Abstract
Key message
Recent updates in JA biosynthesis, signaling pathways and the crosstalk between JA and others phytohormones in relation with plant responses to different stresses.
Abstract
In plants, the roles of phytohormone jasmonic acid (JA), amino acid conjugate (e.g., JA-Ile) and their derivative emerged in last decades as crucial signaling compounds implicated in stress defense and development in plants. JA has raised a great interest, and the number of researches on JA has increased rapidly highlighting the importance of this phytohormone in plant life. First, JA was considered as a stress hormone implicated in plant response to biotic stress (pathogens and herbivores) which confers resistance to biotrophic and hemibiotrophic pathogens contrarily to salicylic acid (SA) which is implicated in plant response to necrotrophic pathogens. JA is also implicated in plant responses to abiotic stress (such as soil salinity, wounding and UV). Moreover, some researchers have recently revealed that JA controls several physiological processes like root growth, growth of reproductive organs and, finally, plant senescence. JA is also involved in the biosynthesis of various metabolites (e.g., phytoalexins and terpenoids). In plants, JA signaling pathways are well studied in few plants essentially Arabidopsis thaliana, Nicotiana benthamiana, and Oryza sativa L. confirming the crucial role of this hormone in plants. In this review, we highlight the last foundlings about JA biosynthesis, JA signaling pathways and its implication in plant maturation and response to environmental constraints.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are threatened by several biotic and abiotic stresses that endanger their life. As sessile beings, evolution has helped plants to develop many mechanisms and behaviors to cope with predators and unfavorable life conditions. Perception of threat by cellular receptors is transduced into signals within seconds. Those signals are called second messengers such as Ca2+, reactive oxygen species (ROS), hydrogen sulfide (H2S), inositol trisphosphate (IP3), nitrogen oxide (NO) which interacts with several proteins to enhance plant response to a given stress (Agurla et al. 2018). ROS-Ca2+ hub have been extensively studied in plants in response to many environmental constraints and controls lots of mechanisms such as osmotic adjustment, hormonal signal transduction (HST), osmoregulation, mineral nutrition, cell elongation and development as well as programmed cell death (Demidchik and Shabala 2017). It has been suggested that ROS-Ca2+ hub/phytohormone signaling simultaneously activates the plant stress signaling cascades (Sewelam et al. 2016). Moreover, ROS, as the main source of signaling in plants under unfavorable conditions, interacts with other second messengers and signaling components such as Mitogen Activated protein Kinases (MAPKs), plant hormones, and transcription factors (Sewelam et al. 2016). In another hand, plant growth, development, and survival are strictly interconnected in a complex biological network that is disturbed by environmental conditions, such as soil condition and salinity, extreme temperatures, and pathogen attacks (Altaf-Ul-Amin et al. 2015). Hence, a relevant number of intermediate compounds, such as ROS and their derivatives, might accumulate in the cytoplasm and disturb signaling pathways especially when they are converted into toxic compounds and affect cell survival (Ayala et al. 2014). Phytohormones which are involved in ROS-Ca2+ mediated signals include abscisic acid (ABA), cytokinins (CKs), auxins (IAA), gibberellins (GA), ethylene (ET), salicylic acid (SA), brassinosteroids (BRs), jasmonic acid (JA) and strigolactones (Pottosin et al. 2014; Deng et al. 2018). All those hormones have different effect and can act with an antagonist or synergistic action in response to a stressor.
Among those phytohormones, JA and its derivatives, such as jasmonyl isoleucine (JA-Ile), methyl jasmonate (MeJA), 12-hydroxyjasmonic acid sulfate (12-HSO4-JA), cis-jasmone, JA-glucosyl ester, JA-Ile methyl ester, jasmonoyl-amino acid, 12-carboxy-JA-IIe, 12-O-glucosyl-JA-IIe, 12-O-glucosyl-JA, JA-Ile glucosyl ester, are known as jasmonates (JAs). JAs are lipid-derived signaling molecules that control many developmental processes. They are implicated in plant response to biotic and abiotic stress and they are detected in almost all plants (Sun et al. 2011). JAs are fatty acids that belong to oxidized lipids family. Those molecules are synthesized from cyclo-pentanones and known as oxylipins (Wasternack and Feussner 2018). The oxylipins signaling molecules are biologically active components produced either through auto-oxidation of poly-unsaturated fatty acids or by the activity of lipoxygenases enzymes or alpha-dioxygenases enzymes (Göbel and Feussner 2009). JAs regulates many crucial biological processes in planta, including stamen and trichome development, stomatal opening, leaf expansion, hook formation, cell cycle regulation, and glucose transport (Wasternack and Hause 2007; Yoshida et al. 2009).
In this review, we intend to describe the most recent updates in JA biosynthesis and signaling pathways, and we focus on the crosstalk between JA and others phytohormones to understand the mechanism of plant responses different stresses.
JA metabolism
JAs are ubiquitous molecules detected in higher plant species (Yang et al. 2019). The endogen level of JA in plants is higher in reproductive organs as well as in flowers but it is very low in roots and mature leaves (Dar et al. 2015). In recent years, the role of JA as an important phytohormone implicated in plant development under normal and stress conditions has emerged (Jang et al. 2019, 2020). Different studies have been conducted on plants belonging to different families such as Brassicacea (the model plant Arabidopsis thaliana), Solanaceae (tobacco: Nicotiana spp., tomato (Lycopersicum esculentum), and monocotyledons (Delker et al. 2006; Kazan and Manners 2013). Many groups described that wounding and systemin increases the endogenous level of JAs including the metabolic precursor, 12-oxo-phytodienoic acid (12-OPDA) and its derivatives JA and MeJA to activate the defense response machinery (Wang et al. 2016).
Reactions in the chloroplast
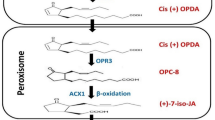
The JA biosynthetic pathway was studied in Arabidopsis (Ishiguro et al. 2001; Schaller 2001; Feussner and Wasternack 2002; Turner et al. 2002). JA is originally synthetized from a ω-3 fatty acid called linoleinc acid (ALA) (C18:39,12,15). Briefly, the reaction starts when phospholipases (PLDs) separates phospholipids such as linolenic (18:2) and α-linolenic (18:3) acid from chloroplast membranes (Bargmann et al. 2009). ALA constitutes the most abundant leaf lipids and can be found as galactolipids forming the main lipid components of chloroplasts: monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) of the chloroplast (Nilsson et al. 2016). Though the first substrate of the pathway is yet to be identified, it has been suggested that an esterification then oxygenation of ALA into galactolipid occurs via 13-lipoxygenase (13-LOX; Fig. 1; (Nilsson et al. 2016)). Moreover, other findings suggest that the ALA could be hydrolyzed then converted into 13-HPL (Wang et al. 2019). Allene oxide synthase (AOS) is the next acting enzyme in the octadecanoid pathway. Allene oxide synthase (AOS) is a cytochrome P450 (CYP) enzyme that belongs to the CYP74A family. AOS converts galactolipids into 12,13-epoxylinolenic acid which is rapidly transformed to cis (+)-12-oxo-phytodienoic acid (OPDA) by allene oxide cyclase (AOC) (Wasternack and Feussner 2018). In tomato, systemin activates a phospholipase A2 (PLA2) that operates the release of LA from membrane lipids after leaves are wounded (Narvaez-Vasquez et al. 1999). For example, systemin a peptide which is involved in the expression of JA-, wound- and insect-inducible proteinase inhibitors (PIs) was found in tomato but not in tobacco or Arabidopsis (Sun et al. 2011). Mechanisms of JA biosynthesis in Arabidopsis are summarized in Fig. 1.
A proposed model of JA biosynthesis in Arabidopsis. AOS enzyme is encoded by a single gene while four genes encode for the cyclase (AOC1–4). Growth and development processes as well as extracellular stimuli activate the octadecanoid pathway for JA biosynthesis with a so-far unknown mechanism. The first step in JA biosynthesis takes place in the choloroplast where we notice a sequential activity of several enzymes involved in the initial steps of JA biosynthesis including lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC) led to the production of 12-oxo-phytodienoic acid (12-OPDA). 12-OPDA is then translocated from chloroplasts via JASSY proteins and penetrates into peroxysome via CTS channels where it is reduced to 3-oxo-2(29[Z]-pentenyl)-cyclopentane-1-octanoic acid (OPC:8). OPC:8 undergoes three rounds of b-oxidation to yield JA. After biosynthesis, JA is conjugates with ILE and transported to the cytoplasm where is could be transported all over the plant or could be inactivated via CYP94B3 proteins
In monocotyledons, JA biosynthetic pathway is almost conserved with dicotyledons. In fact, some works confirmed the involvement of many enzymes such as PLDs in JA biosynthetic (Nahar et al. 2011). Knockdown of PLD enzymes involved in JA biosynthetic pathway such as OsPLDα4 (Os06g0604200) and OsPLDα5 (Os06g0 604300) confers herbivory susceptibility to rice (Qi et al. 2011). As enzymes implicated in the reduction of ODPA are located in the peroxisomes, this cyclopentenone must be exported from chloroplast and transported to peroxisomes. This is ensured by an ABC transporter located in the peroxisomal membrane called COMATOSE (CTS) and a possible implication of an ion trapping mechanism was proposed (Theodoulou et al. 2005). Recently, a novel protein called JASSY was described and it presented an export activity toward OPDA (Guan et al. 2019) and was found in the chloroplast outer envelope (OE) membranes (Simm et al. 2013). JASSY proteins, which are exclusively located in the chloroplastic OE, could have a role in binding and/or transport of hydrophobic molecules as it contains a steroid acute regulatory protein-related lipid transfer (START) domain. JASSY mutants represent an altered phenotype (growth inhibition and decrease in cold stress tolerance) is correlated with a decreased level of transcription factors INDUCER OF CBF EXPRESSION1 (ICE1) and C-REPEAT BINDING FACTOR 3 (CBF3) involved in the cold acclimation response). Moreover, jassy mutant had an increased susceptibility to infection by the necrotrophic pathogen Botrytis cinerea and decreased expression of corresponding marker genes, such as PLANT DEFENSIN1.2 (PDF1.2). In conclusion, after wounding, a lack of JA response (associated with reduced level of JA biosynthesis) and a reduced JA biosynthesis genes activation, as well as transcription factors involved in JA responses, were observed in the jassy mutant (Guan et al. 2019). In jassy mutant plants, ODPA synthesis is normal but its export out of the chloroplast was defected. However, evidence that ODPA pass through a JASSY channel with OPDA-regulated voltage-dependent gating is still missing. Furthermore, little information is available about specificity for OPDA-related compounds (Wasternack and Hause 2019). Thus, a link between JASSY proteins and JA biosynthesis genes was suggested.
Reactions in peroxisomes
The OPDA cyclopentenone component (12-oxo-phytodienoic acid) is scaled down to cyclopentanone using the OPDA reductase (OPDR; 12-oxo-phytodienoic acid reductase). The cyclopentenone ring is reduced by the AOC enzyme (Fig. 1). The (9S,13S)-OPDA [cis-(+)-OPDA] is the only natural component synthetized in nature (Wasternack and Feussner 2018). In another hand, OPDA could act as a signaling molecule and thus activates gene transcription independently of its conversion into JA (Taki et al. 2005). Another JA synthetic pathway was also revealed using Arabidopsis OPDA reductase 3 (OPR3) mutant which revealed another independent JA pathway using 4,5-didehydrojasmonate (Fig. 1; Chini et al. 2018). OPR could also regulate seed germination in Arabidopsis (Dave et al. 2011). In maize, it has been shown that OPR proteins are crucial for plant development and immunity (to pathogens and insects; Dave et al. 2011). Once synthetized, JA (or its active forms, e.g. MeJA) are transported all over the plant and in the phloem and xylem of vascular bundles. This transport is ensured via a protein transporter called GTR1 (Ishimaru et al. 2017; Matsu et al. 2017).
Reactions in cytoplasm
In Arabidopsis, works conducted on jar1-1, a JA-insensitive mutant revealed that the active form of JA was an amino acid conjugate. The isoleucine is the principal amino acid that conjugates with JA to form JA (JA-Ile) synthetized in the cytoplasm (Fig. 1). Other amino acid could rarely conjugate with JA such as valine, leucine, and phenylalanine (Staswick and Tiryaki 2004).
JA catabolism
JA-Ile metabolism is ensured by two major routes: hydrolyzation and oxidation (Fig. 2; Bruckhoff et al. 2016). JA-Ile hydrolyzation allows the liberation JA and Ile from JA-Ile. This is controlled by an aminohydrolase (AMH) enzyme. After that, JA is subjected to a hydroxylation, controlled by a P450 enzyme to generate 12-hydroxy-JA-Ile that could be oxidized later to generate carboxy-derivative. In another hand, JA-Ile could also be subjected to different oxidations of the ω-end of the pentenyl sidechain catalyzed by specific enzyme, CYP94 (Kitaoka et al. 2014). In the first step, JA-Ile is hydroxylated to 12-hydroxy JA-Ile. This can then either undergo further oxidation to produce the 12-carboxy-JA-Ile (Koo et al. 2014), or else can be glycosylated to generate 12-O-glycosyl-JA-Ile (Fig. 2; Haroth et al. 2019).
Catabolism of jasmonic acid (JA). JA could be inactivated via two major pathways: hydrolysation (I) and oxydation (II). Hydrolyzes is unsured by amidohydrolase (AMH) to release Ile and JA. Then JA could be either hydroxylated (to 12-hydroxy-JA), glycosylated, methylated to form methyl JA (Me-JA) or decarboxylated to synthesize cis-jasmone (CJ). JA-Ile could also undergo sequential oxidation reaction to generate 12-carboxy-JA-Ile. JA could also be glycosylated to generate 12-O-glycosyl-JA-Ile. Enzymes that catalyses the reactions are shown in red (colour figure online)
JA conversion into signaling molecules
Me-JA
Me-JA is synthesized via the acceptation of methyl group from S-adenosyl-l-methionine (SAM) to JA. This reaction is catalyzed by the SAM: JA carboxyl methyl-transferase enzyme. This esteric compound formed is volatile in contrast to JA (Sanchez-Aguayo et al. 2004). Me-JA has been detected in plant leaves after an herbivore attack. Interestingly, this molecule is detected only in leaves that have not been predated. Thus, this molecule can serve as an airborne warning molecule signal (Wasternack and Feussner 2018). In plants like potato and tomato, the production of PIN II protease inhibitor is one of the common markers for JA pathway activation (Singh et al. 2020). Those inhibitors act as an anti-feedant compound as it interferes with digestive proteins in the insect gut leading to lower food availability for the insect and hence the insect take more time to grow and complete its life cycle (Singh et al. 2020).
