Abstract
Height relates to plant architecture, lodging resistance, and yield performance. Growth-promoting phytohormones gibberellins (GAs) play a pivotal role in plant height control. Mutations in GA biosynthesis, metabolism, and signaling cascades influence plant height. Moreover, GA interacts with other phytohormones in the modulation of plant height. Here, we first briefly describe the regulation of plant height by altered GA pathway. Then, we depict effects of the crosstalk between GA and other phytohormones on plant height. We also dissect the co-localization of GA pathway genes and established quantitative genetic loci for plant height. Finally, we suggest ways forward for the application of hormone GA knowledge in breeding of crops with plant height ideotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In nature, sessile plants establish their body plan in a fixed position. Unlike animals’ hibernation and escape behavior, plants cannot evade unfavorable conditions. They have evolved intricate mechanisms to properly perceive, decode, and transmit the environmental signals. One strategy for environmental adaptation is the enhancement of plasticity and flexibility in plant architecture. The aerial and underground architecture of higher plants is determined by the shoot apical meristem (SAM) and root apical meristem (RAM), respectively. Plant growth is promoted by cell division and elongation of stem cells in the SAM and RAM. The SAM generates primary shoots, leaves, and floral organs. The axillary meristems (AMs) of shoots form a bud that is dormant or extends to generate a branch (Wang and Li 2008). The maintenance and modulation of SAM and AMs activities enable multiple dimensions of primary and lateral organs’ growth and development, by which the diverse plant architecture is shaped.
Height, key component of plant architecture, has an important bearing on both natural beauty and yield performance. The increase of grain productivity during the ‘Green revolution’ mainly attributes to better agricultural practices and improved plant height. Plant height was improved by the introduction of semi-dwarf trait (Khush 2001). The semi-dwarf habit of high-yielding varieties developed during the ‘Green revolution’ is due to a significant change in plant hormone GA biosynthesis and signaling pathway (Hedden 2003). Additionally, crosstalk between GA and other phytohormones [auxin, brassinosteroids (BRs), ethylene (ET), jasmonates (JAs), and strigolactones (SLs)] functions in plant height control. During the past decades, roles of phytohormone GA in plant height determinant have been unraveled mainly through the study of GA deficiency and overdose mutants. In this review, we not only present the scenarios of GA mutants but also provide quantitative genetic cues on the function of GA in plant height modulation. We focus on cereal crop maize and extend to other plants, including Arabidopsis, rice, wheat, sorghum, and pea, when necessary. Information from this review will strengthen our understanding of molecular bases underlying plant height and provide guidance in breeding of crops with plant height ideotypes.
GA effects on plant height: mutant scenario
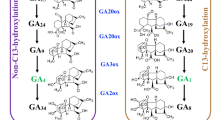
Diverse dwarf mutants related to GA biosynthesis, metabolism, and signaling have been identified. Systematical analysis of these dwarf mutants indicates that GA pathway change may lead to the altered plant height (Fig. 1). Maize dwarf mutants, anther ear1 (an1) and dwarf5 (d5), have defects in terpene synthase (TPS) enzymes involved in the early step of GA biosynthesis (Bensen et al. 1995; Fu et al. 2016). Similarly, mutation in any Arabidopsis TPS gene GA1 or GA2 results in the short stature (Sun and Kamiya 1994; Yamaguchi et al. 1998). Dwarf phenotypes of maize dwarf3 (d3), Arabidopsis ga3, and rice dwarf35 (d35) mutants attribute to lesions in P450 mono-oxygenases (P450s)-mediated steps of GA biosynthesis (Winkler and Helentjaris 1995; Helliwell et al. 1998; Itoh et al. 2004). The dwarfism phenotype is also observed in changes in 2-oxoglutaratedependent dioxygenases (2ODDs) enzymes GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox) important for the final steps of GA biosynthesis. A deficiency in Arabidopsis GA20ox gene GA5 causes the semi-dwarf phenotype (Xu et al. 1995). All mutations in GA3ox genes from three species, including Dwarf1 (D1) from maize, GA4 from Arabidopsis, and Dwarf18 (D18) from rice, result in the dwarfism (Chiang et al. 1995; Itoh et al. 2001; Chen et al. 2014). Another 2ODDs enzyme GA 2-oxidases (GA2oxs) are responsible for GA turnover. Moreover, GA degradation could be achieved by ELONGATED UPPERMOST INTERNODE (EUI) protein through epoxidation. The internode length of recessive eui mutant is significantly increased (Zhu et al. 2006). Methylation modifications by GAMT1 and GAMT2 proteins also lead to GA deactivation. Intriguingly, gross morphology of single or double GAMT1 and GAMT2 mutants is similar to that of wild-type plants (Varbanova et al. 2007).
