Abstract
Key message
Composite potato plants offer an extremely fast, effective and reliable system for studies on gene functions in roots using antisense or inverted-repeat but not sense constructs for gene inactivation.
Abstract
Composite plants, with transgenic roots on a non-transgenic shoot, can be obtained by shoot explant transformation with Agrobacterium rhizogenes. The aim of this study was to generate composite potato plants (Solanum tuberosum) to be used as a model system in future studies on root-pathogen interactions and gene silencing in the roots. The proportion of transgenic roots among the roots induced was high (80–100 %) in the four potato cultivars tested (Albatros, Desirée, Sabina and Saturna). No wild-type adventitious roots were formed at mock inoculation site. All strains of A. rhizogenes tested induced phenotypically normal roots which, however, showed a reduced response to cytokinin as compared with non-transgenic roots. Nevertheless, both types of roots were infected to a similar high rate with the zoospores of Spongospora subterranea, a soilborne potato pathogen. The transgenic roots of composite potato plants expressed significantly higher amounts of β-glucuronidase (GUS) than the roots of a GUS-transgenic potato line event. Silencing of the uidA transgene (GUS) was tested by inducing roots on the GUS-transgenic cv. Albatros event with strains of A. rhizogenes over-expressing either the uidA sense or antisense transcripts, or inverted-repeat or hairpin uidA RNA. The three last mentioned constructs caused 2.5–4.0 fold reduction in the uidA mRNA expression. In contrast, over-expression of uidA resulted in over 3-fold increase in the uidA mRNA and GUS expression, indicating that sense-mediated silencing (co-suppression) was not functional in roots. The results suggest that composite plants offer a useful experimental system for potato research, which has gained little previous attention.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Composite plants consisting of a wild-type (WT) shoot with transgenic roots were initially described in the legume Lotus corniculatus (Hansen et al. 1989). They were obtained by inoculating an apical stem explant with Agrobacterium rhizogenes used for hairy root induction and letting both the roots and the shoot grow. Induction of hairy roots has been reported for over 450 plant species, suggesting that many of them are potentially suitable for production of composite plants (Veena and Taylor 2007). Indeed, composite plants have been described in other legume species, such as Vicia hirsuta (Quandt et al. 1993) and Medicaco truncatula (Boisson-Dernier et al. 2001; Mrosk et al. 2009; Vieweg et al. 2004), and also coffee (Coffea arabica) (Alpizar et al. 2006), tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum) and Nicotiana benthamiana (Collier et al. 2005). These composite plants have been used to analyse gene functions (Boisson-Dernier et al. 2005; Küster et al. 1995), promoter activities (Dalton et al. 2009; Gherbi et al. 2008a, b; Gonzalez-Rizzo et al. 2006; Limpens et al. 2004), metabolites and phytohormone reactions (Ding et al. 2008; Isayenkov et al. 2005; Larson et al. 2001; Sun et al. 2006).
Composite potato plants were generated by Collier et al. (2005) in a study that included also a large number of other species and, therefore, the report provides limited amount of information on properties of the composite potato plants. Hyperbranching and plagiotrophy have been observed in the transgenic, hairy potato roots induced by A. rhizogenes (Pistelli et al. 2010). Such anomalies are observed occasionally in the hairy roots of various plant species as a consequence of an imbalanced hormone regulation due to the modified auxin sensitivity, which in turn is caused by the co-transformed rol genes (Christey 2001; Hashem 2009; Maurel et al. 1991; Shen et al. 1990). Treatments of hairy root cultures with auxin (mostly 1.0 mg/l) have been reported to cause either a very slight decrease (Bais et al. 2001; Gangopadhyay et al. 2011) or a very slight increase (Kim et al. 2012; Yang et al. 2010) in root biomass and very often a rapid disorganisation (Bais et al. 2001; Gangopadhyay et al. 2011; Rhodes et al. 1994). Relative to auxin, cytokinin has been shown to have even a greater impact on the growth of hairy roots. Cytokinin supplementation has been reported to decrease both biomass and elongation of hairy roots in Artemisia annua (Weathers et al. 2005) and to arrest both wild-type and hairy root growth in M. truncatula (Gonzalez-Rizzo et al. 2006). Addition of 1.0 mg/l cytokinin leads to increased lateral branching in hairy roots of A. annua (Weathers et al. 2005) and Hyoscyamus albus (Sauerwein et al. 1992).
The intensity of the reaction on exogenous hormone supply is influenced by the strain of A. rhizogenes used for transformation (Sauerwein et al. 1992).
However, in spite of the changes in hormone level, Mrosk et al. (2009) found no difference between hairy roots and WT roots in the rate of mycorrhization, morphology of the fungal structures or the molecular responses in the root-fungus interaction. Hence, normally developed roots of composite plants may be very useful for studies on root-microbe interactions, including plant-symbiont interactions (Estrada-Navarrete et al. 2006; Gherbi et al. 2008a; Küster et al. 1995; Mrosk et al. 2009; Vijn et al. 1995) and plant-pathogen interactions (Collier et al. 2005; Larson et al. 2001), including soilborne pathogens, vectors of soilborne viruses and the viruses themselves (Lennefors et al. 2006, 2008; Santala et al. 2010; Sasaya et al. 2008).
In the case of potato, resistance to soilborne viruses and other pathogens is one of the most important goals of breeding. Nevertheless, it is limited by the few known natural sources of resistance. Therefore, also transgenic resistance is seen as an important alternative (Simón-Mateo and García 2011). One of the latest strategies is a cisgenic approach employing natural alleles or genetically modified forms of host genes required by pathogens during the infection cycle and using them as resistance genes (Piron et al. 2010). Composite potatoes should be a fast and efficient system to test the influence of the genes on resistance.
It remains to be ascertained whether the roots of composite plants are also suitable to study RNA silencing, which is the most important natural, basal defence mechanism against viruses in plants (Simón-Mateo and García 2011; Smith et al. 2000). RNA silencing can target and degrade the genomes of RNA viruses and the gene transcripts of DNA viruses, as also observed with host gene mRNAs in the process called post-transcriptional gene silencing. Consequently, similar types of gene constructs can be designed to target the viruses and cellular genes with RNA silencing (Smith et al. 2000). Transgenes made of viral genes or viral genome fragments to induce RNA silencing against viruses can be categorized as sense (co-suppression), antisense or inverted-repeat constructs (Smith et al. 2000). However, studies on transgenic resistance to soilborne viruses have provided lines of evidence suggesting that RNA silencing is not particularly efficient in roots (Andika et al. 2005; Germundsson et al. 2002; Kawazu et al. 2009). There are few studies comparing the different types of gene silencing constructs in roots in the same experiments, making it difficult to elucidate which constructs could be more efficient than others. Therefore, efficient systems to test resistance genes in roots are needed, for which composite plants may offer a solution.