The consequence of insect growth delay is that weather changes may occur and hence reduce the survival of the insect (Bayram and Tonga 2018). In addition, Me-JA is one of complex volatiles compounds released during plant infection that could play the role of chemical attractor for insect predators such as wasps as demonstrated for Triticum aestivum (Bayram and Tonga 2018). Moreover, treatment with Me-JA stimulates phytoalexins compounds such as alkaloids terpenoids and glycosteroids which have an antimicrobial activity (Shi et al. 2015), enhances carotenoid biosynthesis as demonstrated for maize (Luo et al. 2019, 2020) or stimulates sucrose degradation. Moreover, MeJA changes enzymatic activity of many enzymes of the phenylpropanoid pathway (Sadeghnezhad et al. 2020). Me-JA also decreased the amino acid biosynthesis rate thus it controls the cell growth rate (Sadeghnezhad et al. 2020).
cis-Jasmone (CJ)
CJ is used in perfume industry as an important ingredient. This natural compound is liberated from flowers of different species such as jasmine, neroli (Citrus bigaradia; Matsu et al. 2017), jonquil (Narcissus jonquilla L.; Dabrowska and Boland 2007), bergamot (Citrus bergamia), and members of the Pittosporum family (Griffiths 2020). After plant infection, CJ is released, and it could act like JA. It can act as an insect repellent or it could participate to attract insect predators (Matthes et al. 2010). In fact, exogenous application of CJ to wheat was effective against cereal aphids and some phytophagous thrips species as well as wheat stem sawflies Bayram and Tonga 2018). Moreover, foliar application of CJ increased volatile organic compounds (VOCs) production as well as the upregulation of genes encoding them. Besides, CJ negatively affect tomato pest Spodoptera exigua reproduction (Disi et al. 2017).
The signaling pathway of JA
The SCFCOI1 protein and JA signaling
Experiments conducted with the Arabidopsis mutant coi1 (coronatine insensitive 1) add knowledge on the comprehension of the JA response pathway (Feys et al. 1994). COI1 was identified as an F-box protein; it is closely related to the auxin receptor/F-box protein TIR1 (Xie et al. 1998). Thus, the authors suggested that it could act as part of an SCF (Skp/Cullin/F-box) E3 ubiquitin ligase to mediate JA signaling (Xie et al. 1998). In fact, COI1 was demonstrated to be associated with components of the SCF complex, including ASK1, ASK2, CUL1, and RBX1 (Xu et al. 2002). Moreover, in Arabidopsis, other works suggest that SCFCOI1 protein targets the repressor proteins of JA signaling for degradation and, therefore, activates JA-modulated changes at transcriptional level. The same founding was also reported in tobacco, tomato, and soybean (Scheer and Ryan 2002; Li et al. 2004).
The AtMYC2: master regulator of JA signaling
MYC2 structure
Several works have classified the basic helix-loop-helix (bHLH) transcription factor (TF) as the greater JA signaling pathway in Arabidopsis (Browse 2009; Chini et al. 2009a; Howe 2010; Pauwels and Goossens 2011; Wager and Browse 2012). The best characterized TF in this group and the most multifunctional in the JA signaling pathway in Arabidopsis is MYC2 (Kazan and Manners 2013). This nucleolar protein (Chini et al. 2009b) presents the general characteristics of bHLH TF protein family (Toledo-Ortiz et al. 2003; Pires and Dolan 2010) such as the conserved bHLH domain at its carboxyl domain. Those domains are formed by two amphipathic alpha-helices consisting of hydrophobic amino acids united by a loop region important to form homo/heterodimers with other TFs like the most closed TFs MYC3 and MYC4 (Fernandez-Calvo et al. 2011). It has been suggested that the C-terminal leucine zipper domain could function as a supplementary dimerization domain to increase the specificity of cooperation with other TFs (Amoutzias et al. 2008). Moreover, the 15–20 mostly basic amino acids are involved in linking to the G-box (5′-CACGTG-3′) found in MYC2 target promoters (Toledo-Ortiz et al. 2003; Pires and Dolan 2010; Fernandez-Calvo et al. 2011; Amoutzias et al. 2008; Carretero-Paulet et al. 2010). In the N-terminal part of the MYC2 TF, a putative transcriptional activation domain (TAD) is found, which is important for transcription initiation (Fernandez-Calvo et al. 2011; Amoutzias et al. 2008; Carretero-Paulet et al. 2010). Moreover, MYC2 and their closely related proteins, MYC3, and MYC4, interplay with the C-terminal part of JAZ proteins (JAS domain) through JID domain (JAZ Interaction Domain) (Fernandez-Calvo et al. 2011; Amoutzias et al. 2008; Carretero-Paulet et al. 2010; Chini et al. 2007). Some works proposed that protein phosphorylation is involved in the modulation of JA signaling (Hiruma et al. 2011). Though MYC2 was shown to be phosphorylated, the identity of kinase protein implicated in this phosphorylation is still unknown (Zhai et al. 2013).
MYC2 and JA response
MYC2 orchestrates the JA pathway by controlling several mechanisms such as plant response to pathogen infection, abiotic stress applications and developmental processes (Kazan and Manners 2013). It tightly regulates the expression of downstream transcriptional activators as well as JAZ repressors that, in turn, act upstream from MYC2. In fact, myc2 mutant Arabidopsis seedlings had reduced expression of JAZ genes comparing with wild-type proteins in response to JA treatment (Chini et al. 2007; Grunewald et al. 2009). Later, it has been demonstrated that MYC2 directly binds to the promoter of JAZ3 (Chini et al. 2007) and JAZ2 (Figueroa and Browse 2012) confirming that the MYC2 TF is a regulator of JAZ gene expression. MYC2-mediated transcriptional activation and JAZ repressors synthesis coordinate to degrade the JAZ proteins (Chini et al. 2007; Demianski et al. 2012). MYC2 TF controls the expression of a high number of JA-responsive genes including TFs (Guo et al. 2012; Dombrecht et al. 2007). In fact, several researchers have identified that MYC2 presented a dual effect on the JA signaling pathways as it acts as a transcriptional activator as well as a repressor in the regulation of JA signaling network (Fig. 3). In fact, it has been demonstrated that a comparative microarray analysis using JA-treated and untreated myc2 and wild-type plants. The analysis identified many JA-responsive genes that are directly or indirectly managed by MYC2 all along JA signaling (Dombrecht et al. 2007). Moreover, bioinformatic analyzes showed that MYC2 interacts with at least 100 proteins as revealed by The Arabidopsis thaliana Promoter Analysis Net, network (AtPAN) (http://atpan.itps.ncku.edu.tw/), conclude more than 100 different MYC2 cooperator (Chen et al. 2012). The most important studied interactome are resumed in Table 1.
A simplified schematic presentation of jasmonate (JA) signaling. After its biosynthesis in plastids, JA is conjugated with aa, especially Ile by JAR under the control of JAR enzymes in the cytoplasm. In some conditions, JAZ interacts with NINJA, TOPLESS (TPL), and histone deacetylase (HAD) proteins to repress JA gene expression via inhibiting MYC2 TF. It also inhibits many negative regulators of JA signaling such as IIId bHLH factors. In presence of extracellular stimuli or in growth and development processes, JA-Ile enhances the CORONATINE INSENSITIVE1 (COI1)/JA ZIM-domain (JAZ) family proteins interaction. This leads to JAZ ubiquitination and degradation with the 26S proteasome. MYC2 TFs interacts with the MED25 subunit of the moderator complex then interacts with the G-box motif of the target promoters, which allows the activation of JA-responsive genes. In another hand, IIId bHLH factors antagonize MYC2 action via competitive binding to the G-box motif and inhibiting JA-responsive genes. Thus, succeeding transcription factors are stimulated, allowing them to enhance JA-responsive early genes activities and JA responses
Jasmonate ZIM-domain proteins: inhibitor of JA response
Eight different JAZ proteins (jasmonate ZIM-domain proteins) were identified in opr3 mutants during JA response (Thines et al. 2007). Those proteins harbor two conserved interaction domains, called ZIM and Jas (Thireault et al. 2015). The Jas domain controls JAZ interaction with COI1 and with other TFs as well (Gimenez-Ibanez et al. 2014). Moreover, the ZIM (TIFY) domain regulates the JAZ dimerization and interaction with other interactives of JAZ (NINJA) proteins which requires TOPLESS (TPL) proteins, transcriptional co-suppressor, via the interaction with the conserved EAR domain (Zhang et al. 2015). Importantly, in vitro and in vivo assays demonstrated that JA-Ile, not other derivatives, mediates direct JAZ1 and COI1 interaction (Thines et al. 2007). In response to JA, many JAZ proteins, such as JAZ3, are fixed with SCFCOI1 to mediate their proteasome degradation (Chini et al. 2007). Moreover, in absence of JA, JAZ directly interacts with AtMYC2 which leads to transcriptional repression of JA-responsive genes (Chini et al. 2007).
Identification of NINJA proteins
As discussed above, physical interplay between JAZ repressor proteins and AtMYC2 regulates JA-responsive gene expression. This requires the interaction between NINJA proteins and Groucho/Tup1-type co-repressor TOPLESS (TPL) and TPL-related proteins (TPRs) with the presence of unknown adaptor protein. TPL and NINJA proteins act as negative regulators in the JA signaling pathway. NINJA acts as a transcriptional repressor whose activity is mediated by a functional TPL-binding EAR repression motif (Pauwels et al. 2010).
SCFCOI1/JAZ/ AtMYC2 complex: the JA Core Signaling Module
Generally, JA-Ile endogenous level is low in normal conditions, but they can rapidly increase in presence of a given stimuli. This increase in sensed by a JA receptor called COI1 which constitute a fundamental component of SCFCOI1. COI1 lie to JAZ proteins and enhances JAZ ubiquitination and degradation through the 26S proteasome pathway. Thus, downstream TFs (e.g.: MYCs) can be released to activate JA responses (Gimenez-Ibanez et al. 2014; Dubois et al. 2018). As a result, many works identified MYC, JAZ, and COI1 protein as the core signal transduction mechanism of JA signaling. Those proteins play a crucial role in plant extension and maturing as well as its reply to environmental stresses (Gimenez-Ibanez et al. 2014; Dubois et al. 2018). JAZ interact with many TFs and specifically regulate many responses (Chini et al. 2016). In Arabidopsis thaliana, when endogenous jasmonate concentration increase, JAZ proteins interact with SCFCOI1 ubiquitin ligase which leads to JAZ degradation by proteasome 26S (Chini et al. 2007). Thus, degradation of JAZ protein liberates COI1 and AtMYC2 to enhance jasmonate-dependent gene expression (Chini et al. 2007; Thines et al. 2007). Defense compounds increases in presence of JAZ-MYC module. Thus, this could stimulate defense response against pathogen infection or inhibit plant growth (Havko et al. 2016). Moreover, COI1/JAZ2/MYC2, 3,4/ANAC19,55,72 and other modules have been identified to enhance plant response to endogenous or exogenous stimuli (Gimenez-Ibanez et al. 2014; Mao et al. 2017). MYCs–JAZs interactions could trigger the hormonal cross talk pathways like ET-mediated cell division throughout ET RESPONSE Transcription factor (ERF) (Gimenez-Ibanez et al. 2014; Zhang et al. 2015; Dubois et al. 2018).
Involvement of JA in plant growth and development
Phytohormones control internal developmental interactions with entourage stimuli to regulate plant maturation and survival. In the last decade, a huge body of studies focused on understanding the role of JA in plant maturation and development. An important number of works have shown that jasmonates are involved in a huge number of plants expanding events, such as primary root growth, leaf senescence, and reproductive development (Wasternack and Hause 2007; Kim et al. 2015). Moreover, JA is implicated into the regulation of various metabolites production, such as phytoalexins and terpenoids.
Effects on seed germination
It has been described that some phytohormones like ABA, IAA, and JA play important roles during seed germination (Xiao et al. 2018). Both ABA and JA inhibit seed germination but their interactions during this process are still elusive (Tang et al. 2020). In Arabidopsis, JA inhibition occurs in a COI1-independent manner (Dave et al. 2011). In Triticum aestivum plants, cold-stimulated germination of seeds resulted in an increase in endogenous level of JA after upregulation of JA biosynthesis-related gene and JA promotes cold-stimulated germination (Xu et al. 2016; Avramova 2017). Recently, a novel SAPK10-bZIP72-AOC’ pathway was identified in rice. In this pathway, ABA stimulates JA biosynthesis to synergically inhibit the germination of rice seeds (Wang et al. 2020). Some authors demonstrated that SAPK10 was activated by auto-phosphorylation, then this molecule stabilizes by phosphorylation bZIP72 TF and promotes it’s binding to the G-box cis-element of AOC promoter to enhance AOC transcription in presence of elevated concentrations of JA. Interestingly, alleviation of ABA sensitivity was recorded after blocking of JA biosynthesis (Wang et al. 2020).