Metabolism, signaling, and interaction atlas of plant hormone GA. For GA metabolism, bioactive GAs are synthesized from precursor GGPP catalyzed by three types of enzymes, including TPSs, P450s, and 2ODDs. Mutations in genes encoding GA biosynthetic and metabolic enzymes influence plant height. Phytohormone GA, GA receptor, and DELLA repressor shape a GA–GID1–DELLA regulatory module in the GA-signaling cascade. GA interacts with other phytohormones (auxin, BRs, ET, JAs, and SLs) in diverse biological processes, including cell elongation, and plant height modulation. BRI1 BRASSINOSTEROID-INSENSITIVE1, BZR1 BRASSINAZOLE RESISTANT1, D14 DWARF14, GGPP trans-geranylgeranyl diphosphate, JAZ JA ZIM-domain, OsEATB rice ERF protein associated with tillering and panicle branching, PIN PIN-FORMED. BR circled by relatively small round represents normal physiological condition, and BR circled by relatively large round represents BR overdose. Arrows indicate positive regulation, and T bars denote negative regulation
DELLA proteins are hub repressors of GA signaling. Gain-of-function mutation in Arabidopsis DELLA gene gibberellin insensitive (gai) leads to the dwarf phenotype (Peng and Harberd 1993). Loss-of-function mutation in rice DELLA gene Slender rice 1 (SLR1) causes dwarfism (Ikeda et al. 2001). In rice, F-box protein GIBBERELLIN-INSENSITIVE DWARF2 (GID2) is involved in the formation of the Skp–Cullin–F-box (SCF) E3 ubiquitin ligase complex that polyubiquitinates DELLA protein for its subsequent degradation by the proteasome. gid2 mutant exhibits a GA-insensitive dwarf phenotype (Sasaki et al. 2003).
GA effects on plant height: phytohormone interaction scenario
The synergy and antagonism between GA and other phytohormones (auxin, BRs, ET, JAs, and SLs) play essential roles in plant height control (Fig. 1).
To coordinate GA metabolism during organ elongation, auxin-responsive auxin response factor (ARF) and auxin/indole-3-acetic acid (Aux/IAA) family members directly up-regulate the expression of multiple GA catabolic genes GA20ox and GA2ox (Frigerio et al. 2006). Additionally, the convergence of auxin and GA cascades can be mediated by PIN-FORMED (PIN) proteins, auxin efflux carriers. In GA-deficient mutant, auxin transport is significantly inhibited because of a degradation of PIN proteins induced by GA deficiency. The reduced auxin transport influences cotyledon differentiation and root gravitropism (Willige et al. 2011). There has been evidence supporting the relationship between auxin transport defects and dwarf phenotypes of maize brachytic2 (br2) and sorghum dwarf3 (dw3) mutants (Multani et al. 2003). It invites the question of whether PIN-mediated auxin–GA interplay functions in plant height control.
Plant hormones, GA and BR, have distinct but also overlapping roles in the regulation of organ elongation and ultimate plant height (Singh and Savaldi-Goldstein 2015). GA and BR pathways are integrated through the crosstalk between BRASSINAZOLE RESISTANT1 (BZR1), transcriptional modulator of the expression of BR-responsive genes, and DELLA protein, negative regulator of GA signaling. DELLA proteins repress regulatory activities of BZR1 by the inhibition of BZR1 binding to the promoters of target genes (Gallego-Bartolomé et al. 2012). In rice, BR triggers growth responses by the modulation of GA homeostasis. For one thing, physiological concentrations of BR enhance GA abundance by the increased expression of GA biosynthetic gene GA3ox to promote growth. For another, BR overdose represses GA biosynthesis and induces GA degradation through up-regulation of GA metabolic gene GA2ox to inhibit cell elongation (Tong et al. 2014). By contrast, BR content and response changes in pea are not accompanied by the altered GA levels (Jager et al. 2005). The interplay between GA and BR pathways may be species specific. The GA–BR interaction need be surveyed in the context of specific species, developmental stages, and physiological condition.