The aim of this study was to develop composite potato plants with transgenic roots on non-transgenic shoots. An important goal was to obtain transgenic hairy roots that do not suffer from anomaly and to test whether they were infected with Spongospora subterranea f.sp. subterranea, a soil microbe transmitting Potato mop top virus (PMTV), to the same extent as the wild-type (WT) roots. Furthermore, we aimed to test silencing of a reporter gene (uidA gene for β-glucuronidase, GUS) with various gene silencing constructs in the transgenic roots and obtain direct evidence for possible differences in their silencing activity in roots.
Materials and methods
Plasmid construction
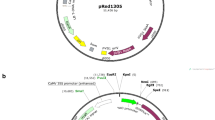
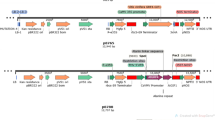
The uidA gene encoding β-glucuronidase (GUS) was amplified with the primers GusFa and GusR (Table 1) to clone uidA in antisense orientation (1,821 nt) to the vector pKOH122 (Holmström 1998; Kreuze et al. 2005). Cloning to pKOH122 was done at the XbaI/BclI site, between the Cauliflower mosaic virus 35S promoter and the terminator region of the nopaline synthase gene (nos) (Fig. 1).
Schematic presentation of GUS (β-glucuronidase) gene silencing cassettes. a Vector pKOH122; b binary vector pKOH; c asGUS silencing construct with uidA gene in antisense orientation; d hpGUS silencing construct with IV2 intron flanked by the sense and antisense fragments (194 nt) derived from the 5′-end of uidA; e srGUS silencing construct with sense fragment (194 nt) derived from the 5′-end of uidA and uidA in antisense orientation, separated by the IV2 intron; f TL-DNA integration vector E40::35S-gusint. AmpR, gene conferring ampicillin resistance; p35S, cauliflower mosaic virus 35S promoter; tnos, nopaline synthase gene terminator sequence; LB and RB, left and right borders of the T-DNA; nptII, coding region of neomycin phosphotransferase; t35S, cauliflower mosaic virus 35S terminator; uidA, coding region of β-glucuronidase; AscI, restriction sites for A; N, NotI; B, BclI; H, BamHI; X, XbaI; S, SbfI are indicated. E40, eco RI fragment 40 from the ARqua1 TL-DNA, (1.5 kb) fragment (Stougaard et al. 1987)
In order to prepare a “GUS hairpin” construct, PCR fragments (194 nt) were amplified from the 5′-end of the uidA gene using primers GusFs and GusRhp (Table 1; sense fragment, 208 nt containing the restriction sites added with the primers) and primers GusFa and GusRhp (Table 1; antisense fragment, 206 nt containing the restriction sites added with the primers). The sense fragment was ligated under the 35S promoter upstream of the IV2 intron [BclI/XbaI-digested 189-nt fragment of the second intron of potato gene ST-LS1 (Eckes et al. 1986; Vancanneyt et al. 1990)] using the NotI/BclI sites, whereas the antisense fragment was inserted between the intron and the nos terminator sequence using the XbaI/BamHI sites in pKOH122 (Fig. 1). A “GUS short-repeat” construct was prepared by replacing the uidA antisense fragment of the “GUS hairpin” construct for the full uidA gene in antisense orientation (Fig. 1).
To obtain binary plasmids asGUS, hpGUS and srGUS, the 35S-(gene fragment)-nos cassettes containing the uidA antisense fragment, “GUS hairpin” or “GUS short repeat”, respectively, where moved from the cloning vector pKOH122 to the binary vector pKOH200 (Holmström 1998; Kreuze et al. 2005) using the AscI/SbfI sites (Fig. 1). Binary vectors were introduced into the competent cells of A. rhizogenes strain ARqua1 (Quandt et al. 1993) by cold shock transformation (Simon et al. 1983).
Bacterial strains and culture media
The strains of A. rhizogenes (Table 2) were maintained on solid yeast extract broth (YEB) medium (Vervliet et al. 1975) and grown in liquid YEB medium overnight at 28 °C before plant transformation. The Agrobacterium tumefaciens strain LBA4404 (Ooms et al. 1982) was cultivated at 28 °C overnight in liquid lysogeny broth medium (Bertani 1951) for plant transformation.
Plant material and growth conditions
Potato cvs. Albatros, Desirée, Sabina, Saturna and Hansa were grown on standard Murashige Skoog (MS) (Murashige and Skoog 1962) as described by Neumann et al. (2005). Composite potatoes were cultivated on slants of potato SP medium (sugar free MS medium with 8.0 g l−1 agar; Duchefa) in Petri dishes at 20 °C under a 12-h photoperiod. Plants of Nicotiana benthamiana were grown in Vermiculite soaked with Hoagland medium (Hoagland and Arnon 1950) at 24/21 °C under a 16-h photoperiod. To obtain root lines hairy roots were individually propagated on solid hormone-free B5 medium according to Stiller et al. (1997) at 20 °C.
Plant transformation
Potato scions were transferred to the ‘Steri Vent’ plant growth containers on to fresh MS medium 3 weeks prior to induction of hairy roots with A. rhizogenes. The plantlets rooted after 2 weeks and were transferred to low light conditions for etiolation for 1 week. Three etiolated potato plantlets were transferred to a Petri dish containing SP medium and inoculated with A. rhizogenes by stab inoculation as described for V. hirsuta (Quandt et al. 1993) in the internodes of potato plantlets. The petri dishes were placed vertically in a growth cabinet (20 °C, 12 h photoperiod). 2 weeks after inoculation transgenic hairy roots emerged from the inoculation site (Fig. 2a, b). About 3 weeks after inoculation when transgenic roots were ca. 1 cm long, the non-transgenic roots growing from basal part of the scion were removed. The composite potatoes were then transferred to fresh SP medium.
