell Sea Gyre isolates Weddell Sea populations from populations of the western Antarctic Peninsula and Scotia Arc Islands.
Materials and methods
Specimen collection
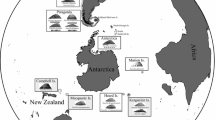
Individuals of L. notorcadensis were collected during research expeditions of RV “Polarstern” to the Scotia Sea (ANT XIX 3–5: ANDEEP I/II & LAMPOS), to the eastern Weddell Sea (ANT XXI-2: BENDEX) and RV “Tangaroa” to the Ross Sea (TAN0402/BIOROSS 2004) (Fig. 1). The sampling included Shag Rocks (SR), Saunders and Southern Thule of the South Sandwich Islands (SSI), Herdman Bank (HB), Discovery Bank (DB), South Orkney Islands (SO), Elephant Island (EI), the eastern Weddell Sea (WS) and the northeastern Ross Sea (RS). When the samples reached the deck, invertebrates were sorted by hand with focus on L. notorcadensis. Specimens were immediately fixed in pre-cooled 96% ethanol, kept at –20°C for at least 48 h and then refrigerated for later molecular analysis. Additionally specimens of Lissarca miliaris (Philippi 1845) were collected in the intertidal of Signy Island/South Orkney islands.
Distribution records and sample sites of L. notorcadensis. White circles sample sites, black circles distribution records, black line Polar Front DB Discovery Bank, EI Elephant Island, HB Herdman Bank, SG South Georgia, SO South Orkney Islands, SR Shag Rocks, SSI South Sandwich Islands (Figure modified from Cope and Linse 2006)
DNA extraction, PCR amplification and DNA sequencing
Genomic DNA was isolated from the entire specimen in small individuals. DNA was extracted with the DNeasy Tissue Extraction Kit (Qiagen, Crawley, West Sussex, United Kingdom) as directed by the manufacturer.
Partial 28S rDNA (domain 2) was amplified using the primers LSU 5 and LSU 3 (Littlewood 1994). The fragment contains the domans 1–3. PCR amplification was performed in a 50 μl reaction volume, containing the following: 50–100 ng of template DNA, 200 μM of each dNTP, 0.5 μM of each primer, 10 × Qiagen PCR Buffer and 5 × Q-solution. Thermal cycling began with a single denaturation step at 94°C for 5 min. Then 40 cycles were performed consisting of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 60 s. A single extension step at 72°C for 7 min was used to complete extension of DNA fragments.
A fragment of the coding COI region was amplified using the primers HCO 1490 and LCO 2198 (Folmer et al. 1994). Thermal cycle conditions were 95°C for 4 min and then 5 cycles of 94°C for 60 s, 45°C for 90 s, and 72°C for 9 s followed by 35 cycles of 94°C for 60 s, 52°C for 60 s, and 72°C for 60 s and finished with 72°C for 5 min.
PCR products were purified with QIAquick Spin Columns (Qiagen). Approximately 200 ng of double stranded PCR product was used in cycle sequencing reactions following the protocol outlined in the DYEnamic ET Dye Terminator Cycle Sequencing kit for MegaBACE DNA (Amersham Biosciences, Little Chalfont, Buckinghamshire, United Kingdom). Unincorporated dyes were removed from sequencing reaction products by using DyeEX 96 Kit (Qiagen) following the manufacturer’s instructions. Reaction products were visualised on a MegaBACE 500 automated DNA sequencer (Amersham Pharmacia, Little Chalfont, Buckinghamshire, United Kingdom).
The sequences were aligned and proofread in CodonCode Aligner Version 1.5.2 (CodonCode Coporation 2006). Additional 28S and COI sequences from GenBank were added as outgroups to the aligned dataset, selected from the sister-taxon Cosa (Philobryidae) and the related families Limopsidae (Empleconia and Limopsis) and Arcidae (Barbatia and Scapharca) (Table 1).
Ambiguous alignment positions associated with gaps in the 28S sequences were omitted from the dataset.