The crosstalk between GA and ET occurs under abiotic stress conditions. Member of APETALA2 (AP2)/ethylene-responsive factor (ERF) family OsEATB down-regulates the expression of GA biosynthetic gene OsCPS2 to suppress ET-induced GA responses in rice, which leads to the inhibition of internode elongation and reduced plant height (Qi et al. 2011). During rice prolonged submergence, transcriptional regulators AP2/ERFs restrain GA biosynthesis and shoot elongation by up-regulation of GA catabolic gene GA2ox (Jung et al. 2010). Phytohormone ET interacts with GA to achieve the tradeoffs between plant growth and defense in adaption to abiotic stress.
GA and JA act antagonistically to modulate seedling growth during plant defense. In JA signaling, JA ZIM-domain (JAZ) repressors sequester MYC2, an important activator of JA responses. JA induces JAZs degradation and delays the degradation of DELLA proteins. The accumulation of DELLA proteins in turn represses JAZs and inhibits GA-promoted shoot elongation, which is beneficial to the release of activator MYC2 and the enhancement of JA-mediated defense responses (Hou et al. 2010; Wild et al. 2012; Yang et al. 2012). Similar to GA–ET interplay, the crosstalk between GA and JA confers resistance through the modulation of plant growth.
A novel class of terpenoid lactones SLs influence plant architecture, including shoot branching and plant height (Al-Babili and Bouwmeester 2015). In rice, the interaction between SL receptor DWARF14 (D14) and GA signaling repressor SLR1 has been reported, suggesting that SL and GA pathways can be integrated by the formation of D14–SLR1 complex (Nakamura et al. 2013). Although the interaction between SL receptor D14 and rice DELLA protein SLR1 has been confirmed, SLs and GAs could function independently during shoot elongation. For example, SLs promote internode elongation of pea shoot independently of GAs (de Saint Germain et al. 2013). GAs are also essential for shoot elongation. More evidence need be collected to unveil the exact role of GA–SL crosstalk in organ elongation and final plant height.
GA effects on plant height: quantitative genetics scenario
Understanding the genetic bases of plant height is not only beneficial to addressing fundamental issues in plant developmental biology but also helpful for the improvement of plant architecture in breeding programs. A number of efforts have been made to elucidate the genetic architecture of plant height. Height in some plants, such as maize and rice, has a quantitative nature (Wang and Li 2005; Peiffer et al. 2014). Quantitative genetics approaches, including linkage mapping and genome-wide association study (GWAS), have been employed to identify quantitative genetic loci for plant height.
Classical linkage mapping or QTL mapping method based on bi-parental crosses is used to reveal genetic loci influencing plant height. Over 200 QTL for maize plant height have been deposited in the MaizeGDB reservoir (Andorf et al. 2016). However, identification of candidate genes underlying these QTL for maize plant height is deemed to be a daunting task mainly because of relatively large confidence interval. Meta-analysis has been proven to be an effective approach to refine QTL interval. We meta-analyzed of QTL for maize plant height. Results indicate that some GA pathway-related genes, GA3ox2 and GA2ox, co-localize with established meta-QTL intervals (Wang et al. 2016b). Intriguingly, the aforementioned GA3ox2 in meta-QTL interval is the gene within maize plant height QTL qPH3.1 that has been map-based cloned (Teng et al. 2013). Of note, GA-deficient dwarf mutant dwarf1 (d1) analysis demonstrates that maize D1 corresponds to GA3ox2 (Chen et al. 2014). Leveraging the separate QTL mapping, integrative QTL meta-analysis, and independent mutant survey information provides new insights into the role of phytohormone GA in plant height determinant.
GWAS or association mapping based on natural populations is employed to dissect the genetic architecture of plant height. Joint linkage–linkage disequilibrium mapping identifies numerous loci influencing maize plant height. Significant association revealed by GWAS co-localizes with putative GA receptor GID1L2 (Peiffer et al. 2014). GWAS analysis of maize plant height in another separate panel identifies association near GA20ox gene essential for the conversion of precursor GA12 to bioactive GAs (Weng et al. 2011). In wheat, GA-related genes within loci for plant height have been confirmed by integrated GWAS and syntenic analysis (Zanke et al. 2014). With marker saturation, phenome advance, and algorithm optimization, novel GA pathway effectors influencing plant height are likely to be unraveled by the GWAS approach.