Composite potato plants cv. Albatros. a, b Root formation on potato scions 2 weeks after induction of hairy roots; c, d histochemical staining of hairy roots of composite potatoes induced by A. rhizogenes strain AR30 carrying E40::35S-gusAint construct (c) and those induced by ARqua1 without a binary vector (d); Blue colour indicates GUS activity. e–h roots 8 weeks after transfer to petri dishes; e, g WT roots; f, h hairy roots on composite potato plants; i mock inoculation on the stem without root induction 8 weeks after treatment (colour figure online)
For generation of uidA expressing potato lines, the binary vector pGUS-INT (Vancanneyt et al. 1990) was introduced into A. tumefaciens strain LBA4404 (Hoekema et al. 1983) and used to transform potato cv. Albatros, as described by Düring et al. (1993). The presence of the transgene in the plant genome of regenerated shoots was demonstrated using primers (Gus-int-fw/rv; Table 1) which amplify a 913-bp fragment located in the coding region of uidA. Transgenic plants were further characterized by Southern blot and flourometric 4-methylumbelliferyl-β-D-glucuronide (MUG) assays. Southern analysis was done according to (Hühns et al. 2008) with DNA digestion by EcoRI or AflII and probed with a uidA PCR fragment of 517 bp produced using the prGUS-fw and prGUS-rv primers (Table 1).
Reaction of composite plants on cytokinin
The reaction of roots of composite plants on cytokinin was analysed according to (Gonzalez-Rizzo et al. 2006) on SP medium in comparison to the response of WT roots. In two repetitions, three to ten composite plants with roots induced by ARqua1 (A. rhizogenes strain transferring only its T-DNA into the plant) or ARnptII (ARqua1 derivate transferring nptII from a binary vector) were grown on SP medium amended with BAP (2 mg l−1) or without BAP supplementation.
Infection of the roots of composite potato plants with S. subterranea f. sp. subterranea
Rooted plantlets of the composite plants of cv. Saturna, and the wild-type (WT) plantlets of Saturna and cv. Hansa, were transferred from the MS medium to stone wool in 80 ml net plastic pots and placed in a hydropond to secure abundant root growth (Merz 1989). Seedlings of N. benthamiana were included for comparison, because N. benthamiana is commonly used in studies on transmission of PMTV by S. subterranea (Germundsson et al. 2002). After 15–20 days the plants had developed 3–4 leaves and an abundant root system had emerged. The WT roots were removed and the composite plants were transferred to 250 ml plastic pots with vermiculite. Sporeballs of S. subterranea (scraped from tuber skin lesions, dried, pulverized and stored in dark at 4 °C until use) were placed into a hole in vermiculite as inoculum and the composite potato planted in the same hole. The plants were transferred to a growth chamber and grown at 18/16 °C (day/night) under a 16-h photoperiod in open plastic containers (980 cm2), five pots in each, and drip-irrigated with nutrient solution (Hoagland and Arnon 1950) added to the water, securing a high water content necessary for sprouting of the resting sporeballs and swimming of the released zoospores. A trial consisted of at least 12 composite plants of a potato cultivar and 15 plants of N. benthamiana.
Twenty-one days after planting root samples were collected, stained with Trypan blue at 60 °C for 3 min and washed in ion-exchanged water. The stained roots of each plant were assessed for presence of zoosporangia of S. subterranea with a microscope at 100 spots chosen at random using Kole’s scale (Kole 1954) for root hair infection, where 0 = no zoosporangia; 1, 2 and 3 indicate very few, several, and many sporangia, respectively; and 4 indicates heavy infection.
GUS assay
GUS activity was demonstrated by histochemical staining (Fig. 2c) or quantified by the fluorometric assay according to Jefferson et al. (1987). For fluorometric assay around 100 mg of root material from 2 to 5 different composite plants was combined to one sample. The background level of fluorescence was determined using samples from non-transgenic material and was subtracted from the values of the GUS-transgenic samples. The total protein content of the plant samples was determined by the Bradford assay (Bradford 1976) and GUS activity calculated in relation to the protein content. The level of GUS activity in the roots induced by the A. rhizogenes strain ARqua1 on the GUS-INT-1 transgenic potato cv. Albatros was used as a standard to which the GUS activity values of other samples were compared.
RNA extraction, cDNA synthesis, qualitative and quantitative RT-PCR
For qualitative reverse transcription PCR (RT-PCR) total RNA was extracted from 70 to 200 mg of propagated hairy roots using a Quiagen RNeasy Plant Mini-Kit (Hilden, Germany) or 10–70 mg of hairy roots using a Machery-Nagel NucleoSpin RNA XS Kit (Düren, Germany). For cDNA synthesis the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas) (St. Leon-Rot, Germany) was used (Hühns et al. 2008). cDNA was produced using specific primers (Table 1) deduced from the respective genomic DNA sequences (rolA, NCBI GenBank K03313.1, Slightom et al. 1986; rolB, X03433, Furner et al. 1986; aux1 and aux2, NCBI GenBank M61151.1, Camilleri and Jouanin 1991) with the following PCR program: 5 min at 94 °C; 9 cycles of 1 min at 94 °C, 1 min at 57 °C, 2 min at 72 °C; 30 cycles of 1 min at 94 °C, 1 min at 57 °C, 2 min at 72 °C with a 10-s time increment per cycle, followed by 10 min at 72 °C for final extension. Products were separated by electrophoresis in 1.5 % agarose gels.
For quantitative RT-PCR (qRT-PCR) total RNA was extracted from the hairy roots of composite potato plants using the Trizol-like reagent (Caldo et al. 2004). RNA concentration and purity were determined spectrophotometrically. RNA (0.5 µg) was diluted to a total volume of 4 µl with nuclease free water. 0.5 µl of RQ1 buffer (Promega, Madison, USA) and 0.5 µl of RQ1 RNase free DNase (Promega, Madison, USA) were added, and the mixture was incubated at 37 °C for 30 min. DNase was inactivated by adding 0.5 µl of RQ1 stop solution.
Non-strand specific qRT-PCR and positive-strand specific qRT-PCR (pss-qRT-PCR) amplifications of uidA transcripts were carried out using a potato gene for actin (GeneBank accession no. X55746.1) as an internal control.
For qRT-PCR, cDNA was synthesized on the DNase treated RNA with Moloney murine leukaemia virus reverse transcriptase (Promega) according to the manufacturer’s instruction using oligo d(T)25N to initiate cDNA synthesis. Subsequently, cDNA was diluted to 50 µl with nuclease-free water.