Inhibition of root growth
Mutations in the co-receptor for JA-Ile COI1 make plants insensitive to the prevention of primary root maturation by JAs (Yan et al. 2009). Moreover, inositol pentakisphosphate (InsP5) enhances COI1–JAZ9 interactions and thus the blocking effect of JAs on root expansion and maturation (Mosblech et al. 2011). In another hand, coronatine-O-methyloxime, a competitive JA antagonist obstruct COI1–JAZs interactions and thus suppress the inhibitory outcome of coronatine on primitive root growth (Monte et al. 2014). In Arabidopsis, most JAZ proteins (13 members) lack the ERF-associated amphiphilic repression (EAR) domain. Thus, they should interact with TPL and TPL-related proteins (TPRs) via the EAR of NINJA proteins to crush JA responses. Interestingly, the non-canonical JAZ proteins, JAZ8 and JAZ13 do not require NINJA protein to interact with TPLs /TPRs but use directly their one EAR domain (Thireault et al. 2015; Chini et al. 2016). Interestingly, overexpressed NINJA proteins or modified JAZ proteins (carrying a deletion, mutation) in Jas domain suppressed the inhibitory effect of JA on primary root growth. But this inhibitory effect was stimulated by the NINJA/TPL or combined mutations in JAZ7, JAZ8, JAZ10, and JAZ13 (Thireault et al. 2015; Thatcher et al. 2016). In Arabidopsis, bHLH type transcription factors (MYC2 and its homologs MYC3/4/5), interact with JAZ proteins (Qi et al. 2015). MYC2/3/4 are in the primary root apex; make place to the primary root growth inhibition by JAs (Gasperini et al. 2015). Moreover, MYC2 inhibits PLETHORA genes (PLT1 and PLT2) expression to minimize the activity of root meristematic cells and suppress primary root growth (Chen et al. 2011). Besides, MYC2 interacts with MED25 subunit to unsure this inhibitory effect by inhibiting RNA polymerase II (Chen et al. 2011). It has been demonstrated that JAZ9 handicaps MYC3–MED25 interactions (Gimenez-Ibanez et al. 2014). MYC2 ubiquitination via PLANT U-BOX PROTEIN10 and its phosphorylation of MYC2 in presence of mitogen-activated protein kinase 3/6 suppresses JA-mediated root maturation inhibition (Chico et al. 2014; Sethi et al. 2014).
In Arabidopsis, ETHYLENE INSENSITIVE (EIN) 3 and EIN3-LIKE1 (EIL1) are primordial TFs acting in ethylene signaling. Those TFs interact with JAZ proteins and positively mediate the JA-dependent primary root growth inhibition as well as JA-induced root hair formation (Zhu et al. 2011). In Arabidopsis, JA stimulates also the lateral root formation as it upregulates ERF109 expression which stimulates ANTHRANILATE SYNTHASE Α1 (ASA1) and YUCCA2 genes implicated in auxin biosynthesizes (Cai et al. 2014). In contrast, JA negatively regulates adventitious root formation via the COI1-MYC2/3/4 cascade (Gutierrez et al. 2012).
Regulation of stomatal closure
Stomata control gas exchange, water loss in plants. It also regulates plant immunity against phytopathogens. Stomatal closure is also mediated by JA. In fact, Methyl-JA/ COI1 activate a plasma membrane H+-ATPase. Membrane depolarization generates an influx of Ca2+ and an efflux of H+ (Yin et al. 2016). Methyl-JA/ COI1 induces also the production of ROS, the stimulation of Cl– channels (which engender an efflux of Cl–) and the stimulation of K+ efflux (via K+ channels), causing turgor guard cell loss and stomatal closure (Yan et al. 2015) Drought and salt stress conditions induce stomatal closure in plants. It has been proven that JA and ABA pathways control stomatal closure in Arabidopsis through the stimulation of OPEN STOMATA1 protein kinase (Yin et al. 2016). Moreover, in drought conditions, plants synthesizes OPDA rather than jasmonate. This molecule is more efficient than JA in such conditions. OPDA interplay with ABA to enhance stomatal closure in Arabidopsis (Savchenko et al. 2014).
Plant infection with Pseudomonas syringae induces also stomatal closure to limit bacterial invasion (Melotto et al. 2006). To protect themselves, many P. syringae strains synthesize coronatine which interferes with COI1-JAZ-MYC signaling and prohibits SA intensification via NAC019/NAC055/NAC072 and JA2L, enhancing stomatal reopening to help bacterial infection (Zhu et al. 2015). Moreover, HopX1 and HopZ1a, effectors of P. syringae, deplete JAZ proteins in a COI1-independent manner, and maintain stomatal apertures to promote infection (Gimenez-Ibanez et al. 2014).
Delay of flowering
In Arabidopsis, vegetative-reproductive maturation transition is inhibited by JA. COI1–JAZ interaction inhibits flowering. In fact, coi1 mutant as well as JAZ1Δ3A transgenic plants presented an early flowering. Moreover, TARGET OF EAT TFs (TOE1 and TOE2), APETALA2/ERF domain TFs type, control plant flowering. In fact, their interaction with JAZ proteins inactivates the transcription of FLOWERING LOCUS T and thus inhibits flowering. In another hand, overexpression of TOE1 and/or TOE2 inhibits coi1 early flowering phenotype (Zhai et al. 2015).
Inhibition of hypocotyl growth
In Arabidopsis, JA inhibits hypocotyl elongation in many light stress conditions such as far-red and blue wavelengths. This is ensured through COI1 (Chen et al. 2013). Studies showed that JA-deficient mutant jar1 plants grown in far-red light conditions represented elongated hypocotyls (Robson et al. 2010). Moreover, coi1 mutants have longer hypocotyls compared to WT plants when they were grown in dark or under fared lights conditions (Chen et al. 2013) or under red light or low R/FR light ratio (Chen et al. 2013; Robson et al. 2010). The same observation was registered for the myc2/jin1 mutant grown under far-red light and a low R/FR light ratio (Robson et al. 2010), but those plants show a shorter hypocotyl under blue light (Yadav et al. 2005). Authors suggest that bHLH type TF MYC2 plays a role in plant response to red or far-red light and positively regulates the repression of hypocotyls expansion, but it negatively controls the inhibition of hypocotyls elongation by blue light. In rice, JA represses coleoptiles maturation and plant height. In Zea mays, JA inhibits ear shoot maturation (Yan et al. 2012; Yang et al. 2012).
Stamen development in Arabidopsis
In Arabidopsis, many male sterile plants were described. Among those mutants, JA-deficient mutants were described. In fact, coi1, lox3 lox4, aos, opr3; fad3 fad7 fad8, defective in anther dehiscence1 (dad1), JAZ1Δ3A and JAZ10.4 mutants as well as CYP94B3 overexpression lines represented incomplete stamen development (Song et al. 2013). Interestingly, exogenous application of JA restores stamen development only in plants deficient in JA biosynthesis but not in JA signaling mutants (Jewell and Browse 2016). Moreover, in coi1 background, re-imposed COI1 expression in many tissues such as filaments epidermis or anthers can retrieve anther dehiscence, filament elongation, and pollen maturation (Jewell et al. 2016). R2R3-MYB TFs such as MYB21, MYB24, and MYB57 interact directly with JAZs (Song et al. 2011). The myb21 myb24 double mutant represents a delayed anther dehiscence, a non-viable pollen grains and short filaments. Coi1-1 plants overexpressing MYB21 proteins represent a restored stamen development (Song et al. 2011). MYB21 and MYB24 connect physically with MYC2, MYC3, MYC4, and MYC5 to control stamen development (Qi et al. 2015). Interestingly, overexpression of MYC5 and MYC3 in coi1-1 plants can restore stamen maturation and productivity (Qi et al. 2015).
Inhibition of petal expansion
In Arabidopsis, JA inhibits petal expansion via COI1 pathway. In fact, coi1 mutants represent larger petals comparing with wild type at plant anthesis. The same observation was registered for Arabidopsis JA-deficient mutants’ aos and opr3 (Reeves et al. 2012; Brioudes et al. 2009). JA represses MYB21 and MYB24 proteins during sexual organs formation which lead to the restriction of petal growth confirming that MYB21 and MYB24 are important for petal expansion (Reeves et al. 2012). Interestingly, aos and coi1 plants showed an increased MYB21 expression level in petals leading to continuous petal extension and thus grand petals (Reeves et al. 2012). Moreover, opr3 plants represent an increased bHLH TF BIGPETALp expression level. This TF controls post-mitotic cell expansion and unsure larger petals and larger cells in this mutant (Brioudes et al. 2009).
Promotion of trichome formation
Trichomes are barriers or sensors that protect plants from herbivore attack. They can also release volatile compounds. In Arabidopsis, it has been demonstrated that defect in JA biosynthesis or perception can block trichome formation after wounding (Yoshida et al. 2009). At low endogen JA level in cells, JAZ proteins physically connect with WD-repeat/bHLH/MYB complexes to inhibit their transcriptional activity (Qi et al. 2011). After wounding or pathogen attack, endogenous JA concentration increase leading to JAZ proteins turn over. In such situations, WD-repeat/bHLH/MYB complexes enhance trichome formation. Subgroup IIIdbHLH TFs inhibits WD-repeat/bHLH/MYB complexes action by binding to the promoters of their putative target genes competitively leading to the inhibition of trichome formation (Nakata et al. 2013).
Inhibition of apical hook formation
COI1-JAZs-MYC2/3/4 cascade inhibits the formation of apical hook of dark grown plants (Song et al. 2014). In dark stress conditions, MYC2, MYC3, and MYC4 TFs are activated by JA and physically interact with and repress the transcriptional activity of EIN3/EIL1. This leads to downregulation of HOOKLESS1 gene that controls apical hook formation and to the inhibition of apical hook curvature (Song et al. 2014; Zhang et al. 2014). Besides, MYC2 activates EIN3 BINDING F-BOX PROTEIN1 expression which leads to EIN3 degradation (Zhang et al. 2014).
Induction of leaf senescence and chlorophyll degradation
JA stimulates leaf senescence in Arabidopsis. This effect occurs in COI1-dependent manner (Qi et al. 2015). Moreover, JAZ7 represses leaf senescence in dark grown plants (Yu et al. 2015). Many NAC TFs members (such as NAC019, NAC055, and NAC072) act downstream of MYC2/3/4 to enhance chlorophyll degradation (Melotto et al. 2006). In another hand, Subgroup IIIdbHLH TFs antagonize MYC2/3/4 actions as they bind competitively to their target promoters and inhibit leaf senescence (Qi et al. 2015). WRKY57 interacts physically with JAZ4/8 and act as a negative regulator of leaf senescence in presence of JA (Jiang et al. 2014). YABBY1 and YABBY3 proteins interact with JAZs leading to chlorophyll degradation promotion (Boter et al. 2015).
Involvement of JA in plant abiotic and biotic stress responses
Role of JA in plant drought and oxidative stress response
JAs and MeJAs are known to take part in various physiological processes. Exogenous application of JAs so far tested on different plants under abiotic stresses particularly salinity, drought, and temperature (low/high) conditions have proved effective in improving plant stress tolerance (Ahmad et al. 2016). Several studies conducted on Arabidopsis, barley, wheat, maize, and pearl millet showed that both ABA and JA positively control plant tolerance to drought stress (Awan et al. 2020; Wang et al. 2021). In rice and Arabidopsis, both ABA and JA-connected genes were controlled in the first drought constraint subjection, but showed different figure under the next drought stress cycles, underlying a potential role of those hormones in stress memory (Li et al. 2019). In pearl millet, water deficit conditions diminish root and shoot length as well as their fresh- and dry weight comparing with control plants (Awan et al. 2020). Exogenous application of ABA and JA weaken drought stress effects on pearl millet plantlet by increasing the fresh and dry weight of seedlings, root length and shoot length with a pronounced effect of JA comparing to ABA (Awan et al. 2020) as also shown for soybean and wheat under drought stress (Ruan et al. 2019). Authors showed that JA have a more pronounced effect of JA comparing with ABA in plant stress alleviation (Awan et al., 2020). This indicates the signaling interaction between ABA and JA in response to environmental stresses.
Moreover, JA intensifies the antioxidant enzyme (SOD, CAT, and APX) activities with a pronounced effect comparing with ABA hormone treatment in pearl millet (Awan et al. 2020). In contrast, in the absence of allene oxide cyclase (AOC) gene, which encodes the enzyme involved in JA biosynthesis, rice showed enhanced tolerance to water deficit stress in term of stomatal conductance, water content, and shoot ABA content compared to the wild type (Dhakarey et al. 2017). Besides, both hormones, ABA and JA, reveal drought stress effects by minimizing oxidative damage (serious oxidative impairment of cell membrane), in the leaves. In fact, exogenously applied ABA and JA improved drought stress in Arabidopsis (de Ollas et al. 2015), wheat (Siddiqi and Husen 2019), and pearl millet (Awan et al. 2020) which positively raise MDA and H2O2 contents in those plants.
JA alleviates negative impacts of heavy metals stress
Heavy metals [manganese (Mn), nickel (Ni), cadmium (Cd), and lead (Pb)] damage the environment and reduce plant growth and maturation (Zhang et al. 2018). Many foundings revealed that JA controls plant responses to heavy metal applications by modulating their antioxidant systems (Noriega et al. 2012; Carvalho et al. 2013; Ali et al. 2018). In tomato, it has been demonstrated that spr2, a JA-deficient mutant accumulated more Cd in roots and leaves comparing with wild-type plants (Zhao et al. 2016). Furthermore, in Vicia faba, external JA supplementation enhanced plant growth and biomass yielding, restores total chlorophyll content and carotenoid concentrations, enhances glutathione reductase activity and proline accumulation (Ahmad et al. 2017), reduces Cd accumulation in roots, shoots, and leaves by stimulating antioxidant activity and inhibiting MDA and H2O2 accumulation (Carvalho et al. 2013; Ahmad et al. 2017). Moreover, JA activated ascorbate or glutathione antioxidant machinery in soybean to inhibit lipid peroxidase activity (Noriega et al. 2012). In Brassica napus, JA minimized the Cd uptake in leaves, which ensured a reduction in membrane damage and MDA content. This facilitated also essential nutrient uptake by roots and increases gas exchange and photosynthetic pigment contents by protecting chloroplasts from damaging effects of Cd (Ali et al. 2018).
Exogenous JA application on Glycine max seeding before NiCl2 stress could enhance tolerance to Ni2+ stress. This positive effect of JA is ensured by protecting total protein contents and controlling the antioxidant machinery (Sirhindi et al. 2015). In Zea mays, external application of JA protects plant from Ni toxicity by stimulating antioxidant enzyme activity (Azeem 2018).