It should be noted that the genetic architecture of plant height in sorghum is different from that in maize. As mentioned above, maize plant height is controlled by numerous loci with minor effects. Moreover, GA pathway-related genes, including GA20ox, GA3ox, GA2ox, and GA receptor gene, are the candidates underpinning quantitative genetic loci for maize plant height (Weng et al. 2011; Teng et al. 2013; Peiffer et al. 2014). In contrast, there are four major loci (DW1, DW2, DW3, and DW4) influencing sorghum plant height (Quinby and Karper 1954). These four major loci for sorghum plant height have been further confirmed by the GWAS results (Morris et al. 2013). Intriguingly, there are not GA pathway-related genes within four sorghum plant height loci DW1–DW4. Recent GA-related mutant analysis indicates that GA-deficient dwarf phenotypes tend to pleiotropically induce culm bending in sorghum. GA-related dwarf traits are not likely to be selected during sorghum breeding due to the close relationship between GA-deficient dwarf phenotypes and negative pleiotropic effects (Ordonio et al. 2014). It is also the reason why none of GA pathway-related genes have been discovered within major loci influencing sorghum plant height.
GA effects on plant height: breeding application scenario
Efficient and sustainable increase of crop yield is an important priority in a changing world (Tester and Langridge 2010). Modification of plant height by the altered GA pathway has been proven to be effective for the improvement of crop yield performance. A landmark example is the development of high-yielding varieties during the ‘Green revolution’. These high-yielding varieties are characteristic of high capacities of nitrogen assimilation and lodging resistance, which mainly attributes to the introduction of semi-dwarf traits. The semi-dwarf habit is due to GA pathway change that is governed by GA biosynthetic gene sd1 in rice and GA signaling gene Rht in wheat (Peng et al. 1999; Monna et al. 2002; Spielmeyer et al. 2002).
With rapid advances in omics-based technologies and computational toolkit, the development of varieties with plant height ideotypes through GA pathway change can be illuminated at multiple layers (Fig. 2). For one thing, dwarf germplasm is collected and grouped into GA-sensitive and GA-insensitive types based on their GA responses. GA mutants with mild mutation effects and breeding potential can be directly adopted in breeding practice. For another, genes underlying dwarf phenotypes due to GA defects can be isolated by transposon tagging and map-based cloning methods. Cloned genes are informative for the identification, transfer, and pyramiding of elite allele. Moreover, interaction network of GA and other phytohormones can be elucidated through systems biology approaches, such as co-expression analysis and meta-analysis of hormone transcriptome (Brenner et al. 2012; Saithong et al. 2015). Hub regulators of GA-involved regulatory network in plant height control can be discovered. In addition, GA-related effectors of plant height control can be dissected by linkage mapping and GWAS approaches. The breeding-favored effectors can be recruited for the improvement and enhancement of germplasm bank specifically tailored to dwarf varieties breeding.
Breeding of crops with plant height ideotypes by altered GA pathway. GA dwarf mutants with mild mutation effects and breeding potential can be directly used in breeding practice. Causative genes underlying GA dwarf phenotypes are identified by transposon tagging and map-based cloning methods. Systems biology approaches, such as co-expression analysis and meta-analysis of hormone transcriptome, are employed for the dissection of hub regulators of GA-mediated network of plant height determinant. GA-related effectors of plant height control can be discovered by linkage mapping and GWAS approaches. To develop crops with plant height ideotypes, isolated causative genes, hub regulators, and effectors are employed for the identification, transfer, and pyramiding of elite allele through MAS and transgene strategies. GWAS genome-wide association study, MAS marker-assisted selection
GA-deficient dwarf mutants tend to exhibit negative pleiotropic effects, such as severely shortened internodes and degenerated inflorescence organs (Wang et al. 2016a). Breeding potential of severe GA-deficient mutants seems marginal. In such case, novel GA-deficient plants with mild mutation effects can be identified through the survey of geographically diverse germplasm. Alternatively, some biotechnological methods, including marker-assisted selection (MAS) and transgene can be used to modify plant height through the regulation of GA pathway (Fig. 2). The advent of CRISPR/Cas9 genome-editing platforms will facilitate the genetic modulation of plant height by altered GA pathway. Editing of Arabidopsis GA signaling gene GAI with the CRISPR/Cas9 system results in a dwarf phenotype (Feng et al. 2013). One of the keys to reshaping plant height via genetic interference of GA pathway is the selection of suitable genes that positively affect height without penalties on yield and other agronomic traits. Some GA pathway genes belong to gene family members, such as GA20oxs and GA3oxs. The expression of GA20oxs and GA3oxs genes can be finely tuned, which is beneficial to the avoidance of undesirable pleiotropy. GA20oxs and GA3oxs could be prioritized for the improvement of plant height.