The cDNA synthesis for pss-qRT-PCR was done according to the protocol of Purcell et al. (2006) with some modifications. A tagged antisense primer (GUStagR) (Table 1) was used to initiate cDNA synthesis on the uidA sense transcripts. Initiation of cDNA synthesis on actin transcripts was done with the antisense primer actinR (Table 1). A mixture (1.0 µl) containing 5 pmol of both primers was added to 5.5 µl of DNase-treated RNA and the samples were incubated at 70 °C for 10 min followed by 3 min cooling on ice. Subsequently, 1× reverse transcriptase buffer, 10 mM dithiotreitol, 0.5 mM dNTPs (Finnzymes, Espoo, Finland), 100 U of Moloney murine leukemia virus reverse transcriptase (Promega, Madison, USA), and 10 U of RNasin (Promega, Madison, USA) were added to a total reaction volume of 10 µl and incubated at 50 °C for 1 h followed by inactivation of the reverse transcriptase at 70 °C for 10 min. To remove the excess primers, the transcription reactions were treated with 2 U of Exonuclease I (Fermentas, Carlsbad, USA) in endonuclease buffer (reaction volume 20 µl) and incubated at 37 °C for 30 min. The enzyme was subsequently inactivated at 80 °C for 15 min. The reaction mix was then diluted to the final volume of 30 µl with nuclease free water.
The potato actin gene-specific primers (actinF and actinR) (Table 1) used for qRT-PCR and pss-qRT-PCR were those described by Vuorinen et al. (2010). The tag primer (Table 1) used as the reverse primer for uidA sense transcript cDNA amplification was adopted from Purcell et al. (2006), whereas the forward primer (GUSF) for the uidA sense transcript cDNA and the uidA primers for non-strand specific qPCR (GUSnsF and GUSnsR) were designed using Primer3 software (Rozen and Skaletsky 2000). Two analytical replicates of each biological replicate were tested. Quantitative PCR was conducted with LightCycler 480 (Roche, Basel, Switzerland) in a 384-well format. Each well contained 1× SYBR Green I master mix (Roche, Basel, Switzerland) with 0.45 pmol of forward and reverse primers and 5 µl of cDNA in a total volume of 15 µl. The PCR program consisted of a pre-incubation step at 95 °C for 5 min followed by 45 cycles of 10 s at 95 °C, 10 s at 60 °C and 10 s at 72 °C. To confirm that each primer pair produced only a specific, single product, a melting curve analysis was done at the end of the program, and amplification products were sequenced without cloning. Analysis of the qRT-PCR data was carried out using the E-method as described (Vuorinen et al. 2010).
Statistical analyses
For the comparisons, the t test or the F test (including the multiple comparison based on the least significant difference, LSD) was used. A P value ≤0.05 was considered significant. The analysis was calculated with IBM SPSS Statistics 19 and Microsoft Excel.
Results
Identification of the optimal combination of the A. rhizogenes strain and potato cultivar
In order to simulate a normal plant, the phenotype, hormone concentrations and sensitivity to hormones in the hairy roots of composite plants should be similar to WT roots. Because previous studies on V. hirsuta indicate that induction of normal roots requires an optimal A. rhizogenes—plant genotype combination (Pistelli et al. 2010; Quandt et al. 1993), we tested the efficiency of hairy root induction of eight strains of A. rhizogenes (Table 2) on four potato cultivars (Abatros, Desirée, Sabina and Saturna). The agropine A. rhizogenes strains induced root growth on 74–100 % of the inoculated shoots, whereas the mannopine and cucumopine strains only induced root growth on 23–77 % and 0–78 % of the inoculated shoots, respectively (Table 3). Hence, in the following, the agropine strains ARqua1 (Quandt et al. 1993), AR30 (Küster et al. 1995) and C58C1::pRi15834::pGUS-INT (Küster et al. 1995) were used for root induction. While ARqua1 and AR30 were most efficient inducers of hairy roots on cvs. Albatros, Saturna and Sabina, C58C1::pRi15834::pGUS-INT proved to be also efficient on Desirée but not on Albatros (Table 3).
All roots of the composite plants were phenotypically similar to WT roots, independent of the strain of A. rhizogenes, and no phenotypic anomaly typical for hairy roots, such as vigorous branching, was observed (Fig. 2e–h). Furthermore, wounding did not stimulate root growth on potato scions (Fig. 2i) indicating that all roots growing on the inoculation site were induced by A. rhizogenes. Accordingly, the presence of the rol RNA could be verified by qualitative RT-PCR in 100 % of the roots analysed (Fig. 3). On protein level, transgene encoded GUS activity could be demonstrated in 95–100 % of the roots (Fig. 2c, d).
Growth of roots expressing rolA and rolB in different combinations with aux genes on hormone free medium. Expression was verified via qualitative RT-PCR in two repetitions. The first box shows the WT roots without rol or aux gene expression leading to no growth. N, number of samples analysed. Error bars indicate standard deviation
When hairy roots were ca. 1 cm long, the WT roots were removed from the basal end of the scions and the composite potatoes were transferred to a fresh medium (Fig. 2h).
Hormone reactions of the transgenic, phenotypically normal hairy roots
Although the roots were phenotypically normal, hormone reactions might be changed due to the expression of the hormone genes located on the T-DNA. Compared to auxin, cytokinin supplementation has been shown to have more impact on hairy root development (Christey 2001; Hashem 2009). Therefore, we decided to analyse the reaction of transformed hairy roots to cytokinin (6-benzylaminopurine, BAP) in comparison with the response of WT roots. After 3 weeks, all roots had grown ca. 4 cm on the medium lacking BAP. In contrast, addition of BAP greatly hampered the growth of WT roots which grew only 0.1 cm in the same period of time. The growth of roots on composite plants was less affected by BAP (Fig. 4), as indicated by growth of ca. 50 % of the roots up to 0.7 cm (ARqua1) or 0.8 cm (ARnptII) during the experiment. 15 hairy roots of potato cv. Saturna induced by ARnptII and three control roots induced by ARqua1with different growth rates were excised and grown in root culture in hormone free medium. The expression of aux and rol genes was analysed via qualitative RT-PCR in order to identify possible correlation to the individual growth rate (Fig. 3). No correlation between the aux and rol gene expression and the growth rate could be observed. The three WT roots showed no expression of rol and aux genes and did not grow as a root culture on hormone-free B5 medium.