Jasmonic acid alleviates also the nefast effect of lead (Pb) stress. Pb minimizes macro and micronutrients uptake by plants roots (Sharma and Dubey 2005). It prevents the penetration of many essential cations (such as K+, Zn2+, Mn2+, Mg2+, Cu2+, Ca2+, Fe3+), and anions (NO3−) in the roots (Bali et al. 2019a). Pb causes also a decline in roots and shoots lengths in many plants such as tobacco ((Alkhatib et al. 2012) and tomato (Bali et al. 2019a). Bali et al. (2018) showed that JA alleviates Pb effects by enhancing tomato growth. External JA application minimizes Pb uptake by tomato roots. Moreover, this hormone helps plants to improve their photosynthetic efficiency and gaseous exchange. Furthermore, JA-treated plants exhibits an increased content of osmolytes (glycine betaine, proline, polyamine, and total carbohydrates). Besides, JA-treated tomato cultivated under Pb stress presented an enhanced accumulation of metal chelating compounds (total thiols, non-protein thiols, and protein thiols) (Bali et al. 2018, 2019b).
Role of JA in plant resistance against nematodes
Recently, the infection of watermelon plants (Citrullus lanatus L.) by a soil-dwelling, microscopic nematode called Meloidogyne incognita was investigated. Plants infection leads to apparition of galls on roots which can enhance plant sensitivity to infection by other pathogens (Kyndt et al. 2013). After nematodes invasion, plants activate different defense strategies such as ROS, hormones, and activation of appropriate defense-related genes (Melillo et al. 2006; Martìnez-Medina et al. 2017; Song et al. 2017). In Arabidopsis thaliana, ROS production is mediated by RbohD and RbohF genes after nematodes infections leading to galls formation and cell death (Siddique et al. 2014). Moreover, SA, JA, ET, BRs, and ABA are involved in plant response to nematodes (Nahar et al. 2011; Kammerhofer et al. 2015; Kyndt et al. 2017; Song et al. 2017). Notably, JA is implicated in systemic defense induction of rice (Nahar et al. 2011), watermelon (Yang et al. 2018) and tomato (Sun et al. 2010) against nematodes. Recently, it has been demonstrated that implication of JA in plant defense against Root Knot nematode is related to the alterations of antioxidative protection and photosynthetic processes (Bali et al. 2017). Moreover, application of exogenous SA alleviates plant infections by nematodes (Molinari et al. 2014; Martìnez-Medina et al. 2017). In another hand, biotrophic and necrotrophic pathogens infections are positively regulated by red lights in several plants (Mutar and Fattah 2013; Yang et al. 2015). Recent founding showed that the incidence of Root Knot nematodes in watermelon was inhibited by diurnal red lights comparing to white lights. This plant defense was closely related to a remarkable increase of many genes (AOS, PRI, ICS, WRKY70, and LOX) in association with a pronounced increase in SA, JA, and H2O2 contents in various tissues of watermelon plants (Yang et al. 2018). JA and SA participate to plant defense against nematodes such as M. incognita (Kammerhofer et al. 2015). This pathogen forces plants to activate SA pathways and to inhibit JA pathway in leaves to allow a successful pathogen invasion (Martìnez-Medina et al. 2017). Interestingly, JA has a higher accumulation in roots after root knot nematodes infection which is transported later to the leaves which helps plants to trigger their defense against nematodes (Zhang and Baldwin 1997). Furthermore, plant treatment by red lights enhances JA accumulation in roots and leaves (Yang et al. 2018) confirming the importance of JA signaling in rhizo-bacteria-induced systemic resistance (Fujimoto et al. 2011). Besides, studies conducted on tomato and watermelon confirms that exogenous MeJA application reduced root knot nematodes infections (Fujimoto et al. 2011; Yang et al. 2018). Moreover, due to the antagonistic roles between SA and JA, red lights inhibit SA accumulation in leaves (Yang et al. 2018) but stimulated its accumulation in roots after nematode infection underlying the positive effect of red lights on SA accumulation on roots (Branch et al. 2004; Yang et al. 2018). Meanwhile, red lights stimulate JA accumulation in roots demonstrating a positive regulation of JA biosynthesis genes by red lights; such results could indicate a synergic interaction (van Wees et al. 2000; Yang et al. 2018). This is shown at the molecular level by transcript accumulation of ICS gene (for SA biosynthesis) and LOX and AOS genes (for JA biosynthesis). In parallel, red lights induces accumulation of WRKY70 transcript and H2O2 particles after root knot nematode infections in both roots and shoots of watermelon (Yang et al. 2018) suggesting a pivotal role of WRKY70 TFs and ROS signaling in red lights-induced plant defense against Root Knot nematodes in watermelon plants. Accumulation of WRKY70 in roots only could suggest that this TF acts as a point of convergence between JA-mediated and SA-mediated signals in plant defense (Li et al. 2004; Yang et al. 2018).
Interaction between JA and other plant growth regulators
Plant genomes encodes for plant growth regulators which are implicated in plant growth and development and are also implicated in plant response to biotic and abiotic stresses. Thousands of works have described biosynthesis of those endogenous organic compounds (Liu et al. 2017; Chandler and Werr 2015; Kazan 2015; Li et al. 2019). They also underline the way to modulate plant response at molecular and physiological levels. Interactions between phytohormones are crucial for prompt and efficient plant responses to biotic and abiotic stress factors. The endogenous concentrations of those natural compounds are low, and it changes depending of the applied stress. Those observations suggest that plant hormones interact with each other to modulate plant development as revealed for IAA/CKs interplay (Liu et al. 2017; Chandler and Werr 2015). Some works demonstrated that JA interacts with salicylic acid (Liu et al. 2016a), ethylene (Kazan 2015; Li et al. 2019), GA (Wasternack and Hause 2007), IAA (Chen et al. 2011), BR (Choudhary et al. 2012) and ABA (Aleman et al. 2016) to unsure plant response to biotic and abiotic stress. Moreover, it has been demonstrated that stress imposition involves usually the activation/inhibition of a high number of genes as a response to the biosynthesis of phytohormones. For example, in response to drought stress ABA, IAA, JA, GA, SA, and ET orchestrate to upregulate almost 859 genes in Arabidopsis as revealed by transcriptomic studies (Ding et al. 2013). In this section, we describe the crosstalk between JA and other phytohomones GA, CKs, SA, ABA, BR, ET, and auxin to modulate plant growth and development.
The JA–GA interaction
GAs are crucial hormones controlling many aspects of plant growth and development, such as leaf expansion, stem elongation, root development, seed germination, and stamen and flower development (Um et al. 2018). In another hand, several studies suggest that GA is crucial for growth inhibition in stress conditions (Liu and Hou 2018). In unfavorable stress conditions, plants inhibit their growth by activating their defense systems and inhibiting developmental programs. Many works suggest an antagonistic role between GA and JA to coordinate plant development under normal and stress conditions (Jang et al. 2017). This is ensured by the direct interaction between DELLA (GA signaling repressor) and JAZ (JA signaling repressor) proteins (Fig. 4; Yang et al. 2012; Hou et al. 2010). It has recently shown that rice OsJAZ9 acts as the key JAZ protein in mediating crosstalk between JA and GA (Um et al. 2018). It interacts with a DELLA protein called SLENDER RICE 1 (SLR1). Overexpression of OsJAZ9 enhanced rice GA response. Thus, we can speculate that JA acts as an essential hormone that modulates plant growth under stress conditions due to its antagonistic interaction with GA.
Schematic representation of antagonistic action between JA and GA. a In Free JA conditions, the GA concentrations in cell increase to unsure growth programs. The direct interaction between JAZs and DELLA proteins is abolished by degradation of DELLAs, and JAZ proteins inhibit the activity of MYC2 TF. This activates PIF TFs to interact with responsive gene inducing GA responses. b In GA-free situation, JA concentration increases. Ile-JA/JAZ1/COI1 complex is released by the ubiquitation and degradation of JAZs by proteosome 26. Thus, the MYC2 TF is activated and enhanced transcription of JA-induced genes
The JA–CKs interaction
Cytokinin (CKs) control a plethora of aspects related to plant growth and development by maintaining stem cell integrity as well as cell proliferation. In higher plants, zeatin is the most abundant form of CKs and occurs as two isomers, trans-zeatin (tZ), the active form and cis-zeatin (cZ); less active form. Trans-zeatin is synthetized under the control of several hormones such as Isopentenyl transferases (IPTs), and cytochrome P450 CYP735A1 and CYP735A2 (Kieber and Schaller 2018). JA and JA-dependent stress response genes affect the expression of CKs gene response (Argueso et al. 2009). CKs deficient plant mutants are tolerant to stress and respond to stress application in a manner like transgenic plants having high JA concentrations (Qi et al. 2014). In higher plants, CKs receptors, called histidine phosphotransferase-proteins transcription factors, regulates CKs responses in plants. Many works suggest that JA and CKs act antagonistically controlling various plant development aspects. In fact, in soybean, the CKs induced callus Growth is inhibited by JA (Ueda and Kato 1982). Furthermore, JA has an antagonistic effect on CKs as JA inhibits the expression of genes implicated in chlorophyll development (Liu et al. 2016b). In another hand, it has been demonstrated that JA positively controls xylem differentiation in Arabidopsis roots and eliminates the specific procambium response to CKs application (Jang et al. 2017). In contrast, CKs acts as a negative regulator of xylem differentiation in Arabidopsis roots. Taken together, those results approve the idea that CKs interacts antagonistically with JA in xylem development in Arabidopsis roots. Interestingly, myc2 mutant did not show extra xylem formation in response to exogenous JA. Moreover, the expression of CKs signaling inhibitor, AHP6, was inhibited in this mutant. Those results suggest that MYC2 transcription factor is involved in this process by promoting AHP6 expression. Nevertheless, it has been demonstrated that JA and CKs antagonistically interact in the plant response to circadian stress (Nitschke et al. 2016). In fact, plants with reduced or defective CKs levels present a JA-dependent cell death phenotype in response to circadian modification, in contrast to wild types (Nitschke et al. 2016).
The JA–auxin interaction
Auxin is a crucial phytohormone implicated in plant growth and development (Figueiredo and Köhler 2018). In plants, the predominant form is called IAA (Indole-3-acetic-acid). IAA biosynthesis could take place through two different pathways: tryptophan-dependent and -independent pathways (Wang et al. 2019). The JA–IAA interaction controls many aspects of plant physiology and development such as production of secondary metabolites, tendril coiling and cell elongation (Saniewski et al. 2002). Unfortunately, those interactions have never been demonstrated at the molecular level. The most studied aspect of JA–IAA interaction is the regulation of plant roots development. In fact, the apical roots growth was inhibited by JA. This was demonstrated by wild-type plants treated by JA. They form much shorter roots comparing with non-treated plants. In contrast, JA defective mutant plants presents roots with similar length comparing with wild type mainly in presence of JA treatment (Jang et al. 2017). Meanwhile, auxin is crucial hormone implicated in the control of root growth. Auxin deficient mutants or signaling mutants develop shorter roots in comparison with wild-type plants as demonstrated for (trp2-12) and auxin resistant 3 (arx3-1) (Zhang et al. 2019). Those findings suggest that JA–IAA interaction could mediate root growth inhibition as revealed by (Chen et al. 2011). The group showed that JA inhibits root growth by reducing root meristem activity as exogenous JA application inhibits PLETHORAs (PLTs) expression, auxin-responsive transcription factors controlling cell proliferation (Mahonem et al. 2014). Interestingly, PLTs expression level was not modified in JA signaling mutants, such as coi1-1 and myc2. Authors suggest then that COI1 could be implicated in controlling root phenotype, whereas MYC2 directly binds to the PTLS promoters and inhibits their expression. This suggests that auxin and JA antagonistically interact in the regulation of apical root growth (Fig. 5).
The development of floral organs is another physiological process controlled by JA–IAA interaction (Reeves et al. 2012). An interaction between IAA and JA modulates petals and stamens development under the control of R2R3 MYB transcription factors especially MYB21 and MYB24 TFs which are considered as key regulators of petal and stamen growth (Reeves et al. 2012). Moreover, ARF6 and ARF8 TFs control the regulation of expression of MYB21 and MYB24 JA-responsive TFs (Fig. 5). Nevertheless, the regulation of leaves senescence is also controlled by IAA–JA interaction. In fact, JA acts as a positive regulator of leaf senescence as JAZ7 proteins suppress dark-induced leaf senescence and MYCs TFs, such as MYC2, promote senescence by activating the senescence-associated genes expression as well as chlorophyll degradating-related genes, demonstrating that JA stimulates leaf senescence pathway in a COI1-dependent manner (Yu et al. 2015). In this pathway, JAZ4, JAZ8 and WRKY57 act as negative regulators contrarily to JAZ7 and the auxin signaling repressor IAA29 functions as a positive regulator. Moreover, WRKY57 negatively regulates senescence-associated gene expression by interacting with JAZ4/8 and IAA29. Authors speculate that competition between the WRKY57–JAZ4/8 WRKY57-IAA29 implies the adequate response to the physiological stimulus. Thus, an antagonistic JA–IAA interaction is involved in leaf senescence (Fig. 5; Jiang et al. 2014).
JA–Brassinosteroids crosstalk
Brassinosteroids (BR) are implicated in different aspects of plant growth and development. They are also involved in the modulation of JA signaling and JA-dependent growth inhibition.
In contrast to JA, BR promotes above-ground plant growth. Thus, JA-BR crosstalk is insured by the balance between plant growth and defense resistance (Yang et al. 2019). For example, low BR concentrations induce OsDI1 and OsDWARF expression at early and late stages of BR biosynthesis, which activate the defense pathway (Yang et al. 2019). Moreover, DWARF4 encodes a key enzyme responsible for BR biosynthesis and a leaky mutation of DWARF4 restored JA sensitivity in the coi1 mutant background and showed JA hypersensitivity in the wild-type background. Furthermore, expression of DWARF4 was down regulated by JA in a COI1-dependent manner, and exogenous BR treatment attenuated the effects of JA on root growth inhibition (Ren et al. 2009). These results indicate that a BR–JA interaction is involved in the modulation of JA signaling. A low BR concentration activates BR signaling pathways such as BR receptor BRI1, BR-related kinase BAK1, and BR-related TFs. This induces the activation of several downstream genes such as BES1 and BZR1, which activate plant response to many abiotic stresses (Yang et al. 2019).