GA pathway change not only affects plant height but also influences biotic and abiotic stress tolerance, floral organ morphogenesis and fertility, and quality traits (Achard et al. 2008; Plackett et al. 2012; Saville et al. 2012; Voorend et al. 2016; Wu et al. 2016). In the breeding relevant context, diverse roles of GA can be a two-edged sword. For one thing, dwarf germplasm with unfavorable pleiotropy due to GA deficiency or overdose may limit its breeding application. For another, functional diversity of phytohormone GA holds promise for improved plant architecture, stress tolerance, fertility, quality, and yield benefits through the modification of GA cascade.
Conclusion
Height, key component of plant architecture, is related to lodging resistance, photosynthetic capacity, harvest index, and ultimate yield performance. Ample evidence supports the role of phytohormone GAs in plant height control. Systematic characterization of GA pathway mutants in diverse species indicates that changes in the biosynthesis, metabolism, and signaling cascades of GA influence plant height. Some GA pathway genes belong to gene family members, such as GA20oxs, GA3oxs, and GA2oxs. Deficient in one member of GA20oxs, GA3oxs, and GA2oxs may not result in the altered phenotype due to functional redundancy. In such case, double, triple, and quadruple mutant populations need be developed to untangle their exact roles and the combinatorial interplay among GA20oxs, GA3oxs, and GA2oxs in plant height control. GA2ox enzymes are responsible for GA degradation. GA turnover can also be achieved through epoxidation and methylation modifications. Novel mechanisms of GA degradation may be discovered by forward and reverse genetic approaches. Interaction between GA and other phytohormones adds the complexity of GA-mediated regulatory network of plant height. For example, GA–BR interaction varies among species, developmental stages, and physiological condition. In such case, function of the interplay between GA and other phytohormones in plant height modulation need be surveyed in species-specific, developmental, and physiological contexts. In addition, regulatory hubs and edges of GA-mediated network could be identified through the systems biology methods, such as co-expression analysis and meta-analysis of hormone transcriptome. In some plants, plant height is quantitative, meaning that it is controlled by numerous loci with minor effects. Two complementary approaches, linkage mapping and GWAS, have been employed to identify loci for plant height. GA pathway genes co-localize with identified loci for plant height. Intriguingly, roles of some GA pathway genes in plant height have been confirmed by the mutant research and QTL meta-analysis. The integration of QTL mapping, QTL meta-analysis, and mutant survey provides complementary information for the understanding of the function of GA in plant height determinant. Moreover, combined linkage mapping and GWAS is helpful to the discovery of novel GA pathway genes relevant for plant height control. During the ‘Green revolution’, high-yielding varieties were developed by the introduction of semi-dwarf traits. The semi-dwarf habit mainly attributes to the altered GA pathway. However, the GA dwarf mutants tend to exhibit negative pleiotropic effects, which limits their breeding application. Thus, novel GA mutants with mild mutation effects and breeding potential need be isolated through the survey of geographically diverse germplasm. Moreover, diverse omics-based methods could be used to identify GA effectors for plant height control. Identified GA effectors can be engineered by some genome-editing techniques, such as CRISPR/Cas9 to reshape plant height. Additionally, isolated GA effectors could be served for the identification, transfer, and pyramiding of elite allele to boost crop productivity and food security, which is an important priority to cope with a constantly changing world.
Author contribution statement
YW conceived the topic. JZ, WL, and DD collected background information. JZ and WL contributed to the figures. YW wrote the manuscript.