Influence of the cytokinin 6-benzylaminopurine (BAP) on growth of the non-transgenic roots as compared with the hairy roots induced by A. rhizogenes. Values indicate the percentage of roots which showed any detectable elongation during the experiment (3 weeks). Saturna niV, non-transformed potato cv. Saturna with adventitious roots; ARqua1, hairy roots of cv. Saturna induced by A. rhizogenes carrying no transgene. ARnptII, hairy roots of cv. Saturna induced by A. rhizogenes with a binary vector carrying the coding region for neomycin phosphotransferase. a–c Significant differences of means were analysed by post hoc test by the program SPSS and are indicated by different letters on the columns; N, number of plants tested. Error bars indicate standard deviation. Any outliers are marked with a circle and extreme outliers with an asterisk
Influence of the transformation vector on the percentage of roots expressing the transgene
Previous studies have shown that the proportion of uidA expressing (GUS+) roots of V. hirsuta could be increased when the uidAint gene (uidA carrying an intron) was integrated into the TL-DNA of the Ri Plasmid instead of the T-DNA of a binary vector (Horn et al. 2014 submitted). The efficiency of the system was determined by comparing the proportion of GUS+ roots induced with either C58C1::pRi15834::pGUS-INT containing uidAint gene in a binary vector (Quandt 1994) or AR30 (Küster et al. 1995), an ARqua1 derivative with uidAint gene integrated in the TL-DNA. On cvs. Albatros and Desirée AR30 induced 95–98 % roots expressing GUS, while C58C1::pRi15834::pGUS-INT induced only 87–88 % GUS+ roots (Table 4). On cv. Saturna AR30 induced less (81 %) GUS+ roots than C58C1::pRi15834::pGUS-INT (91 %). As expected the negative control ARqua1, that does not carry the uidA gene, induced no GUS+ roots (Table 4).
Infection of the roots of composite potato plants with S. subterranea
The potato powdery scab pathogen S. subterranea is a soil micro-organism that infects roots, stolons and tubers of potato. The propensity of the composite potato hairy roots for infection with S. subterranea was studied using a bioassay, in which the resting spores of the pathogen were added onto the growth medium of the composite potato plants of cv. Saturna, including non-transgenic plants of cv. Saturna with non-transformed roots as controls. Potato cv. Hansa and N. benthamiana plants susceptible to S. subterranea were included as additional controls (Andersen et al. 2002). The three independent experiments carried out with composite plants of cv. Saturna each included at least 12 composite and control potato plants. Results showed that while infection rates expressed as the proportion of infected plants were similar, the infection intensity measured on Kole’s scale (Kole 1954) was higher in the potato plants than in the N. benthamiana control plants (Fig. 5). No differences were found either in the proportion of infected plants or in the infection intensity between hairy roots of composite cv. Saturna and the non-transformed roots of Saturna and Hansa (Fig. 5).
Infection rate and the intensity of infection in hairy roots of composite potato cv. Saturna and in non-transgenic roots inoculated with S. subterranea f. sp. subterranea. A total of at least 12 plants of a type were inoculated in one experiment. Each experiment was repeated at least once. Infection rate (scale to the left) is expressed as the percentage of plants containing infected roots. Infection intensity was assessed using Kole’s scale (0–4, scale to the right). N. benthamiana plants were included for comparison. comp. Saturna, composite plants of potato cv. Saturna with hairy roots induced by AR30; Saturna niV, potato cv. Saturna with non-transgenic roots; Hansa niV, potato cv. Hansa with non-transgenic roots; N, number of experiments. a–c Significant differences of means were determined using post hoc test and are indicated by different letters on the columns. Error bars indicate standard error. Means that differ are indicated with different letters above the columns
Post-transcriptional silencing of uidA in the hairy roots of composite potato plants
In order to test the silencing of uidAint in hairy roots, a GUS+ event was produced via transformation with A. tumefaciens carrying a binary vector (pGUS-INT) (Table 2) with a uidA gene interrupted by an intron (Vancanneyt et al. 1990). Out of ten transformants, a single event with high (2.0 ± 0.6 pmol min−1 µg protein−1) and stable GUS activity in roots was selected for further studies. Southern analysis confirmed that this event (designated as Albatros GUS-INT-1) had only a single copy of the uidA in its genome. In order to trigger post-transcriptional silencing of uidA in the transgenic event Albatros GUS-INT-1, it was transformed with strains of A. rhizogenes either carrying the uidA gene on the T-DNA (strain AR30) or a binary plant transformation vector containing various uidA-derived constructs that do not express GUS but uidA-homologous single-stranded antisense (strain ARasGUS) or double-stranded RNA (strains ARhpGUS and ARsrGUS) (Table 2; Fig. 1). Strains of A. rhizogenes lacking any transgene on the T-DNA (ARqua1) or carrying a binary vector containing the neomycin phosphotransferase gene (ARnptII) were included as controls. GUS activity was measured using the MUG assay, calculated relative to the total protein content of the sample, and compared with the GUS activity of hairy roots induced with ARqua1 on Albatros GUS-INT-1 (activity = 100 %).
Following transformation of WT cv. Albatros, GUS activity was detected only in the hairy roots of the composite plants induced by transformation with the A. rhizogenes strain AR30, whereas transformation with any other strain resulted in no detectable GUS activity (Fig. 6), as expected.
Silencing of the GUS activity in hairy roots induced on the uidA-transgenic potato cv. Albatros GUS-INT-1. Hairy roots were induced by A. rhizogenes either lacking a transgene (ARqua1) or containing uidAint in the T-DNA (AR30). Furthermore, A. rhizogenes strains with binary vectors carrying antisense (ARasGUS), short repeat (ARsrGUS) or hairpin (ARhpGUS) constructs of uidAint gene, or just the coding region for neomycin phosphotransferase (ARnptII) were used (Table 2). GUS activities are shown relative to the activity detected in the roots of Albatros GUS-INT-1 ARqua1 (activity = 100 %). N number of samples analysed. a–b Significant differences of means determined by post hoc test are indicated with different letters on top of the columns. Error bars indicate standard deviation. Outliers are marked with a circle and extreme outliers with an asterisk
Transformation of the event GUS-INT-1 with ARqua1 (no transgene) and ARnptII resulted in similar levels of GUS activity reflecting the levels typical for this transgenic cultivar (Fig. 6). Compared with ARqua1 and ARntpII, transformation with AR30 resulted in 3-fold higher GUS activity, thus being even higher than observed for the hairy roots on WT cv. Albatros (Fig. 6). Finally, the hairy roots induced with strains ARasGUS, ARsrGUS and ARhpGUS carrying antisense, short-repeat or hairpin constructs of uidAint gene, respectively, exhibited significantly less GUS activity (31–44 %) than the hairy roots in plant transformed with ARqua1 or ARntpII (Fig. 6). The most pronounced reduction of GUS activity was observed in the hairy roots induced with strain ARhpGUS (30.9 ± 14.6 %).