The JA–SA crosstalk
After stress perception, JA rapidly regulates plant gene defense (Genva et al. 2019). The A. thaliana coronatine insensitive 1 (coi1) mutants presented a blocked JA response (Feys et al. 1994). COI1 is a JA receptor that belongs to the F-box protein family. It acts as an E3-ubiquitin ligase-mediated proteolysis of target proteins (Hiruma et al. 2011) such as JASMONATE ZIM-DOMAIN (JAZ) proteins. JA is essentially involved in plant response against necrotrophic pathogens. SA mediates plant resistance against biotrophic and hemibiotrophic pathogens (Fu et al. 2012). JA signaling modulates many NAC Tfs activities (such as ANAC019/055/072) to block SA accumulation through modulation of multiple NAC TFs. Briefly, physical interaction between MYC2 and several NACs promoter activates their transcription. Expressed NAC TFs inhibits the expression of SA biosynthesis gene called ISOCHORISMATE SYNTHASE 1 (ICS1) but stimulate the expression of a SA methylation gene, BENZOIC ACID/SA CARBOXYL METHYLTRANSFERASE 1 (BSMT1) (Zheng et al. 2012). In addition, many other components are involved in SA-JA crosstalk such as MYC2, TGAs, and PDF 1.2 (Gatz 2013); mitogen-activated protein kinase (MAPK) (Suarez-Rodriguez et al. 2010), redox regulators glutathione (GRX) and thioredoxin (TRX) (Sopel and Gary 2011) and WRKY70 (Shim et al. 2013). GRXs genes can block TGA-mediated JA response gene expression, such as ORA59 confirming SA–JA antagonism (Zander et al. 2010). MPK4 is a positive regulator of GRX480 (SA signaling pathway) and acts as a negative regulator of MYC2 (JA signaling pathway) (Wasternack and Hause 2007). Meanwhile, TRX enzyme transforms NPR1 polymers to monomers in presence of SA. Those monomers such as GRX480 are transported to the nucleus and specifically bind to TGAs, which also directly regulate the expression of PR1 (Fu et al. 2012; Gatz 2013; Zander et al. 2010). A schematic presentation of JA–SA crosstalk component is presented in Fig. 6.
JA-SA crosstalk in plants in response to pathogen infection. Plant infection by a pathogen leads to the increase in the endogenous SA concentration and the activation of NONEXPRESSOR OF PR GENES1 (NPR1), which induces WRKY70 transcriptional activity. In turn, WRKY70 promotes PR1 gene expression and thus enhancing pathogen response
In addition to its crucial role in ensuring plant defense against pathogens, other important roles of SA in regulating many physiological processes and responding to adverse environmental situations have emerged. SA is implicated in plant response to salinity (Khan et al. 2012), light (Tuteja and Gill 2013) and cold (Gornik et al. 2014). In fact, plant treatment with JA or SA improves plant resistance to chilling (Gornik et al. 2014). Moreover, MeJA and MeSA present a protective effect against chilling injury in pomegranates fruits (Sayyari et al. 2010). SA and JA are also implicated in plant tolerance to drought (Ilyas et al. 2017). In another hand, several studies concentrated on the role of SA and JA in plant tolerance to salinity. In fact, some researches demonstrated that JA and SA protect plants from negative effects of salinity by the induction of gene encoding effective proteins expression (Khan et al. 2012). Moreover, it has been found that exogenous SA and JA application decreased Na+ concentrations in soybean cells under different salt stress levels and that JA has a greater effect on Na+ reduction than SA (Farhangi-Abriz and Ghassemi-Golezani 2018). Therefore, a regulatory protein called glutaredoxin GRX480 is a key protein in JA and SA signaling. This protein mediates redox regulation of proteins by catalyzing disulfide transitions (Meldau et al. 2012). In Nicotiana attenuata plants, researches demonstrated that mitogen-activated protein kinase 4 (MPK4) have a dual role in plant signaling. In fact, in response to light stress, this kinase acts as a negative regulator during the SA signaling pathway, but it plays a positive role during the JA signaling pathway (Tuteja and Gill 2013). To coordinate regulation between SA and JA, cells involve several factors such as GRX480 or MAPK4. Moreover, exogenous application of SA and/or JA can enhance abiotic stress responses (Tuteja and Gill 2013).
JA–ABA crosstalk
Upon external stimuli application on plants, an immense signaling network of events occurs in cells. Ca2+ sensors act in the first line of defense to decode and then transducer signals through the suitable stress hormone pathways to activate the appropriate signaling cascade. ABA is a well-studied phytohormone implicated in plant response to abiotic stress. Many studies described that ABA and JA presents an antagonistic role in plant growth and development (Chen et al. 2011). In another hand, those hormones act synergistically in response to environmental stress and MYC2 unsure that crosstalk (Chen et al. 2011). In A. thaliana as well as tobacco plants, PYRABACTIN RESISTANCE1-Like proteins (PYLs) known as ABA receptor proteins control metabolic reprogramming via JA signaling pathway. Thus, JA/ABA crosstalk can control plant metabolism and growth (Per et al. 2018). When endogenous ABA concentration increases in response to external (salt stress, drought…) or internal (growth and development) conditions, ABA interacts with many members of the PYL/RCAR ABA receptor family to inhibit 2C protein phosphatases (PP2C proteins) thus to initiate signal transduction. Recently, a direct interaction between the ABA receptor PYL6 RCAR9 and MYC2 TF has been demonstrated (Aleman et al. 2016). This interaction is enhanced in presence of ABA. Moreover, ABA receptor PYL6 forms a complex with JAZ6 and JAZ8 to stimulate the transcriptional activity of MYC2. In fact, MYC2 stimulate VSP2 expression, inhibits PTL1 and PTL2 expression and root development.
JA–ET crosstalk in plants
Many researchers found that JA and ET could antagonize or coordinate to regulate plant stress response (Zhu and Lee 2015). In fact, MYC2 (JA signaling) and (EIN3-like 1 (EIL1) and ET INSENSITIVE3 (EIN3) involved in ET signaling pathway are implicated in JA/ET crosstalk (Zhang et al. 2014). Recent findings showed that when plants are exposed to a necrotrophic or hemibiotrophic pathogen attacks, JA signaling pathway act in synergy with the ET signaling pathway to activate defense proteins such as PDF1.2 expression by activating ERF1 and ORA59 expression. Thus, JA and ET control plant response to nectrophic pathogens through JAZs-MYC2 and EIN3/EIL1 (Pieterse 2012). Moreover, JA enhances the degradation of JAZ, so MYC2 is activated to regulate ORA59/ERF1 expression and VSP2, a wound responsive gene to respond to herbivorous insect attacks (Yang et al. 2019). Besides, JAZ inhibits EIL2/EIN3 transcriptional activity downstream ORA59/ERF1 that targets the promoter of PLANT DEFENSIN 1.2 (PDF1.2) and induces its expression thereby resisting the infection of necrotrophic pathogens and hemibiotrophic pathogens (Pieterse 2012). In Arabidopsis, JA inhibits apical hook formation via activation of MYC2:3:4 TFs. ET stabilizes EIN3/ EIL1 complex leading to the inhibition of the transcriptional function of MYCs, and it attenuates MYC-mediated plant defenses against insect attack (Song et al. 2014).
In the lights of those findings, we can conclude that hormonal interactions play a critical role for insuring plant physiology and growth (Yang et al. 2019). The cross talk of JA with other hormones (IAA, GA, CKs, BR, JA ABA and SA) ensures plant growth and defense. Direct JA–GA, JA–IAA, JA–BR, and JA–CKs interactions play important role in modulating physiological role such as root and floral organs development and can help plant to optimize their growth and development in stressful conditions. However, this interaction is almost antagonistic but in some cases it is synergic. In fact, despite JA–GA interaction antagonistically control stem elongation, it plays a positive role to control stamen development (Qi et al. 2014). In the same way of idea, JA–IAA interaction negatively regulates apical roots growth but positively regulate lateral root growth (Cai et al. 2014).
Conclusion and prospects
Jasmonic acid and its derivatives have pivotal roles in the establishing plant defense and resistance in response to environmental stimuli. The mechanisms of action of JA are specific when plants are subjected to different environmental constraints due to the diversification of plant hormone interplay. Several genes (e.g. JAZ, AOS1, AOC, LOX2, and COI1) and TFs (MYC2 and bHLH) are involved in the core JA signaling pathway as activators or repressors, thereby modulating the responses to stimuli received by the plants from the surrounding environment. The JAZ-MYC module has a key role in the JA signaling pathway through the assimilation of regulatory TFs and associated genes. Plant hormone signaling systems are complicated and unstable, especially under complex environmental conditions. Over the years, JA crosstalk with other phytohormones has been elucidated. However, information in plants is still limited. Crosstalk nodes between those hormones have to be investigated. In presence of abiotic constraints, JA orchestrate plant stress responses by stimulating the plant’s defense apparatus, which mainly involves phytohormones and other defensive compounds such as antioxidative enzymes. JA functions synergistically and antagonistically with ABA, ET, SA, and other plant hormones to accomodate environmental constraints. The process is usually accompanied by the regulation of plant maturation and development hormones. Future studies can spot how plant distinguish between different environmental signals to activate a specific signaling pathway to respond to stress as well as to synthesis Jas and TFs/JAZ repressors interactions during JA action. Future studies can also clarify the molecular control of JA interaction with its transporter to control it’s moving through plants. Furthermore, crosstalk between JA and other phytohormones is not linear and researches should focus on the spatio-temporal relationships between hormones using new chemicals that act as targets of JA in vivo or using high-throughput nucleotide sequencing techniques as well CRISPR-CAS technology to generate specific mutant that allows underlying mechanisms of plant tolerance to unfavorable environments. Thus, understanding the signal transduction mechanism of JA under environmental stress is essential to promote plant resistance, with potential improvements in plant survival and crop yield.