Phylogenetic analysis
Phylogenetic reconstruction was conducted using maximum parsimony (MP) optimality criterion and maximum likelihood (ML) in PAUP*b10 (Swofford 2002). A heuristic search was conducted with maximum trees held set to 1,000, 50 random additions, tree bisection reconnection (TBR) branch swapping, with five trees held at each step and with “MulTrees” turned off. One thousand bootstrap pseudoreplicates were conducted to assess the reliability of the reconstructed nodes. We used an iterative optimisation approach to find an appropriate substitution model for ML analyses: the data were used to construct a neighbour joining tree in PAUP*b10. From this tree, parameters were estimated and used for likelihood analysis (heuristic search, one random addition, TBR). Parameters were re-estimated and the process repeated until the likelihood scores converged. The tree topology was assessed by a likelihood search with 500 bootstrap pseudo-replicates. Bayesian phylogenetic reconstruction was conducted using Mr Bayes v3.1.2 (Ronquist and Huelsenbeck 2003, 2005). A six parameter model was used with a gamma distribution and proportion of invariant sites (GTR + I + Γ) with flat priors. 50,000,000 generations with four chains were run, sampling every 50th generation.
Pair-wise distances were calculated in MEGA v 3 (Kumar et al. 2004). The evolutionary relationships among COI haplotypes were reconstructed using statistical parsimony in TCS v1.21 (Clement et al. 2000). In order to explore protein evolution, sequences were translated into amino acids and phylogenies reconstructed in MEGA.
Results
Partial sequences of the 28S rDNA and COI genes were generated to examine the phylogenetic relationships and population structure of L. notorcadensis. For the first 28S rDNA gene segment we determined sequences of 36 L. notorcadensis and one L. miliaris and added Genbank sequences of one phylobryid, four limopsids and one arcid as outgroup taxa. The aligned 28S rDNA dataset comprised 806 characters of which 106 were variable and 55 were parsimony informative. Within L. notorcadensis only 16 characters varied. Average base frequencies were pi(A)21.5%, pi(C) = 24.3%, pi(G) = 33.2% and pi(T) = 20.9%.
Within Lissarca there was not enough resolution to identify any structure using ML, MP or Bayesian analyses. However, between Lissarca and the included phylobryids and limopsids, the three analysis techniques yielded congruent results.
The ML Trees were rooted with the arcid Barbatia to reveal the phylogenetic position of Lissarca within the Limopsidae and Phylobryidae (Fig. 2). The topology of the tree shows that neither of the sister families is monophyletic but supports Lissarca as a monophyletic taxon.
For the COI segment we obtained sequences of 27 L. notorcadensis and added Genbank sequences of one phylobryid, one limopsid and two arcids as outgroup taxa. The length of the aligned COI dataset consisted of 337 nucleotides of which 169 were variable and 93 were parsimony informative. Within L. notorcadensis, the COI fragment revealed 27 variable sites and thirteen haplotypes forming six haplotype groups (Table 1). Of the 27 detected nucleotide substitutions, 21 were third codon, 5 second codon and 1 first codon. The unusual ratio, due to a comparatively high number of second codon changes, prompted us to investigate the geographic associations of second position variation. Of the five second position changes, three of them separate WS and RS from the SA. Within the Weddell Sea/Ross Sea group there were ten variable sites, all third positions. Within the Scotia arc group there were nine variable sites; one first position, two second position and six third position changes. The second position changes form two groups; ESA and SO form one group while SR and EI form a second.
Tree topologies from MP, ML and Bayesian analyses were congruent and revealed two major, highly supported lineages in L. notorcadensis; lineage A comprising individuals from the banks and islands of the Scotia Sea, and lineage B individuals from the Weddell and Ross Seas (Fig. 3). Within Lineage A, four well supported haplotype groups (HG) were identified: HG1 includes individuals from around the South Sandwich Islands, Discovery Bank and Herdman Bank. HG2 contains a single haplotype from South Orkneys, HG3 contains a single haplotype from Elephant Island, and HG4 contains two haplotypes from Shag Rocks near South Georgia. Lineage B consists of two strongly supported haplotype groups, Weddell Sea and Ross Sea with three haplotypes each. Pair-wise genetic differences highlight within-lineage variabilities (Fig. 4). The East Scotia Arc (ESA) group is clearly different from the Elephant Island and Shag Rock groups. The statistical parsimony networks in Fig. 5 graphically display the genetic and geographic associations of haplotypes (Fig. 5).