The hairy roots were tested for uidA mRNA expression levels by qRT-PCR using sense strand specific primers. Results from three biological replicates and two technical (analytical) replicates of each biological replicate indicated that uidA transcripts were detected only in the roots induced with A. rhizogenes strain AR30 on WT cv. Albatros (Fig. 7), as expected, which indicated that pss-qRT-PCR was highly strand-specific and did not detect the transcripts of the other uidA-derived constructs, which were detected using strand non-specific primers in qRT-PCR. Melting curve analysis and sequencing of the amplification products verified target-specificity. The uidA transcript levels were similar in the hairy roots induced with strains ARqua1 and ARntpII on Albatros GUS-INT-1 (Fig. 7) and reflected the typical levels of uidA expression in the roots of the uidA-transgenic cultivar. Transformation of Albatros GUS-INT-1 with strain AR30 resulted in ca. 6-fold increase in the uidA transcript levels in the hairy roots, as compared with the roots induced with ARqua1 or ARntpII. These results further confirmed that over-expression of the uidA sense transcripts in the uidA-transgenic roots did not cause RNA silencing (co-suppression). On the other hand, induction of hairy roots with ARasGUS, ARsrGUS and ARhpGUS led to 2.5–4 fold reduction in the uidA mRNA expression levels, indicating that these constructs did cause silencing of the uidA transgene (Fig. 7).
Relative abundance of the uidA mRNA determined by qRT-PCR in hairy roots induced on non-transgenic potato cv. Albatros and uidA-transgenic cv. Albatros GUS-INT-1 using A. rhizogenes that lacks (ARqua1) or contains (AR30) the uidAint gene in the T-DNA. Furthermore, A. rhizogenes strains with binary vectors carrying antisense (ARasGUS), short repeat (ARsrGUS) or hairpin (ARhpGUS) constructs of uidAint gene, or only the coding region for neomycin phosphotransferase (ARnptII) were used (Table 2). The uidA mRNA expression levels (numbers above columns) are shown relative to one biological replicate of Albatros GUS-INT-1 ARntpII. Error bars indicate the standard deviation of the expression levels (n = 3). Significant differences were tested by F test and Tukey’s test for least significant difference (p ≤ 0.05) using log 2 transformed fold values and are indicated with different letters above the columns (a–f)
Discussion
Composite plants can be used experimentally to simulate WT plants and have proven useful for, e.g., analysis of gene functions (Estrada-Navarrete et al. 2006; Mrosk et al. 2009; Quandt et al. 1993). The system is advantageous because large numbers of independent transformation events (transgenic roots) can be cultured in a small space under highly controlled conditions and, hence, sources of uncontrolled experimental variability including positional and environmental effects are minimized. The transgenic hairy roots of the composite potato plants generated in this study were phenotypically indistinguishable from WT roots (Fig. 2e–h), in contrast to the roots on composite potato plants described by Collier et al. (2005) which exhibited some hyperbranching and plagiotrophy. Furthermore, all strains of A. rhizogenes used in our study induced phenotypically normal roots, in contrast to the studies of Quandt et al. (1993) and Thimmaraju et al. (2008) who described phenotypic changes in hairy roots of. V. hirsuta and Beta vulgaris L., respectively, associated with specific strains of A. rhizogenes.
It is important to assure that the transgenic hairy roots are not only phenotypically but also physiologically similar to the non-transgenic roots. It is well known that the responses to growth hormones such as auxin (Maurel et al. 1991; Rhodes et al. 1994; Shen et al. 1988; Spanò et al. 1988; Spena 1993; Thimmaraju et al. 2008; Yamada 1993) or cytokinin (e.g., Bais et al. 2001; Rhodes et al. 1994; Sauerwein et al. 1992; Vanhala et al. 1998) or the hormone levels (Hashem 2009; Konieczny et al. 2011; Thimmaraju et al. 2008) may differ in the hairy roots, as compared with WT roots. In addition, the intensity of reaction on hormones may also depend on the A. rhizogenes used for transformation (Sauerwein et al. 1992).
In our study hairy roots expressing one or both of the aux1 or aux2 genes were comparable with the roots expressing none of them (Fig. 3), indicating that expression of these genes is no prerequisite for hairy root induction in potato. This result is in line with other studies (Alpizar et al. 2008; Batra et al. 2004) which showed that expression of auxin genes does not play an important role for hairy root induction. However, unlike hairy roots of M. truncatula that do not differ from WT roots in their response to BAP application (Gonzalez-Rizzo et al. 2006), the potato hairy roots induced by A. rhizogenes showed decreased sensitivity to cytokinin (Fig. 4). Therefore, the hairy roots induced on potato seem to be unsuitable for experiments that depend on varied cytokinin levels.
Utility of the system also depends on the proportion of transgenic roots among the roots induced on composite plants. In the present study, no adventitious WT roots were formed at mock inoculation sites of the potato explants (Fig. 2i). At RNA level, rol gene expression could be demonstrated in 100 % of the roots examined (Fig. 3), verifying that all roots are transgenic. This is in accordance with the fact that nearly 100 % of the roots induced on cvs. Albatros and Desirée by AR30 were GUS-positive (GUS+), proving that the gene of interest is also present in the root when it is integrated into the TL-DNA. This simplifies the system significantly. Nevertheless, the efficiency of the transfer of the gene of interest may be reduced when binary vectors are used since in this case the T-DNA present on the vector has to be integrated in addition to the TL-DNA. This became obvious when hairy roots, induced on cvs. Albatros and Desirée, were analysed for the uidA expression level. Here, the 100 % frequency of transgenic roots obtained with the TL-DNA was reduced to 80 % when a binary vector was used (Table 4). This result is consistent with studies on other species (Alpizar et al. 2006; Stougaard et al. 1987). However, in potato cvs. Sabina and Saturna the percentage of GUS+ roots was similar (approx. 80 % with TL-DNA and 80–90 % for the binary vector) irrespective of the vector. Therefore, selection of the cultivar seems to be crucial in this respect. The 80–90 % of GUS+ roots obtained with a binary vector in our system is higher than transformation efficiencies reported previously (40–70 %) for other species (Boisson-Dernier et al. 2001; Collier et al. 2005; Quandt et al. 1993; Stougaard et al. 1987). Hence, the usage of binary vectors might be more appropriate in potato then in other crops.
An additional advantage of the potato roots induced by A. rhizogenes was that they showed higher expression and activity of GUS than the roots of transgenic plants (transformed by A. tumefaciens; Fig. 6). Kumar et al. (2006) also reported a 6-fold higher expression level of a transgene-encoded protein in the hairy roots of potato as compared with the roots of transgenic potato plants.