References
Agurla S, Gahir S, Munemasa S, Murata M, Raghavendra AS (2018) Mechanism of stomatal closure in plants exposed to drought and cold stress. Adv Exp Med Biol 1081:215–232. https://doi.org/10.1007/978-981-13-1244-1_12
Ahmad P, Rasool S, Gul A, Sheikh SA, Akram NA, Ashraf M, Kazi AM, Gucel S (2016) Jasmonates: multifunctional roles in stress tolerance. Front Plant Sci 7:813. https://doi.org/10.3389/fpls.2016.00813
Ahmad P, Alyemeni MN, Wijaya L, Alam P et al (2017) Jasmonic acid alleviates negative impacts of cadmium stress by modifying osmolytes and antioxidants in faba bean (Vicia faba L.). Arch Agron Soil Sci 63(13):1889–1899
Aleman F, Yazaki J, Lee M, Takahashi Y, Kim AY, Li Z, Kinoshita T, Ecker JR, Schroeder JI (2016) An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: a putative link of ABA and JA signaling. Sci Rep 6:28941. https://doi.org/10.1038/srep28941
Ali E, Hussain N, Shamsi IH, Jabeen Z, Siddiqui MH, Lx J (2018) Role of jasmonic acid in improving tolerance of rapeseed (Brassica napus L.), to Cd toxicity. J Zhejiang Univ Sci B 19:130–146. https://doi.org/10.1631/jzus.B1700191
Alkhatib R, Maruthavanan J, Ghoshroy S, Steiner R, Sterling T, Creamer R (2012) Physiological and ultrastructural effects of lead on tobacco. Biol Plant 56:711–716. https://doi.org/10.1007/s10535-012-0241-9
Altaf-Ul-Amin M, Katsuragi T, Sato T, Kanaya S (2015) A glimpse to background and characteristics of major molecular biological networks. Biomed Res Int. https://doi.org/10.1155/2015/540297
Amoutzias GD, Robertson DL, Van de Peer Y, Oliver SG (2008) Choose your partners, dimerization in eukaryotic transcription factors. Trends Biochem Sci 33:220–229. https://doi.org/10.1016/j.tibs.2008.02.002
Argueso CT, Ferreira FJ, Kieber JJ (2009) Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant Cell Environ 32:1147–1160. https://doi.org/10.1111/j.1365-3040.2009.01940.x
Avramova Z (2017) The jasmonic acid signaling and abscissic acid signaling pathways during one, but not repeated dehydratation stress: a nonspecific ‘paniky’ or a meaningful response? Plant Cell Env 40:1704–1710. https://doi.org/10.1111/pce.12967
Awan SA, Khan I, Rizwan M, Zhang X, Brestic M, Khan A, El-Sheikh MA, Alyemeni MN, Ali S, Huang L, Huang L (2020) Exogenous abscisic acid and jasmonic acid restrain polyethylene glycol-induced drought by improving the growth and antioxidative enzyme activities in pearl millet. Physiol Plant. https://doi.org/10.1111/ppl.13247
Ayala A, Muñoz MF, Argüelles S (2014) Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. https://doi.org/10.1155/2014/360438
Azeem U (2018) Ameliorating Nickel stress by Jasmonic acid treatment in Zea mays L. Russ Agric Sci 44:209–215
Bali S, Kaur P, Sharma A, Ohri P, Bhardwaj R, Alyemeni MN et al (2017) Jasmonic acid-induced tolerance to root-knot nematodes in tomato plants through altered photosynthetic and antioxidative defense mechanisms. Protoplasma 255:471–484. https://doi.org/10.1007/s00709-017-1160-6
Bali S, Kaur P, Kohli SK, Ohri P, Thukral AK, Bhardwaj R, Wijaya L, Alyemeni MN, Ahmad P (2018) Jasmonic acid induced changes in physio-biochemical attributes and ascorbate-glutathione pathway in Lycopersicon esculentum under lead stress at different growth stages. Sci Total Environ 645:1344–1360. https://doi.org/10.1016/j.scitotenv.2018.07.164
Bali S, Jamwal VL, Kaur P, Kohli SK, Ohri P, Gandhi SG, Bardwaj R, Al-Huqail AA, Sidiqui MH, Ali HM, Ahmad P (2019a) Role of P-type ATPase metal transporters and plant immunity induced by jasmonic acid against Lead (Pb) toxicity in tomato. Ecotoxicol Environ Saf 174:283–294. https://doi.org/10.1016/j.ecoenv.2019.02.084
Bali S, Lakshmi V, Sukhmeen J, Kohli K, Kaur P, Tejpal R, Bhalla V, Gandhi SG, Bhardwi R, Al-Huqail AA, Sidiqui MH, Ali HM, Ahmad P (2019b) Jasmonic acid application triggers detoxification of lead (Pb) toxicity in tomato through the modifications of secondary metabolites and gene expression. Chemosphere. https://doi.org/10.1016/j.chemosphere.2019.06.188
Bargmann BOR, Laxalt AM, Ter Riet B, Testerink C, Merquiol E, Mosblech A, Leon-Reyes A, Pieterse CMJ, Haring MA, Heilmann I, Bartels D, Munnik T (2009) Reassessing the role of phospholipase D in the Arabidopsis wounding response. Plant Cell Environ 32:837–850. https://doi.org/10.1111/j.1365-3040.2009.01962.x
Bayram A, Tonga A (2018) cis-Jasmone treatments affect pests and beneficial insects of wheat (Triticum aestivum L.): the influence of doses and plant growth stages. Crop Prot 105:70–79. https://doi.org/10.1016/j.cropro.2017.11.011
Boter M, Golz JF, Giménez-Ibañez S, Fernandez-Barbero G, Franco-Zorrilla JM, Solano R (2015) FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell 27:3160–3174. https://doi.org/10.1105/tpc.15.00220
Branch C, Hwang CF, Navarre DA, Williamson VM (2004) Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Mol Plant Microbe Interact 17:351–356. https://doi.org/10.1094/MPMI.2004.17.4.351
Brioudes F, Joly C, Szécsi J, Varaud E, Leroux J, Bellvert F, Bertrand C, Bendahmane M (2009) Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J 60:1070–1080. https://doi.org/10.1111/j.1365-313X.2009.04023.x
Browse J (2009) Jasmonate passes muster, a receptor and targets for the defense hormone. Annu Rev Plant Biol 60:183–205. https://doi.org/10.1146/annurev.arplant.043008.092007
Bruckhoff V, Haroth S, Feussner K, Konig S, Brodhun F, Feussner I (2016) Functional characterization of CYP94-genes and identification of a novel jasmonate catabolite in flowers. PLoS ONE 11(7):e0159875. https://doi.org/10.1371/journal.pone.0159875
Cai XT, Xu P, Zhao PX, Liu R, Yu LH, Xiang CB (2014) Arabidopsis ERF109 mediates crosstalk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat Commun 5:5833. https://doi.org/10.1038/ncomms6833
Carretero-Paulet L, Galstyan A, Roig-Villanova I, MartínezGarcía JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153:1398–1412. https://doi.org/10.1104/pp.110.153593
Carvalho RF, Campos ML, Azevedo RA (2013) The role of phytochromes in stress tolerance. Salt stress plants. Springer, Berlin, pp 283–299. https://doi.org/10.1111/j.1744-7909.2011.01081.x
Chandler JW, Werr W (2015) Cytokinin–auxin crosstalk in cell type specification. Trends Plant Sci 20:291–300. https://doi.org/10.1016/j.tplants.2015.02.003
Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J et al (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23:3335–3352. https://doi.org/10.1105/tpc.111.089870
Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J et al (2012) The Arabidopsis Mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24:2898–2916. https://doi.org/10.1105/tpc.112.098277
Chen J, Sonobe K, Ogawa N, Masuda S, Nagatani A, Kobayashi Y, Ohta H (2013) Inhibition of Arabidopsis hypocotyl elongation by jasmonates is enhanced under red light in phytochrome B dependent manner. J Plant Res 126:161–168. https://doi.org/10.1007/s10265-012-0509-3
Chico JM, Fernández-Barbero G, Chini A, Fernández-Calvo P, Díez-Díaz M, Solano R (2014) Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26:1967–1980. https://doi.org/10.1105/tpc.114.125047
Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR et al (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448:666–671. https://doi.org/10.1038/nature06006
Chini A, Boter M, Solano R (2009a) Plant oxylipins, COI1/ JAZs/MYC2 as the core jasmonic acid-signaling module. FEBS J 276:4682–4692. https://doi.org/10.1111/j.1742-4658.2009.07194.x
Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R (2009b) The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J 59:77–87. https://doi.org/10.1111/j.1365-313X.2009.03852.x
Chini A, Gimenez-Ibanez S, Goossens A, Solano R (2016) Redundancy and specificity in jasmonate signaling. Curr Opin Plant Biol 33:147–156. https://doi.org/10.1016/j.pbi.2016.07.005
Chini A, Monte I, Zamarreno AM, Hamberg M, Lassueur S, Reymond P et al (2018) An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat Chem Biol 14:171. https://doi.org/10.1038/nchembio.2540
Choudhary SP, Yu JQ, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2012) Benefits of brassinosteroid crosstalk. Trends Plant Sci 17:594–605. https://doi.org/10.1016/j.tplants.2012.05.012
Dabrowska P, Boland W (2007) iso-OPDA: An Early Precursor of cis-Jasmone in Plants? ChemBioChem 8:2281–2285. https://doi.org/10.1002/cbic.200700464
Dar TA, Uddin M, Khan MMA, Hakeem KR, Jaleel H (2015) Jasmonates counter plant stress: a review. Environ Exp Bot 115:49–57. https://doi.org/10.1016/j.envexpbot.2015.02.010
Dave A, Hernandez ML, He ZS, Andriotis VME, Vaistij FE, Larson TR et al (2011) 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23:583–599. https://doi.org/10.1105/tpc.110.081489
de Ollas C, Arbona V, GóMez-Cadenas A (2015) Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant Cell Environ 38(10):2157–2170. https://doi.org/10.1111/pce.12536
Delker C, Stenzel I, Hause B, Miersch O, Feussner I, Wasternack C (2006) Jasmonate biosynthesis in Arabidopsis thaliana—enzymes, products, regulation. Plant Biol 8:297–306. https://doi.org/10.1055/s-2006-923935
Demianski AJ, Chung KM, Kunkel BN (2012) Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol Plant Pathol 13:46–57. https://doi.org/10.1111/j.1364-3703.2011.00727.x
Demidchik V, Shabala S (2017) Mechanisms of cytosolic calcium elevation in plants: the role of ion channels, calcium extrusion systems and NADPH oxidase-mediated ‘ROS-Ca2+ Hub.’ Funct Plant Biol 45(2):9–27. https://doi.org/10.1071/FP16420
Deng X, Wang J, Li Y, Wu S, Yang S, Chao J, Chen Y, Zhang X, Shi M, Tian W (2018) Comparative transcriptome analysis reveals phytohormone signaling, heat shock module and ROS scavenger mediate the cold-tolerance of rubber tree. Sci Rep 8:4931. https://doi.org/10.1038/s41598-018-23094-y
Dhakarey R, Raorane ML, Treumann A, Peethambaran PK, Schendel RR, Sahi VP, Hause B, Bunzel M, Henry A, Kohli A, Riemann M (2017) Physiological and Proteomic Analysis of the Rice Mutant cpm2 Suggests a Negative Regulatory Role of Jasmonic Acid in Drought Tolerance. Front Plant Sci 8:1903
Ding Y, Liu N, Virlouvet L, Riethoven JJ, Fromm M, Avramova Z (2013) Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Boil 13:229. https://doi.org/10.3390/ijms21041446
Disi JO, Zebelo S, Ngumbi E, Fadamiro HY (2017) cis-Jasmone primes defense pathways in tomato via emission of volatile organic compounds and regulation of genes with consequences for Spodoptera exigua oviposition. Arthropod Plant Interact 11:591–602. https://doi.org/10.1007/s11829-017-9503-y
Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM et al (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19:2225–2245. https://doi.org/10.1105/tpc.106.048017
Dubois M, Van den Broeck L, Inzé D (2018) The pivotal role of ethylene in plant growth. Trends Plant Sci 23(4):311–323. https://doi.org/10.1016/j.tplants.2018.01.003
Farhangi-Abriz S, Ghassemi-Golezani K (2018) How can salicylic acid and jasmonic acid mitigate salt toxicity in soybean plants? Ecotox Environ Saf 147:1010–1016. https://doi.org/10.1016/j.ecoenv.2017.09.070
Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM et al (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23:701–715. https://doi.org/10.1105/tpc.110.080788
Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53:275–297. https://doi.org/10.1146/annurev.arplant.53.100301.135248
Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6:751–759. https://doi.org/10.1105/tpc.6.5.751
Figueiredo DD, Köhler C (2018) Auxin: a molecular trigger of seed development. Genes Dev 32:479–490. https://doi.org/10.1101/gad.312546.118
Figueroa P, Browse J (2012) The Arabidopsis JAZ2 promoter contains a G-box and thymidine-rich module that is necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol 53:330–343. https://doi.org/10.1093/pcp/pcr178
Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, Dong X (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486:228–232. https://doi.org/10.1038/nature11162
Fujimoto T, Tomitaka Y, Abe H, Tsuda S, Futai K, Mizukubo T (2011) Expression profile of jasmonic acid-induced genes and the induced resistance against the root-knot nematode (Meloidogyne incognita) in tomato plants (Solanum lycopersicum) after foliar treatment with methyl jasmonate. J Plant Physiol 168:1084–1097. https://doi.org/10.1016/j.jplph.2010.12.002
Gasperini D, Chételat A, Acosta IF, Goossens J, Pauwels L, Goossens A, Dreos R, Alfonso E, Farmer EE (2015) Multilayered organization of jasmonate signaling in the regulation of root growth. PLoS Genet 11:e1005300. https://doi.org/10.1371/journal.pgen.1005300
Gatz C (2013) From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. MPMI 26:151–159. https://doi.org/10.1094/MPMI-04-12-0078-IA
Genva M, Akong FO, Andersson MX, Deleu M, Lins L, Fauconnier ML (2019) New insights into the biosynthesis of esterified oxylipins and their involvement in plant defense and developmental mechanisms. Phytochem Rev 18:343–358. https://doi.org/10.1007/s11101-018-9595-8
Gimenez-Ibanez S, Boter M, Fernández-Barbero G, Chini A, Rathjen JP, Solano R (2014) The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol 12:e1001792. https://doi.org/10.1371/journal.pbio.1001792
Göbel C, Feussner I (2009) Methods for the analysis of oxylipins in plants. Phytochem 70:1485–1503. https://doi.org/10.1016/j.phytochem.2009.07.040
Górnik K, Badowiec A, Weidner S (2014) The effect of seed conditioning, short-term heat shock and salicylic, jasmonic acid or brasinolide on sunflower (Helianthus annuus L.) chilling resistance and polysome formation. Acta Physiol Plant 36:2547–2554. https://doi.org/10.1007/s11738-014-1626-5
Griffiths G (2020) Jasmonates: biosynthesis, perception and signal transduction. Essays Biochem. https://doi.org/10.1042/EBC20190085
Grunewald W, Vanholme B, Pauwels L, Plovie E, Inzé D, Gheysen G, Goossens A (2009) Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep 10:923–938. https://doi.org/10.1038/embor.2009.103