Molecular phylogeny of single gene analysis of the COI mtDNA gene for L. notorcadensis. Tree was produced by ML analysis. ML and MP support values are bootstrap values of 1,000 replicates, MP values are in bold, ML values are in italics. Bayesian support values (posterior probabilities) are underlined. A lineage A, B lineage B, HG haplotype group
Discussion
The work presented here has detected population structure within L. notorcadensis and explored unresolved issues in the systematics of the Philobryidae and Limopsidae. Based on shell and soft part morphology, Tevez (1977) suggested the ancestry of the Philobryidae to be traced to the Limopsidae. On a molecular level, the relationships between these two families have not been adequately studied. Matsumoto (2003), in his phylogeny of the Pteriomorpha using the CO1 gene, cited Cosa waikikia (Dall, Bartsch and Rehder 1939) and the limopsid Empleconia cumingii (Adams 1863) as examined species in his dataset but only the latter was used in his analysis. The Matsumoto 28S and CO1 sequences for Cosa and Empleconia were included in our analyses. As the position of the philobryid Cosa within the Limopsidae questions the monophyly of these families, only further analyses including more species of the philobryid genera, like Adacnarca, Philobrya and Cratis, will resolve this question.
Our primary goal was to establish a model to address the three a priori hypotheses relating to (1) the underestimation of biodiversity in Antarctic and sub-Antarctic Mollusca, (2) the Scotia Arc is a source of biodiversity, and (3). the Weddell Sea gyre isolates Weddell Sea populations from those of the Scotia Sea.
The 28S data set was not informative enough to confidently distinguish geographic differences, the CO1 results clearly identified two deeply diverged lineages separating continental Antarctic from Scotia Sea samples, and six geographically explicit haplotype groups indicating discrete populations, one each in the Weddell and Ross Seas, and four in the Scotia Sea. These lineages are also supported by phenotypic data. Cope and Linse (2006) found significant difference in the shell length/height ratios between Scotia Sea specimens and Weddell/Ross Sea specimens. Whether or not the populations from the Weddell Sea and Ross Sea are typical of a linked circum-Antarctic distribution is not tested here. Even so, the Antarctic Peninsula appears to be associated with a barrier for the Antarctic lineage sampled, so the samples we examined from the Weddell Sea and the Ross Sea are likely to be at the extremes of the range of this population and thus should reflect the greatest genetic distance. However, the variation within the Scotia Sea lineage exceeds that within the continental Antarctic Lineage (Fig. 4). A study on the isopod Glyptonotus antarcticus Eights, 1852 found a strikingly similar pattern regarding the genetic versus the geographic distances between Ross Sea, Weddell Sea and Antarctic Peninsula populations (Held and Wägele 2005). Using mitochondrial 16S sequences from specimens collected in the eastern Weddell Sea, at Elephant Island, Adelaide Island/Antarctic Peninsula and in the Ross Sea, Held and Wägele (2005) found 11 unique haplotypes forming four haplotype groups. One group contained the haplotypes from Elephant Island and Adelaide Island and another the Ross Sea individuals while two haplotype groups occurred in the Weddell Sea. The pair-wise genetic differences between the Ross Sea group and the Weddell Sea groups were smaller than those between the former groups and the Elephant/Adelaide Island group. As suggested by Held and Wägele (2005) in the case of the isopod, we find it likely that the continental Antarctic and Scotia Sea populations may in fact denote cryptic species and supports the premise of the first hypothesis.
Within the Scotia Sea lineage genetic variation is congruent with the general morphological differences found by Cope and Linse (2006) between Shag Rock and other Scotia Sea specimens. The depth of the divergence between these groups lends support for the second hypothesis. The diversity present among different island groups of the Scotia Arc may be strongly affected by deep water separating habitats and preventing migration. This was found to be the case with the octopod Pareledone turqueti where deep water between South Georgia and Shag Rocks presented a barrier to geneflow resulting in profound population structure (Allcock et al. 2007). As L. notorcadensis is a brooding species and restricted to shelf upper slope waters (Cope and Linse 2006), the deep water is likely to prevent or substantially restrict migration between shallow water habitats. It is interesting to note the pattern of divergence, with Elephant Island specimens more similar to Shag Rock than South Orkney and East Scotia Arc specimens despite greater geographic distance of deep waters separating Elephant Island and Shag Rock relative to that between South Orkney. To further investigate the patterns, processes and depth of divergence between these putative populations, a more intense geographical sampling regime is required together with a multilocus genetic analysis approach.
Our third hypothesis stated that the Weddell Sea gyre partially acted as a barrier to geneflow between Eastern Weddell Sea populations and those of the Antarctic Peninsula and the Scotia arc. Using mitochondrial 16S sequence data Raupach and Wägele (2006) found strong differentiation between Eastern and Western Weddell Sea samples in the isopod Acanthaspidia drygalski Vanhöffen, 1914. This pattern is similar to that of the isopod Glyptonotus antarcticus mentioned above (Held and Wägele 2005) which does indeed indicate restricted geneflow between the east and west Weddell Sea. Interestingly, divergence between groups within the Weddell Sea was greater than that between the Weddell Sea and the Ross Sea. Our results clearly indicate that Weddell Sea samples are genetically distinct from those of the Scotia arc. However, as there is a similar difference between the Ross Sea and Scotia arc we find no specific evidence supporting the premise that the Weddell Sea gyre is the sole mechanism preventing geneflow.