The hairy roots of composite potato plants were infected with S. subterranea f. sp. subterranea, the potato powdery scab pathogen and vector of PMTV, to the same rate as WT potato roots. These results are encouraging in terms of future use of the composite potato plants for studies on resistance to S. subterranea and the vector-transmitted PMTV. Currently, experiments are carried out by growing plants in soil or using hydroponic or aeroponic culture systems, but none of these methods allow easy, non-invasive, real-time monitoring of root-microbe or root-virus interactions. The transgenic roots of composite potato plants offer a possibility to produce large numbers of transgenic roots for study, e.g., by silencing them for specific host genes encoding proteins putatively needed for infection by PMTV or the vector, inoculate them and study them under highly controlled conditions invasively or noninvasively.
Studies on transgenic plants expressing virus-derived sequences suggest that RNA silencing, the basal antiviral defence mechanism, is less efficient in roots than leaves (Andika et al. 2005; Germundsson et al. 2002; Kawazu et al. 2009). In our previous studies, the roots of transgenic N. benthamiana plants expressing the coat protein gene sequence of PMTV were less resistant to infection with PMTV than the leaves, regardless of whether mechanical inoculation of leaves or S. subterranea mediated inoculation of roots was used (Germundsson et al. 2002). More recently, similar results have been obtained in other plant-soilborne virus interactions using sense (Andika et al. 2005) and inverted-repeat (Kawazu et al. 2009) constructs of the viral coat protein encoding sequence. Furthermore, in non-transgenic plants of N. benthamiana inoculated with Beat necrotic yellow vein virus (BNYVV), viral RNA-derived small interfering RNAs (siRNAs) accumulated in lower levels in roots than in leaves (Andika et al. 2005).
In this study, we addressed the question as to whether different RNA silencing constructs might differ or not in their efficiency to induce post-transcriptional gene silencing in roots and utilized the composite potato plants for the purpose (Figs. 6, 7). Inverted-repeat and hairpin constructs express transcripts that fold to double-stranded RNA (dsRNA), and antisense RNA can generate dsRNA by binding to the homologous positive sense gene transcript or viral ssRNA genome. In these cases, transgene expression can readily produce dsRNA that triggers the dsRNA-specific Dicer-like enzymes of the host and antiviral defence. In contrast, over-expressed sense RNAs must be first recognized by a cellular RNA-dependent RNA polymerase (RdRp), which will synthesize a complementary strand and hence create dsRNA required for recognition by Dicer (Qu et al. 2005; Vazquez et al. 2004). Results of our study clearly showed that those silencing constructs, which can generate uidA-specific dsRNA without involvement of the cellular RdRp, reduced expression of the uidA transgene significantly in the roots of composite potato plants (Fig. 7). In contrast, no sense-mediated silencing (co-suppression) was observed in roots over-expressing uidA mRNA, but accumulation of uidA mRNA and GUS showed a 3-fold increase in these roots. Collectively, results show that gene co-suppression is debilitated in roots, which may also explain why over-expression of virus-derived genes as transgenes in roots of transgenic plants confers little if any detectable resistance to single-stranded positive-sense RNA viruses, such as PMTV, BNYVV and Mirafiori lettuce big-vein associated virus (MLBVV) (Andika et al. 2005; Germundsson et al. 2002; Kawazu et al. 2006).
Since it was discovered that antisense, inverted-repeat and hairpin constructs (Schwab et al. 2006) offer more reliable and efficient silencing of target genes than the use of sense constructs (co-suppression), sense-mediated silencing has been used rarely in plants. This may be one reason why the insensitivity of roots to sense-mediated silencing has gained little attention in previous studies.
Taken together, the high transformation rates achieved with binary vectors and the specificity of A. rhizogenes for initiating adventitious root production at infected, wounded sites on the stem suggest that the composite potato plants offer a superior system compared with, e.g., tomato or tobacco (N. tabacum) with transformation rates as low as 30 and 60 %, respectively, Collier et al. (2005). The high proportions of transgenic roots on composite potato plants obtained in our study are comparable with those generated on composite L. corniculatus plants in previous studies (Stougaard et al. 1987). According to our study, antisense, inverted-repeat and hairpin constructs can be used to induce gene silencing in hairy roots of composite plants. It is possible to induce systemic silencing of a gene in a non-silenced shoot by grafting it on to a silenced rootstock (Palauqui et al. 1997; Sonoda and Nishiguchi 2000) or by expressing the silencing construct only in roots (Mohanpuria et al. 2011). Therefore, the composite potato plants described in this study may be useful also for the study of systemic antiviral silencing or induced systemic resistance (ISR) (Pieterse et al. 1998) caused by infection of the roots by viruses or microbes, respectively.
References
Alpizar E, Dechamp E, Espeout S, Royer M, Lecouls AC, Nicole M, Bertrand B, Lashermes P, Etienne H (2006) Efficient production of Agrobacterium rhizogenes transformed roots and composite plants for studying gene expression in coffee roots. Plant Cell Rep 25(9):959–967
Alpizar E, Dechamp E, Lapeyre-Montes F, Guilhaumon C, Bertrand B, Jourdan C, Lashermes P, Etienne H (2008) Agrobacterium rhizogenes-transformed roots of coffee (Coffea arabica): conditions for long-term proliferation, and morphological and molecular characterization. Ann Bot 101(7):929–940. doi:10.1093/aob/mcn027
Andersen B, Nicolaisen M, Nielsen S (2002) Alternative hosts for potato mop-top virus, genus Pomovirus and its vector Spongospora subterranea f. sp. subterranea. Potato Res 45(1):37–43. doi:10.1007/bf02732217
Andika IB, Kondo H, Tamada T (2005) Evidence that RNA silencing-mediated resistance to Beet necrotic yellow vein virus is less effective in roots than in leaves. Mol Plant Microbe Interact 18(3):194–204. doi:10.1094/MPMI-18-0194
Bais HP, Sudha G, George J, Ravishankar GA (2001) Influence of exogenous hormones on growth and secondary metabolite production in hairy root cultures of Cichorium intybus L. cv. Lucknow local. In Vitro Cell Develop Biol Plant 37(2):293–299. doi:10.1007/s11627-001-0052-8