Guan L, Denkertb N, Eisaa A, Lehmanna M, Sjutsa I, Weiberga A, Solla J, Meineckeb M, Schwenkert S (2019) JASSY, a chloroplast outer membrane protein required for jasmonate biosynthesis. Proc Natl Acad Sci USA 116:10568–10575. https://doi.org/10.1073/pnas.1900482116
Guo J, Pang Q, Wang L, Yu P, Li N, Yan X (2012) Proteomic identification of MYC2-dependent jasmonate-regulated proteins in Arabidopsis thaliana. Proteome Sci 10:57
Gutierrez L, Mongelard G, Flokova K et al (2012) Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24:2515–2527. https://doi.org/10.1105/tpc.112.099119
Haroth S, Feussner K, Kelly AA, Zienkiewicz K, Shaikhqasem A, Herrfurth C et al (2019) The glycosyl transferase UGT76E1 significantly contributes to 12-O-glucopyranosyl-jasmonic acid formation in wounded Arabidopsis thaliana leaves. J Biol Chem 294:9858–9872. https://doi.org/10.1074/jbc.RA119.007600
Havko NE, Major IT, Jewell JB, Attaran E, Browse J, Howe GA (2016) Control of carbon assimilation and partitioning by jasmonate: an accounting of growth-defense tradeoffs. Plants 5:7. https://doi.org/10.3390/plants5010007
Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, SchulzeLefert P, Takano Y (2011) Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J 67:980–992. https://doi.org/10.1111/j.1365-313X.2011.04651.x
Hou X, Lee LYC, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19:884–894. https://doi.org/10.1016/j.devcel.2010.10.024
Howe GA (2010) Ubiquitin ligase-coupled receptors extend their reach to jasmonate. Plant Physiol 154:471–474. https://doi.org/10.1104/pp.110.161190
Ilyas N, Gull R, Mazhar R, Saeed M, Kanwal S, Shabir S, Bibi F (2017) Influence of salicylic acid and jasmonic acid on wheat under drought stress. Commun Soil Sci Plant Anal 48:2715–2723. https://doi.org/10.1080/00103624.2017.1418370
Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13:2191–2209. https://doi.org/10.1105/tpc.010192
Ishimaru Y, Oikawa T, Suzuki T, Takeishi S, Matsuura H, Takahashi K et al (2017) GTR1 is a jasmonic acid and jasmonoyl-L-isoleucine transporter in Arabidopsis thaliana. Biosci Biotechnol Biochem 81:249–255. https://doi.org/10.1080/09168451
Jang G, Chang SH, Um TY, Lee S, Kim JK, Do Choi Y (2017) Antagonistic interaction between jasmonic acid and cytokinin in xylem development. Sci Rep 7:10212. https://doi.org/10.1038/s41598-017-10634-1
Jang G, Yoon Y, Choi YD (2019) Jasmonic acid modulates xylem development by controlling expression of PIN-FORMED 7. Plant Signal Behav 14(9):1637664. https://doi.org/10.1080/15592324.2019.1637664
Jang G, Yoon Y, Choi YD (2020) Crosstalk with jasmonic acid integrates multiple responses in plant development. Int J Mol Sci 21:305. https://doi.org/10.3390/ijms21010305
Jewell JB, Browse J (2016) Epidermal jasmonate perception is sufficient for all aspects of jasmonate-mediated male fertility in Arabidopsis. Plant J 85:634–647. https://doi.org/10.1111/tpj.13131
Jiang Y, Liang G, Yang S, Yu D (2014) Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26:230–245. https://doi.org/10.1105/tpc.113.117838
Kammerhofer N, Radakovic Z, Regis JM, Dobrev P, Vankova R, Grundler FM et al (2015) Role of stress-related hormones in plant defence during early infection of the cyst nematode Heterodera schachtii in Arabidopsis. New Phytol 207:778–789. https://doi.org/10.1111/nph.13395
Kazan K (2015) Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci 20:219–229. https://doi.org/10.1016/j.tplants.2015.02.001
Kazan K, Manners JM (2013) MYC2: The Master in Action. Mol Plant 6:686–703. https://doi.org/10.1093/mp/sss128
Khan MIR, Syeed S, Nazar R, Anjum NA (2012) An insight into the role of salicylic acid and Jasmonic acid in salt stress tolerance. Phytohormones and abiotic stress tolerance in plants. Springer, Berlin, pp 277–300. https://doi.org/10.1007/978-3-642-25829-9
Kieber JJ, Schaller GE (2018) Cytokinin signaling in plant development. Development 145:149344. https://doi.org/10.1242/dev.149344
Kim J, Chang C, Tucker ML (2015) To grow old: regulatory role of ethylene and jasmonic acid in senescence. Front Plant Sci 6:20. https://doi.org/10.3389/fpls.2015.00020
Kitaoka N, Kawaide H, Amano N, Matsubara T, Nabeta K, Takahashi K et al (2014) CYP94B3 activity against jasmonic acid amino acid conjugates and the elucidation of 12-O-beta-glucopyranosyl-jasmonoyl-L-isoleucine as an additional metabolite. Phytochem 99:6–13. https://doi.org/10.1016/j.phytochem.2013.12.019
Koo AJ, Thireault C, Zemelis S, Poudel AN, Zhang T, Kitaoka N et al (2014) Endoplasmic reticulum-associated inactivation of the hormone jasmonoyl-L-isoleucine by multiple members of the cytochrome P450 94 family in Arabidopsis. J Biol Chem 289:29728–29738. https://doi.org/10.1074/jbc.M114.603084
Kyndt T, Vieira P, Gheysen G, De Almeida-Engler J (2013) Nematode feeding sites: unique organs in plant roots. Planta 238:807–818. https://doi.org/10.1007/s00425-013-1923-z
Kyndt T, Nahar K, Haeck A, Verbeek R, Demeestere K, Gheysen G (2017) Interplay between carotenoids, abscisic acid and jasmonate guides the compatible rice-Meloidogyne graminicola interaction. Front Plant Sci 8:951. https://doi.org/10.3389/fpls.2017.00951
Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16:126–143. https://doi.org/10.1105/tpc.017954
Li N, Han X, Feng D, Yuan D, Huang LJ (2019) Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering? Int J Mol Sci 20:671
Liu X, Hou X (2018) Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Front Plant Sci 9:251. https://doi.org/10.3389/fpls.2018.00251
Liu L, Li H, Zeng H, Cai Q, Zhou X, Yin C (2016a) Exogenous jasmonic acid and cytokinin antagonistically regulate rice flag leaf senescence by mediating chlorophyll degradation, membrane deterioration, and senescence-associated genes expression. J Plant Growth Regul 35:366–376. https://doi.org/10.3390/ijms21010305
Liu L, Sonbol FM, Huot B, Gu Y, Withers J, Mwimba M, Yao J, He SY, Dong X (2016b) Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat Commun 7:13099. https://doi.org/10.1038/ncomms13099
Liu J, Moore S, Chen C, Lindsey K (2017) Crosstalk complexities between auxin, cytokinin, and ethylene in Arabidopsis root development: from experiments to systems modeling, and back again. Mol Plant 10:1480–1496. https://doi.org/10.1016/j.molp.2017.11.002
Luo J, Xia W, Cao P, Xiao Z, Zhang Y, Liu M, Zhan C, Wang N (2019) Integrated transcriptome analysis reveals plant hormones jasmonic acid and salicylic acid coordinate growth and defense responses upon fungal infection in poplar. Biomolecules 9:12. https://doi.org/10.3390/biom9010012
Luo H, He WW, Li DJ, Bao YH, Riaz A, Xiao YD et al (2020) Effect of methyl jasmonate on carotenoids biosynthesis in germinated maize kernels. Food Chem 307:125525. https://doi.org/10.1016/j.foodchem.2019.125525
Mahonem AP, Ten Tusscher K, Siligato R, Smetana O, Diaz-Trivino S, Salojarvi J, Wachsman G, Prasad K, Heidstra R, Scheres B (2014) PLETHORA gradient formation mechanism separates auxin responses. Nature 515:125–129. https://doi.org/10.1038/nature13663
Mao YB, Liu YQ, Chen DY, Chen FY, Fang X, Hong GJ, Wang LJ, Wang JW, Chen XY (2017) Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat Commun 8:13925. https://doi.org/10.1038/ncomms13925
Martìnez-Medina A, Fernandez I, Lok GB, Pozo MJ, Pieterse CM, Van Wees SC (2017) Shifting from priming of salicylic acid- to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol 213:1363–1377. https://doi.org/10.1111/nph.14251
Matsu R, Amanu R, Takahashi K, Tagushi Y, Saburi W, Mori H, Kondo N, Matsuda K, Matsuura H (2017) Elucidation of the biosynthetic pathway of cis-jasmonate in Lasiodiplodia theobromae. Sci Rep 7:6688. https://doi.org/10.1038/s41598-017-05851-7
Matthes MC, Bruce TJA, Ton J, Verrier PJ, Pickett JA, Napier JA (2010) The transcriptome of cis-jasmone-induced resistance in Arabidopsis thaliana and its role in indirect defence. Planta 232:1163–1180. https://doi.org/10.1007/s00425-010-1244-4
Meldau S, Ullman-Zeunert L, Govind G, Bartram S, Baldwin IT (2012) MAPK-dependent JA and SA signaling in Nicotiana attenuate affects plant growth and fitness during competition with conspecifics. BMC Plant Biol 12:213. https://doi.org/10.1186/1471-2229-12-213
Melillo MT, Leonetti P, Bongiovanni M, Castagnone-Sereno P, Bleve-Zacheo T (2006) Modulation of reactive oxygen species activities and H2O2 accumulation during compatible and incompatible tomato-root-knot nematode interactions. New Phytol 170:501–512. https://doi.org/10.1111/j.1469-8137.2006.01724.x
Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126:969–980. https://doi.org/10.1016/j.cell.2006.06.054
Molinari S, Fanelli E, Leonetti P (2014) Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol Plant Pathol 15:255–264. https://doi.org/10.1111/mpp.12085
Monte I, Hamberg M, Chini A, Gimenez-Ibanez S, García-Casado G, Porzel A, Pazos F, Boter M, Solano R (2014) Rational design of a ligand-based antagonist of jasmonate perception. Nat Chem Biol 10:671–676. https://doi.org/10.1038/nchembio.1575
Mosblech A, Thurow C, Gatz C, Feussner I, Heilmann I (2011) Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J 65:949–957. https://doi.org/10.1111/j.1365-313X.2011.04480.x
Mutar SS, Fattah FA (2013) Red light-induced systemic resistance to root knot nematodes in tomato. J Biol Agric Healthc 3:110–114. https://doi.org/10.3389/fpls.2018.00899
Nahar K, Kyndt T, De Vleesschauwer D, Höfte M, Gheysen G (2011) The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol 157:305–316. https://doi.org/10.1104/pp.111.177576
Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M (2013) A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/ JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25:1641–1656. https://doi.org/10.1105/tpc.113.111112
Narvaez-Vasquez J, Florin-Christensen J, Ryan CA (1999) Positional specificity of a phospholipase A activity induced by wounding, systemin, and oligosaccharide elicitors in tomato leaves. Plant Cell 11:2249–2260. https://doi.org/10.1105/tpc.11.11.2249
Nilsson AK, Fahlberg P, Johansson ON, Hamberg M, Andersson MX, Ellerstrom M (2016) The activity of hydroperoxide lyase 1 regulates accumulation of galactolipids containing 12-oxo-phytodienoic acid in Arabidopsis. J Exp Bot 67:5133–5144. https://doi.org/10.1093/jxb/erw278
Nitschke S, Cortleven A, Iven T, Feussner I, Havaux M, Riefler M, Schmülling T (2016) Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 28:1616–1639. https://doi.org/10.1105/tpc.16.00016
Noriega G, Cruz DS, Batlle A, Tomaro M, Balestrasse K (2012) Heme oxygenase is involved in the protection exerted by Jasmonic acid against Cadmium stress in Soybean roots. J Plant Growth Regul 31:79–89. https://doi.org/10.1007/s00344-011-9221-0
Pauwels L, Goossens A (2011) The JAZ proteins, a crucial interface in the jasmonate signaling cascade. Plant Cell 23:3089–3100. https://doi.org/10.1105/tpc.111.089300
Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E et al (2010) NINJA connects the co-repressor TOPLESS to jasmonate signaling. Nature 464:788–791. https://doi.org/10.1038/nature08854
Per TS, Iqbal M, Khan R, Anjum NA, Masood A, Hussain SJ (2018) Jasmonates in plants under abiotic stresses: Crosstalk with other phytohormones matters. Environ Exp Bot 145:104–120. https://doi.org/10.1016/j.envexpbot.2017.11.004
Pieterse CMJ (2012) Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front Plant Sci 2:1–12. https://doi.org/10.3389/fpls.2011.00047
Pires N, Dolan L (2010) Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol 27:862–874. https://doi.org/10.1093/molbev/msp288
Pottosin I, Velarde-Buendía AM, Bose J, Zepeda-Jazo I, Shabala S, Dobrovinskaya O (2014) Crosstalk between reactive oxygen species and polyamines in regulation of ion transport across the plasma membrane: implications for plant adaptive responses. J Exp Bot 65:1271–1283. https://doi.org/10.1093/jxb/ert423
Qi J, Zhou G, Yang L, Erb M, Lu Y, Sun X, Cheng J, Lou Y (2011) The chloroplast-localized phospholipases D α4 and α5 regulate herbivore-induced direct and indirect defenses in rice. Plant Physiol 157:1987–1999. https://doi.org/10.1104/pp.111.183749
Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, Xie D (2014) Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26:1118–1133. https://doi.org/10.1105/tpc.113.121731
Qi T, Huang H, Song S, Xie D (2015) Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27:1620–1633. https://doi.org/10.1105/tpc.15.00116
Reeves PH, Ellis CM, Ploense SE et al (2012) A regulatory network for coordinated flower maturation. PLoS Genet 8:e1002506. https://doi.org/10.1371/journal.pgen.1002506
Ren C, Han C, Peng W, Huang Y, Peng Z, Xiong X, Zhu Q, Gao B, Xie D (2009) A leaky mutation in DWARF4 reveals an antagonistic role of brassinosteroid in the inhibition of root growth by jasmonate in Arabidopsis. Plant Physiol 151:1412–1420. https://doi.org/10.1104/pp.109.140202
Robson F, Okamoto H, Patrick E, Harris SR, Wasternack C, Brearley C, Turner JG (2010) Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22:1143–1160. https://doi.org/10.1105/tpc.109.067728
Ruan J, Zhou Y, Zhou M, Yan J, Khurshid M, Weng W et al (2019) Jasmonic acid signaling pathway in plants. Inter J Mol Sci 20(10):2479. https://doi.org/10.3390/ijms20102479
Sadeghnezhad E, Sharifi M, Zare-Maivan H, Chashmi NA (2020) Time-dependent behavior of phenylpropanoid pathway in response to methyl jasmonate in Scrophularia striata cell cultures. Plant Cell Rep 39:227–243. https://doi.org/10.1007/s00299-019-02486-y
Sanchez-Aguayo I, Rodrıguez-Galan JM, Garcıa R, Torreblanca J, Pardo JM (2004) Salt stress enhances xylem development and expression of S-adenosyl-L-methionine synthase in lignifying tissues of tomato plants. Planta 220:278–285. https://doi.org/10.1007/s00425-004-1350-2
Saniewski M, Ueda J, Miyamoto K (2002) Relationships between jasmonates and auxin in regulation of some physiological processes in higher plants. Acta Physiol Plant 24:211. https://doi.org/10.1007/s11738-002-0013-9
Savchenko T, Kolla VA, Wang CQ et al (2014) Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol 164:1151–1160. https://doi.org/10.1104/pp.113.234310