An interesting question relates to the genetic differences between haplotypes from the geographically well-separated Weddell and Ross Seas that are smaller than those between the neighbouring and connected Weddell and Scotia Seas in our study and in that of Held and Wägele (2005). Not only molecular data show close relations between the Weddell and the Ross Seas. The most recent biogeographic analysis of bivalves and shelled gastropods at various systematic levels showed that the relationships between these two seas are closer than either ‘s relationship to the Scotia Sea islands (Linse et al. 2006). The processes behind these patterns are yet unknown. Possible explanations lie either in the past of the Antarctic continent or in its more recent oceanography. A linking seaway between the Weddell and Ross Seas may have been present during the Eocene to mid-Miocene (50–15 Ma BP), connecting the two and separating them from the West Antarctic, microcontinental fragments that later on formed the Antarctic Peninsula and Scotia arc islands (Lawver and Gahagan 2003). Scherer et al. (1998) proposed a Pleistocene collapse of the West Antarctic Ice Sheet providing seaways between the Weddell and Ross Seas which may have facilitated geneflow between these populations. The present large-scale features in Antarctic oceanography are the clockwise Antarctic circumpolar current (ACC) system, separating the warmer water masses north of the Polar Front from the colder Southern Ocean, the continental, anti-clockwise counter current and the Weddell Gyre (Fahrbach et al. 1994; Orsi et al. 1993, 1995). The continental counter current used as a migration tool could explain the lower genetic variability between the Weddell and Ross Seas under the assumption that related haplotypes occur along the East Antarctic coastline. The strong flowing ACC, initiated in the Early Oligocene (30 Ma BP) (Lawver and Gahagan 2003), is likely to be important for the closer haplotype relationships within the Scotia Sea.
In this study we have identified strong genetic structure in the brooding bivalve L. notorcadensis between Antarctic and sub-Antarctic groups, and within the sub-Antarctic specimens sampled throughout the Scotia arc. We have found evidence for cryptic species and it may be that more detailed phylogeographic studies with intensive sampling and multiple single copy loci could uncover the true nature of the genetic structure and identify the processes underlying the patterns.
References
Allcock AL, Breirley AS, Thorpe JP, Rodhouse PG (1997) Restricted geneflow and evoltuionary divergence between geographically separated populations of the Antarctic octopus Pareledone turqueti. Mar Biol 129:97–102
Allcock AL, Strugnell JM, Prodoehl P, Piatkowski U, Vecchione M (2007) A new species of Pareledone (Cephalopoda: Octopodidae) from the Antarctic Peninsula. Polar Biol doi:10.1007/s00300-006-0248-9
Allegrucci G, Carchini G, Todisco V, Convey P, Sbordoni V (2006) A molecular phylogeny of Antarctic chironomidae and its implications for biogeographical history. Polar Biol 29:320–326
Avise JC (2004) Molecular markers. Sinauer, Sunderland, p 541
Barker PF, Burrell J (1977) The opening of the Drake passage. Mar Geol 25:15–34
Boenigk J, Pfandl K, Garstecki T, Harms H, Novarino G, Chatzinotas A (2006) Evidence for geographic isolation and signs of endemism within a protistan morphospecies. Appl Environ Microbiol 72(8):5159–5164
Brey T, Hain S (1992) Growth, reproduction and production of Lissarca notorcadensis (Bivalvia, Philobryidae) in the Weddell Sea, Antarctica. Mar Ecol Prog Ser 82:219–226
Brey T, Clarke A (1993) Population dynamics of marine benthic invertebrates in Antarctic and subantarctic enviroments: are there unique adaptaions? Antarct Sci 5(3):253–266
Brey T, Starmans A, Magiera U, Hain S (1993) Lissarca notorcadensis (Bivalvia: Philobryidae) living on I sp. (Echinoidea:Cidaridae):population dynamics in limited space. Polar Biol 13:89–95