References
Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20(8):2117–2129
Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66:161–186
Andorf CM, Cannon EK, Portwood JL 2nd, Gardiner JM, Harper LC, Schaeffer ML, Braun BL, Campbell DA, Vinnakota AG, Sribalusu VV, Huerta M, Cho KT, Wimalanathan K, Richter JD, Mauch ED, Rao BS, Birkett SM, Sen TZ, Lawrence-Dill CJ (2016) MaizeGDB update: new tools, data and interface for the maize model organism database. Nucleic Acids Res 44(D1):D1195–D1201
Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7(1):75–84
Brenner WG, Ramireddy E, Heyl A, Schmülling T (2012) Gene regulation by cytokinin in Arabidopsis. Front Plant Sci 3:8
Chen Y, Hou M, Liu L, Wu S, Shen Y, Ishiyama K, Kobayashi M, McCarty DR, Tan BC (2014) The maize DWARF1 encodes a gibberellin 3-oxidase and is dual localized to the nucleus and cytosol. Plant Physiol 166(4):2028–2039
Chiang HH, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7(2):195–201
de Saint Germain A, Ligerot Y, Dun EA, Pillot JP, Ross JJ, Beveridge CA, Rameau C (2013) Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiol 163(2):1012–1025
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23(10):1229–1232
Frigerio M, Alabadí D, Pérez-Gómez J, García-Cárcel L, Phillips AL, Hedden P, Blázquez MA (2006) Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 142(2):553–563
Fu J, Ren F, Lu X, Mao H, Xu M, Degenhardt J, Peters RJ, Wang Q (2016) A tandem array of ent-kaurene synthases in maize with roles in gibberellin and more specialized metabolism. Plant Physiol 170(2):742–751
Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 109(33):13446–13451
Hedden P (2003) The genes of the Green Revolution. Trends Genet 19(1):5–9
Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JA, Peacock WJ, Dennis ES (1998) Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA 95(15):9019–9024
Hou X, Lee LY, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19(6):884–894
Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13(5):999–1010
Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M (2001) Cloning and functional analysis of two gibberellin 3 beta -hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci USA 98(15):8909–8914
Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54(4):533–547
Jager CE, Symons GM, Ross JJ, Smith JJ, Reid JB (2005) The brassinosteroid growth response in pea is not mediated by changes in gibberellin content. Planta 221(1):141–148
Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol 152(3):1674–1692
Khush GS (2001) Green revolution: the way forward. Nat Rev Genet 2(10):815–822
Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y (2002) Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res 9(1):11–17
Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, Harriman J, Glaubitz JC, Buckler ES, Kresovich S (2013) Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA 110(2):453–458
Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS (2003) Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302(5642):81–84
Nakamura H, Xue YL, Miyakawa T, Hou F, Qin HM, Fukui K, Shi X, Ito E, Ito S, Park SH, Miyauchi Y, Asano A, Totsuka N, Ueda T, Tanokura M, Asami T (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat Commun 4:2613
Ordonio RL, Ito Y, Hatakeyama A, Ohmae-Shinohara K, Kasuga S, Tokunaga T, Mizuno H, Kitano H, Matsuoka M, Sazuka T (2014) Gibberellin deficiency pleiotropically induces culm bending in sorghum: an insight into sorghum semi-dwarf breeding. Sci Rep 4:5287
Peiffer JA, Romay MC, Gore MA, Flint-Garcia SA, Zhang Z, Millard MJ, Gardner CA, McMullen MD, Holland JB, Bradbury PJ, Buckler ES (2014) The genetic architecture of maize height. Genetics 196(4):1337–1356
Peng J, Harberd NP (1993) Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. Plant Cell 5(3):351–360
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400(6741):256–261
Plackett AR, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, Jikumaru Y, Benlloch R, Nilsson O, Ruiz-Rivero O, Phillips AL, Wilson ZA, Thomas SG, Hedden P (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24(3):941–960
Qi W, Sun F, Wang Q, Chen M, Huang Y, Feng YQ, Luo X, Yang J (2011) Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol 157(1):216–228
Quinby JR, Karper RE (1954) Inheritance of height in sorghum. Agron J 46(5):211–216
Saithong T, Saerue S, Kalapanulak S, Sojikul P, Narangajavana J, Bhumiratana S (2015) Gene co-expression analysis inferring the crosstalk of ethylene and gibberellin in modulating the transcriptional acclimation of cassava root growth in different seasons. PLoS One 10(9):e0137602
Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, Matsuoka M (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299(5614):1896–1898
Saville RJ, Gosman N, Burt CJ, Makepeace J, Steed A, Corbitt M, Chandler E, Brown JK, Boulton MI, Nicholson P (2012) The ‘Green revolution’ dwarfing genes play a role in disease resistance in Triticum aestivum and Hordeum vulgare. J Exp Bot 63(3):1271–1283
Singh AP, Savaldi-Goldstein S (2015) Growth control: brassinosteroid activity gets context. J Exp Bot 66(4):1123–1132
Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA 99(13):9043–9048
Sun TP, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6(10):1509–1518
Teng F, Zhai L, Liu R, Bai W, Wang L, Huo D, Tao Y, Zheng Y, Zhang Z (2013) ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J 73(3):405–416
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327(5967):818–822
Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C (2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26(11):4376–4393
Varbanova M, Yamaguchi S, Yang Y, McKelvey K, Hanada A, Borochov R, Yu F, Jikumaru Y, Ross J, Cortes D, Ma CJ, Noel JP, Mander L, Shulaev V, Kamiya Y, Rodermel S, Weiss D, Pichersky E (2007) Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19(1):32–45
Voorend W, Nelissen H, Vanholme R, De Vliegher A, Van Breusegem F, Boerjan W, Roldán-Ruiz I, Muylle H, Inzé D (2016) Overexpression of GA20-OXIDASE1 impacts plant height, biomass allocation and saccharification efficiency in maize. Plant Biotechnol J 14(3):997–1007
Wang Y, Li J (2005) The plant architecture of rice (Oryza sativa). Plant Mol Biol 59(1):75–84
Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59:253–279
Wang Y, Lu W, Chen Y, Deng D, Ding H, Bian Y, Yin Z, Zhu Y, Zhao J (2016a) Revealing physiological and genetic properties of a dominant maize dwarf Dwarf11 (D11) by integrative analysis. Mol Breed 36(3):31
Wang Y, Xu J, Deng D, Ding H, Bian Y, Yin Z, Wu Y, Zhou B, Zhao Y (2016b) A comprehensive meta-analysis of plant morphology, yield, stay-green, and virus disease resistance QTL in maize (Zea mays L.). Planta 243(2):459–471
Weng J, Xie C, Hao Z, Wang J, Liu C, Li M, Zhang D, Bai L, Zhang S, Li X (2011) Genome-wide association study identifies candidate genes that affect plant height in Chinese elite maize (Zea mays L.) inbred lines. PLoS One 6(12):e29229
Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24(8):3307–3319
Willige BC, Isono E, Richter R, Zourelidou M, Schwechheimer C (2011) Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23(6):2184–2195
Winkler RG, Helentjaris T (1995) The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. Plant Cell 7(8):1307–1317
Wu Y, Wang Y, Mi XF, Shan JX, Li XM, Xu JL, Lin HX (2016) The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet 12(10):e1006386
Xu YL, Li L, Wu K, Peeters AJ, Gage DA, Zeevaart JA (1995) The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA 92(14):6640–6644
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251
Yamaguchi S, Sun TP, Kawaide H, Kamiya Y (1998) The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol 116(4):1271–1278
Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, Lee CM, Thomashow MF, Yang Y, He Z, He SY (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109(19):E1192–E1200
Zanke CD, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, Argillier O, Stiewe G, Hinze M, Neumann K, Ganal MW, Röder MS (2014) Whole genome association mapping of plant height in winter wheat (Triticum aestivum L.). PLoS One 9(11):e113287
Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, Zhu X, Mander LN, Kamiya Y, Yamaguchi S, He Z (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18(2):442–456
Acknowledgements
We apologize for not being able to cite many relevant original papers owing to space limitations. This work was supported by the National Natural Science Foundation of China (31571671 and 31201213), the National Key Research and Development Program of China (2016YFD0101002), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Jiangsu Government Scholarship for Overseas Studies.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Neal Stewart.
Rights and permissions
About this article
Cite this article
Wang, Y., Zhao, J., Lu, W. et al. Gibberellin in plant height control: old player, new story. Plant Cell Rep 36, 391–398 (2017). https://doi.org/10.1007/s00299-017-2104-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2104-5