Sayyari M, Babalar M, Kalantari S, Martínez-Romeroa D, Guilléna F, Serranob M, Valeroa D (2010) Vapour treatments with methyl salicylate or methyl jasmonate alleviated chilling injury and enhanced antioxidant potential during postharvest storage of pomegranates. Food Chem 124:964–970. https://doi.org/10.1016/j.foodchem.2010.07.036
Schaller F (2001) Enzymes of the biosynthesis of octadecanoid-derived signaling molecules. J Exp Bot 52:11–23
Scheer JM, Ryan CA Jr (2002) The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc Natl Acad Sci USA 99:9585–9590. https://doi.org/10.1073/pnas.132266499
Sethi V, Raghuram B, Sinha AK, Chattopadhyay S (2014) A mitogen activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell 26:3343–3357. https://doi.org/10.1105/tpc.114.128702
Sewelam N, Kazan K, Schenk PM (2016) Global Plant Stress Signaling: Reactive Oxygen Species at the Crossroad. Front Plant Sci 7:187. https://doi.org/10.3389/fpls.2016.00187
Sharma P, Dubey RS (2005) Lead toxicity in plants. Braz J Plant Physiol 17:35–52. https://doi.org/10.1590/s1677-04202005000100004
Shi J, Ma CY, Qi DD, Lv HP, Yang T, Peng QH et al (2015) Transcriptional responses and flavor volatiles biosynthesis in methyl jasmonate-treated tea leaves. BMC Plant Biol 15:233. https://doi.org/10.1186/s12870-015-0609-z
Shim JS, Jung C, Lee S, Min K, Lee YW, Choi Y, Lee JS, Song JT, Kim JK, Choi YD (2013) AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J 73:483–495. https://doi.org/10.1111/tpj.12051
Siddiqi KS, Husen A (2019) Plant response to jasmonates: current developments and their role in changing environment. Bull Nat Res Centre 43(1):1–11. https://doi.org/10.1186/s42269-019-0195-6
Siddique S, Matera C, Radakovic ZS, Shamim Hasan M, Gutbrod P, Rozanska E (2014) Parasitic worms stimulate host NADPH oxidases to produce reactive oxygen species that limit plant cell death and promote infection. Sci Signal 7:ra33. https://doi.org/10.1126/scisignal.2004777
Simm S, Papasotiriou DG, Ibrahim M, Leisegang MS, Müller B, Schorge T, Karas M, Mirus O, Sommer MS, Schleiff E (2013) Defining the core proteome of the chloroplast envelope membranes. Front Plant Sci 4:11. https://doi.org/10.3389/fpls.2013.00011
Singh S, Singh A, Kumar S, Mittal P, Singh IK (2020) Protease inhibitors: recent advancement in its usage as a potential biocontrol agent for insect pest management. Insect Sci 27:186–201. https://doi.org/10.1111/1744-7917.12641
Sirhindi G, Mir MA, Sharma P, Gill SS, Kaur H, Mushtaq R (2015) Modulatory role of jasmonic acid on photosynthetic pigments, antioxidants and stress markers of Glycine max L. under nickel stress. Physiol Mol Biol Plants 21:559–565. https://doi.org/10.1007/s12298-015-0320-4
Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23:1000–1013. https://doi.org/10.1105/tpc.111.083089
Song S, Qi T, Huang H, Xie D (2013) Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol Plant 6:1065–1073. https://doi.org/10.1093/mp/sst054
Song S, Huang H, Gao H et al (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26:263–279. https://doi.org/10.1105/tpc.113.120394
Song LX, Xu XC, Wang FN, Wang Y, Xia XJ, Shi K et al (2017) Brassinosteroids act as a positive regulator for resistance against root-knot nematode involving RESPIRATORY BURST OXIDASE HOMOLOG-dependent activation of MAPKs in tomato. Plant Cell Environ 41:1113–1125. https://doi.org/10.1111/pce.12952
Sopel HS, Gary JL (2011) Redox-based protein modifications: the missing link in plant immune signaling. Curr Opin Plant Biol 14:358–364. https://doi.org/10.1016/j.pbi.2011.03.007
Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16:2117–2127. https://doi.org/10.1105/tpc.104.023549
Suarez Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-Activated Protein Kinase Signaling in Plants. Annu Rev Plant Biol 61:621–649. https://doi.org/10.1146/annurev-arplant-042809-112252
Sun YC, Cao HF, Yin J, Kang L, Ge F (2010) Elevated CO2 changes the interactions between nematode and tomato genotypes differing in the JA pathway. Plant Cell Environ 33:729–739. https://doi.org/10.1371/journal.pone.0019751
Sun JQ, Hong-Ling J, Li CY (2011) Systemin/jasmonate-mediated systemic defense signaling in tomato. Mol Plant 4:607–615. https://doi.org/10.1093/mp/ssr008
Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, Takamiya K, Shibata D, Kobayashi Y, Ohta H (2005) 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol 139:1268–1283. https://doi.org/10.1104/pp.105.067058
Tang G, Ma J, Hause B, Nick P, Riemann M (2020) Jasmonate is required for the response to osmotic stress in rice. Environ Exp Bot 175:104047. https://doi.org/10.1016/j.envexpbot.2020.104047
Thatcher LF, Cevik V, Grant M, Zhai B, Jones JD, Manners JM, Kazan K (2016) Characterization of a JAZ7 activation-tagged Arabidopsis mutant with increased susceptibility to the fungal pathogen Fusarium oxysporum. J Exp Bot 67:2367–2386. https://doi.org/10.1093/jxb/erw040
Theodoulou FL, Job K, Slocombe SP, Footitt S, Holdsworth M, Baker A, Larson TR, Graham IA (2005) Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisome. Plant Physiol 137:835–840. https://doi.org/10.1104/pp.105.059352
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF COI1 complex during jasmonate signaling. Nature 448:661–665. https://doi.org/10.1038/nature05960
Thireault C, Shyu C, Yoshida Y, St Aubin B, Campos ML, Howe GA (2015) Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J 82:669–679. https://doi.org/10.1111/tpj.12841
Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–1770. https://doi.org/10.1105/tpc.013839
Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell 14:S153–S164. https://doi.org/10.1105/tpc.000679
Tuteja N, Gill SS (2013) Salicylic acid: a novel plant growth regulator-role in physiological processes and abiotic stresses under changing environments. Climate change and plant abiotic stress tolerance. Wiley-VCH, Weinheim, pp 939–990. https://doi.org/10.1002/9783527675265.ch36
Ueda J, Kato J (1982) Inhibition of cytokinin-induced plant growth by jasmonic acid and its methyl ester. Physiol Plant 54(3):249–252. https://doi.org/10.1111/j.1399-3054.1982.tb00255.x
Um TY, Lee HY, Lee S, Chang SH, Chung PJ, Oh KB, Kim JK, Jang G, Choi YD (2018) JASMONATE ZIM-DOMAIN PROTEIN 9 interacts with SLENDER RICE 1 to mediate the antagonistic interaction between jasmonic and gibberellic acid signals in rice. Front Plant Sci 9:1866. https://doi.org/10.3389/fpls.2018.01866
van Wees SC, De Swart EA, Van Pelt JA, Van Loon LC, Pieterse CM (2000) Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci USA 97:8711–8716. https://doi.org/10.1073/pnas.130425197
Wager A, Browse J (2012) Social network, JAZ protein interactions expand our knowledge of jasmonate signaling. Front Plant Physiol 3:41. https://doi.org/10.3389/fpls.2012.00041
Wang Y, Yuan G, Yuan S, Duan W, Wang P, Bai J, Zhang F, Gao S, Zhang L, Zhao C (2016) TaOPR2 encodes a 12-oxo-phytodienoic acid reductase involved in the biosynthesis of jasmonic acid in wheat (Triticum aestivum L.). Biochem Biophys Res Commun 470:233–238. https://doi.org/10.1016/j.bbrc.2016.01.043
Wang K, Guo Q, Froehlich JE, Hersh HL, Zienkiewicz A, Howe GA et al (2019) Two abscisic acid-responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana. Plant Cell 30:1006–1022. https://doi.org/10.1105/tpc.18.00250
Wang Y, Hou Y, Qi J, Wang H, Wang S, Tang S, Tong X, Zhang J (2020) Abscissic acid promotes jasmonic acid biosynthesis via a “SAPK10-bZIP72-AOC” pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol 228:1336–1353. https://doi.org/10.1111/NPH.16774
Wang X, Li Q, Xie J, Huang M, Cai J, Zhou Q, Dai T, Jiang D (2021) Abscisic acid and jasmonic acid are involved in drought priming-induced tolerance to drought in wheat. Crop J 9:120–132. https://doi.org/10.1016/j.cj.2020.06.002
Wasternack C, Feussner I (2018) The oxylipin pathways: biochemistry and function. Ann Rev Plant Biol 69:363–386. https://doi.org/10.1146/annurev-arplant-042817-040440
Wasternack C, Hause B (2007) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058. https://doi.org/10.1093/aob/mct067
Wasternack C, Hause B (2019) The missing link in jasmonic acid biosynthesis. Nat Plants 5:776–777
Xiao HM, Cai WJ, Ye TT, Ding J, Feng YQ (2018) Spatio-temporal profiling of abscisic acid, indoleacetic acid and jasmonic acid in single rice seed during seed germination. Anal Chim Acta 1031:119–127. https://doi.org/10.1016/j.aca.2018.05.055
Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Sci 280:1091–1094. https://doi.org/10.1126/science.280.5366.1091
Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCFCOI1 ubiquitin ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14:1919–1935. https://doi.org/10.1105/tpc.003368
Xu Q, Truong TT, Barrero JM, Jacobsen JV, Hocart CH, Gubler F (2016) A role for jasmonates in the release of dormancy by cold stratification in wheat. J Exp Bot 67:3497–3508. https://doi.org/10.1093/jxb/erw172
Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17:1953–1966. https://doi.org/10.1105/tpc.105.032060
Yan J, Zhang C, Gu M et al (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21:2220–2236. https://doi.org/10.1105/tpc.109.065730
Yan Y, Christensen S, Isakeit T, Engelberth J, Meeley R, Hayward A, Emery RJ, Kolomiets MV (2012) Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 24:1420–1436. https://doi.org/10.1105/tpc.111.094151
Yan S, McLamore ES, Dong S, Gao H, Taguchi M, Wang N, Zhang T, Su X, Shen Y (2015) The role of plasma membrane H+-ATPase in jasmonate-induced ion fluxes and stomatal closure in Arabidopsis thaliana. Plant J 83:638–649. https://doi.org/10.1111/tpj.12915
Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109:E1192–E1200. https://doi.org/10.1073/pnas.1201616109
Yang YX, Wang MM, Yin YL, Onac E, Zhou GF, Peng S et al (2015) RNA-seq analysis reveals the role of red light in resistance against Pseudomonas syringae pv. Tomato DC3000 in tomato plants. BMC Genom 16:120. https://doi.org/10.1186/s12864-015-1228-7
Yang YX, Wu C, Ahammed GJ, Wu C, Yang Z, Wan C, Chen J (2018) Red light-induced systemic resistance against root-knot nematode is mediated by a coordinated regulation of salicylic acid, jasmonic acid and redox signaling in watermelon. Front Plant Sci 9:899. https://doi.org/10.3389/fpls.2018.00899
Yang J, Duan G, Li C, Liu L, Han G, Zhang Y, Wang C (2019) The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front Plant Sci 10:1349. https://doi.org/10.3389/fpls.2019.01349
Yin Y, Adachi Y, Nakamura Y, Munemasa S, Mori IC, Murata Y (2016) Involvement of OST1 protein kinase and PYR/PYL/RCAR receptors in methyl jasmonate-induced stomatal closure in Arabidopsis guard cells. Plant Cell Physiol 57:1779–1790. https://doi.org/10.1093/pcp/pcw102
Yoshida Y, Sano R, Wada T, Takabayashi J, Okada K (2009) Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Develop 136:1039–1048. https://doi.org/10.1242/dev.030585
Yu J, Zhang Y, Di C, Zhang Q, Zhang K, Wang C, You Q, Yan H, Dai SY, Yuan JS (2015) JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. J Exp Bot 67:751–762. https://doi.org/10.1093/jxb/erv487
Zander M, La Camera S, Lamotte O, Métraux JP, Gatz C (2010) Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J 61:200–210. https://doi.org/10.1111/j.1365-313X.2009.04044.x
Zhai Q, Yan L, Tan D, Chen R, Sun J, Gao L, Dong MQ, Wang Y, Li Ch (2013) Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet 9(4):e1003422. https://doi.org/10.1371/journal.pgen.1003422
Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C (2015) Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27:2814–2828. https://doi.org/10.1105/tpc.15.00619
Zhang ZP, Baldwin IT (1997) Transport of [2-14C] jasmonic acid from leaves to roots mimics wound-induced changes in endogenous jasmonic acid pools in Nicotiana sylvestris. Planta 203:436–441
Zhang X, Zhu Z, An F, Hao D, Li P, Song J, Yi C, Guo H (2014) Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26:1105–1117. https://doi.org/10.1105/tpc.113.122002
Zhang F, Yao J, Ke J, Zhang L, Lam VQ, Xin XF, Zhou XE, Chen J, Brunzelle J, Griffin PR, Zhou M, Xu HE, Melcher K, He SY (2015) Structural basis of JAZ repression of MYC transcription factors in jasmonate signaling. Nature 525(7568):269–273. https://doi.org/10.1038/nature14661
Zhang X, Li M, Yang HH, Li XX, Cui ZJ (2018) Physiological responses of Suaeda glauca and Arabidopsis thaliana in phytoremediation of heavy metals. J Environ Manag 223:132–139. https://doi.org/10.1016/j.jenvman.2018.06.025
Zhang Y, He P, Ma X, Yang Z, Pang C, Yu J, Wang G, Friml J, Xiao G (2019) Auxin-mediated statolith production for root gravitropism. New Phytol 224:761–774. https://doi.org/10.1111/nph.15932
Zhao SY, Ma QF, Xu X, Li GZ, Hao L (2016) Tomato Jasmonic acid-deficient mutant spr2 seedling response to Cadmium stress. J Plant Growth Regul 35:603–610. https://doi.org/10.1007/s00344-015-9563-0
Zheng XY, Weaver NS, Weiqing Z, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11:587–596. https://doi.org/10.1016/j.chom.2012.04.014
Zhu Z, Lee B (2015) Friends or foes: new insights in jasmonate and ethylene co-actions. Plant Cell Physiol 56(3):414–420. https://doi.org/10.1093/pcp/pcu171
Zhu Z, An F, Feng Y, Li P, Xue L, Mu A, Jiang Z, Kim JM, To TK, Li W et al (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108:12539–12544. https://doi.org/10.1073/pnas.1103959108
Zhu X, Chen J, Xie Z, Gao J, Ren G, Gao S, Zhou X, Kuai B (2015) Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J 84:597–610. https://doi.org/10.1111/tpj.13030
Funding
This review received no external funding.
Author information
Authors and Affiliations
Contributions
MG and FB wrote the manuscript. AS and ML read and revised the manuscript. All the authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This review does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Aryadeep Roychoudhury.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ghorbel, M., Brini, F., Sharma, A. et al. Role of jasmonic acid in plants: the molecular point of view. Plant Cell Rep 40, 1471–1494 (2021). https://doi.org/10.1007/s00299-021-02687-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-021-02687-4