Clarke A (2000) Evolution in the cold. Antarct Sci 112:257
Clarke A, Johnston NM (2003) Antarctic marine benthic diversity. Oceanogr Mar Biol 41:47–114
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9(10):1657–1660
Cope T, Linse K (2006) Morphological differences in Lissarca notorcadenis Melvill and Standen, 1907 from the Scotia, Weddell and Ross Seas. Deep-Sea Res II 53:903–911
Dell RK (1990) Antarctic Mollusca: with special reference to the fauna of the Ross Sea. Bull Roy Soc NZ 27:1–311
Fahrbach E, Rohardt G, Krause G (1992) The Antarctic Coastal Current in the southeastern Weddell Sea. Polar Biol 12:171–182
Fahrbach E, Rohardt G, Schroeder M, Strass V (1994) Transport and structure of the Weddell Gyre. Annales Geophysicae 12:840–855
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3:294–299
Frankham R, Ballou JD, Briscoe DA (2004) A Primer of conservation genetics. University Press, Cambridge pp 234
Frati F, Spinsant G, Dallai R (2001) Genetic variation of mtCOII gene sequences in the collembolan Isotoma klovstadi from Victoria Land, Antarctica: evidence of population differentiation. Polar Biol 12:934–940
Freckman DW, Virginia RA (1997) Low-diversity Antarctic soil nematode communities: distribution and response to disturbance. Ecology 78(2):363–369
Griffiths HJ, Linse K, Crame JA (2003) SOMBASE—Southern Ocean Mollusc Database: a tool for biogeographic analysis in diversity and ecology. Org Divers Evol 3(3):207–213
Gutt J, Sirenko BI, Arntz WE, Smirnov IS, De Broyer C (2000) Biodiversity of the Weddell Sea: macrozoobenthic species (demrsal fish included) sampled during the expedition ANT XIII/3 (EASIZ I) with RV ‘Polarstern’. Ber Polarforsch 372
Hain S (1990) Die beschalten benthischen Mollusken (Gastropoda und Bivalvia) des Weddellmeeres, Antarktis. Berichte zur Polarforschung 70:1–181
Held C (2000) Phylogeny and biogeography of serolid isopods (Crustacea, Isdopoda, Serolidae) and the use of ribosomal expension segments in molecular systematics. Mol Phylogenet Evol 15(2):165–178
Held C, Wägele JW (2005) Cryptic speciation in the giant Antarctic isopod Glyptonotus antarcticus (Isopoda: Valvifera: Chaertiliidae). Scientia Marina 69(2):175–181
Held C, Leese F (2006) The utility of fast evolving molecular markers for studying speciation in the Antarctic benthos. Polar Biol 30:513–521
Hofmann EE, Klinck JM, Locarnini RA, Fach B, Murphy E (1998) Krill transport in the Scotia Sea and environs. Antarct Sci 10:406–415
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lawver LA, Gahagan LM (2003) Evolution of Cenozoic seaways in the circum-Antarctic region. Paleogeogr Paleoclimatol Paleoecol 198(1–2):11–37
Linse K, Griffiths HJ, Barnes DKA, Clarke A (2006) Biodiversuty and biogeography of Antarctic and sub-Antarctic Mollusca. Deep-Sea Res II 53:985–1008
Littlewood DTJ (1994) Molecular phylogenetics of cupped oysters based on partial 28S rDNA gene sequences. Mol Phylogenet Evol 3:221–229
Lörz AN, Maas EW, Linse K, Fenwick GD (2007) Epimeria schiaparelli sp. nov., an amphipod crustacean (family Epimeriidae) from the Ross Sea, Antarctica, with molecular characterisation of the species complex. Zootaxa 1402:23–37
Maslen NR, Convey P (2006) Nematode diversity and distribution in the southern maritime Antarctic—clues to history? Soil Biol Biochem 38(10):3141–3151
Matsumoto M (2003) Phylogenetic analysis of the subclass. Pteriomorphia (Bivalvia) from mtDNA COI sequences. Mol Phylogenet Evol27(33):429–440
Orsi AH, Nowlin WD, Whitworth T (1993) On the circulation and stratification of the Weddell Gyre. Deep-Sea Res 40:169–203
Orsi AH, Whitworth T, Nowlin WD (1995) On the meridional extent and fronts of the Antarctic Circumpolar Current. Deep-Sea Res Part I-Oceanogr Res Pap 42(5):641–673
Page TJ, Linse K (2002) More evidence of speciation and dispersal across the Antarctic Polar Front through molecular systematics of Southern Ocean Limatula (Bivalvia: Limidae). Polar Biol 25:818–826
Pawlowski J, Fahrni JF, Brykczynska U, Habura A, Bowser SS (2002) Molecular data reveal high taxonomic diversity of allogromiid Foraminifera in Explorers Cove (McMurdo Sound, Antarctica). Polar Biol 25(2):96–105
Powell AWP (1951) Antarctic and sub-Antarctic Mollusca: Pelecypoda and Gastropoda. Discov Rep 26:49–196
Prezant RS, Showers M, Winstead RL, Cleveland C (1992) Reproductive ecology of the Antarctic bivalve Lissarca notorcadensis (Philobryidae). Am Malacol Bull 9(2):173–186
Raupach MJ, Wägele JW (2006) Distinguishing cryptic species in Antarctic Asellota (Crustacea: Isopoda)—a preliminary study of mitochondrial DNA in Acanthaspidia drygalskii. Antarct Sci 18:191–198
Raupach MJ, Held C, Wägele JW (2004) Multiple colonization of the deep sea by the Asellota (Crustacea: Peracarida: Isopoda). Deep-SEa Res II 51:1787–1795
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinform 19:1572–1574
Ronquist F, Huelsenbeck JP (2005) Bayesian analysis of molecular evolution using MrBayes. In: Nielsen R (eds) Statistical methods in molecular evolution. Springer, New York
Scherer RP, Aldahan A, Tulaczyk S, Possnert G, Engelhardt H, Kamb B (1998) Pleistocene collapse of the West Antarctic Ice sheet. Science 281:82–85
Stary J, Block W (1998) Distribution and biogeography of oribatid mites (Acari: Oribatida) in Antarctica, the sub-Antarctic islands and nearby land areas. J Nat Hist 32(6):861–894
Stein M, Heywood RB (1994) Antarctic environment—physical oceanography: the Antarctic Peninsula and Southwest Atlantic region of the Southern Ocean. In: El-Sayed SZ (eds) Southern Ocean ecology. CPU, Cambridge pp 11–24
Stevens MI, Hogg ID (2003) Long-term isolation and recent range expansion revealed for the endemic springtail Gomphiocephalus hodgsoni from southern Victoria Land, Antarctica. Mol Ecol 12:2357–2369
Stevens MI, Hogg ID (2006) Contrasting levels of mitochondrial DNA variability between mites (Penthalodidae) and springtails (Hypogastruridae) from the Trans-Antarctic Mountains suggest long-term effects of glaciation an dlife history on substitution rates, and speciation processes. Soil Biol Biochem 38:3171–3180
Stevens MI, Greenslade P, Hogg ID, Sunnucks P (2006) Southern hemisphere springtails: could any have survived glaciation of Antarctica? Mol Biol Evol 23:874–882
Swofford D (2002) Paup*. Phylogenetic Analysis using Parsimony (*and other methods). (Version 4.10beta). Sinauer Associates, Sunderland, Massachusetts
Tevesz MJS (1977) Taxonomy and ecology of the Philobryidae and Limopsidae (Mollusca: Pelecypoda). Postilla 171:1–64
Wagner DL, Liebherr JK (1992) Flightlessnes in insects. Trends Ecol Evol 7:216–220
Whitworth T, Nowlin WD, Orsi AH, Locarnini RA, Smith SG (1994) Weddell Sea Shelf Water in the Bransfield Strait and Weddell-Scotia Confluence. Deep-Sea Res Part I-Oceanogr Res Pap 41(4):629–641
Acknowledgment
We are grateful to the cruise leaders, captains, officers and crews of PFS Polarstern (ANT XIX-4, ANT XIX-5 and ANT XXI-2) and of RV Tangaroa (TAN0402) who enabled us to collect the samples for this study. Thanks are due to S. Lockhardt for access to ANDEEP I cidaroid sea urchins hosting Lissarca and to H. Griffiths for providing the map. NERC (NER/M/S/2003/00102) funded the core research programme. The FRST Program CO1X0502 supported the work on the Ross Sea samples held by the NIWA Marine Invertebrate Collection. This paper is a contribution to British Antarctic Survey core project ‘BIOPEARL’, ANDEEP publication no 71 and linked with the SCAR ‘EBA’ programme.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Linse, K., Cope, T., Lörz, AN. et al. Is the Scotia Sea a centre of Antarctic marine diversification? Some evidence of cryptic speciation in the circum-Antarctic bivalve Lissarca notorcadensis (Arcoidea: Philobryidae). Polar Biol 30, 1059–1068 (2007). https://doi.org/10.1007/s00300-007-0265-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-007-0265-3