Batra J, Dutta A, Singh D, Kumar S, Sen J (2004) Growth and terpenoid indole alkaloid production in Catharanthus roseus hairy root clones in relation to left- and right-termini-linked RiT-DNA gene integration. Plant Cell Rep 23(3):148–154. doi:10.1007/s00299-004-0815-x
Bertani G (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. The. J Bacteriol 62(3):293–300
Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14(6):695–700
Boisson-Dernier A, Andriankaja A, Chabaud M, Niebel A, Journet EP, Barker DG, de Carvalho-Niebel F (2005) MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol Plant Microbe Interact 18(12):1269–1276
Bradford MM (1976) Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem 72(1–2):248–254
Caldo RA, Nettleton D, Wise RP (2004) Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell 16(9):2514–2528. doi:10.1105/tpc.104.023382
Camilleri C, Jouanin L (1991) The TR-DNA region carrying the auxin synthesis genes of the Agrobacterium rhizogenes agropine-type plasmid pRiA4: nucleotide sequence analysis and introduction into tobacco plants. Mol Plant Microbe Interact 4(2):155–162
Christey MC (2001) Use of ri-mediated transformation for production of transgenic plants. In Vitro Cell Dev Biol Plant 37(6):687–700. doi:10.1007/s11627-001-0120-0
Collier R, Fuchs B, Walter N, Kevin Lutke W, Taylor CG (2005) Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant J 43(3):449–457. doi:10.1111/j.1365-313X.2005.02454.x
Dalton DA, Boniface C, Turner Z, Lindahl A, Kim HJ, Jelinek L, Govindarajulu M, Finger RE, Taylor CG (2009) Physiological roles of glutathione S-transferases in soybean root nodules. Plant Physiol 150(1):521–530
Ding YL, Kalo P, Yendrek C, Sun JH, Liang Y, Marsh JF, Harris JM, Oldroyd GED (2008) Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20(10):2681–2695
Düring K, Porsch P, Fladung M, Lörz H (1993) Transgenic potato plants resistant to the phytopathogenic bacterium Erwinia carotovora. Plant J 3(4):587–598
Eckes P, Rosahl S, Schell J, Willmitzer L (1986) Isolation and characterization of a light-inducible, organ-specific gene from potato and analysis of its expression after tagging and transfer into tobacco and potato shoots. Mol Gen Genet 205(1):14–22. doi:10.1007/bf02428027
Estrada-Navarrete G, Alvarado-Affantranger X, Olivares JE, Díaz-Camino C, Santana O, Murillo E, Guillén G, Sánchez-Guevara N, Acosta J, Quinto C, Li D, Gresshoff PM, Sánchez F (2006) Agrobacterium rhizogenes transformation of the Phaseolus spp.: a tool for functional genomics. Mol Plant Microbe Interact 19(12):1385–1393
Furner IJ, Huffman GA, Amasino RM, Garfinkel DJ, Gordon MP, Nester EW (1986) An Agrobacterium transformation in the evolution of the genus Nicotiana. Nature 319(6052):422–427
Gangopadhyay M, Dewanjee S, Chakraborty D, Bhattacharya S (2011) Role of exogenous phytohormones on growth and plumbagin accumulation in Plumbago indica hairy roots and conservation of elite root clones via synthetic seeds. Ind Crops Prod 33(2):445–450. doi:10.1016/j.indcrop.2010.10.030
Germundsson A, Sandgren M, Barker H, Savenkov EI, Valkonen JP (2002) Initial infection of roots and leaves reveals different resistance phenotypes associated with coat protein gene-mediated resistance to Potato mop-top virus. J Gen Virol 83(Pt 5):1201–1209
Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Peret B, Laplaze L, Franche C, Parniske M, Bogusz D (2008a) SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc Natl Acad Sci USA 105(12):4928–4932
Gherbi H, Nambiar-Veetil M, Zhong C, Felix J, Autran D, Girardin R, Vaissayre V, Auguy F, Bogusz D, Franche C (2008b) Post-transcriptional gene silencing in the root system of the actinorhizal tree Allocasuarina verticillata. Mol Plant Microbe Interact 21(5):518–524
Gonzalez-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18(10):2680–2693
Hansen J, Jorgensen JE, Stougaard J, Marcker KA (1989) Hairy roots—a short cut to transgenic root nodules. Plant Cell Rep 8(1):12–15
Hashem EA (2009) Estimation of the endogenous auxins and cytokinins in hairy roots incited on Solanum dulcamara plants by Ri plasmid of Agrobacterium rhizogenes. Aust J Basic Appl Sci 3(1):142–147
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn 347:1–32
Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA (1983) A binary plant vector strategy based on separation of vir-region and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303(5913):179–180
Holmström KO (1998) Engineering plant adaptation to water stress. Acta Universitatis Agriculturae Sueciae, Agraria Ph.D. thesis 84
Horn P, Schlichting A, Hammesfahr U, Thiele-Bruhn S, Kriete G, Baum C, Leinweber P, Broer I (2014) Influence of transgenic plants on soil: fast and sensitive in vivo studies under controlled environmental conditions to substitute long-term field trials. J Biotechnol (submitted)
Hühns M, Neumann K, Hausmann T, Ziegler K, Klemke F, Kahmann U, Staiger D, Lockau W, Pistorius EK, Broer I (2008) Plastid targeting strategies for cyanophycin synthetase to achieve high-level polymer accumulation in Nicotiana tabacum. Plant Biotechnol J 6(4):321–336
Isayenkov S, Mrosk C, Stenzel I, Strack D, Hause B (2005) Suppression of allene oxide cyclase in hairy roots of Medicago truncatula reduces jasmonate levels and the degree of mycorrhization with Glomus intraradices. Plant Physiol 139(3):1401–1410
Jefferson RA, Kavanagh TA, Bevan MW (1987) Beta-Glucuronidase (GUS) as a sensitive and versatile gene fusion marker in plants. J Cell Biochem:57–57
Kawazu Y, Fujiyama R, Sugiyama K, Sasaya T (2006) A transgenic lettuce line with resistance to both lettuce big-vein associated virus and Mirafiori lettuce virus. J Am Soc Hortic Sci 131(6):760–763
Kawazu Y, Fujiyama R, Noguchi Y (2009) Transgenic resistance to Mirafiori lettuce virus in lettuce carrying inverted repeats of the viral coat protein gene. Transgenic Res 18(1):113–120. doi:10.1007/s11248-008-9200-9
Kim YS, Li X, Park WT, Uddin MR, Park NI, Kim YB, Lee MY, Park SU (2012) Influence of media and auxins on growth and falvone production in hairy root cultures of baikal skullcap, Scutellaria baicalensis. Plant OMICS 5(1):24–27
Kole AP (1954) A contribution to the knowledge of Spongospora subterranean, the cause of powdery scab of potatoes. Tijdschrift over Plantenziekten 60:65
Konieczny R, Obert B, Bleho J, Novák O, Heym C, Tuleja M, Müller J, Strnad M, Menzel D, Šamaj J (2011) Stable transformation of Mesembryanthemum crystallinum (L.) with Agrobacterium rhizogenes harboring the green fluorescent protein targeted to the endoplasmic reticulum. J Plant Physiol 168(7):722–729. doi:10.1016/j.jplph.2010.10.013
Kreuze JF, Savenkov EI, Cuellar W, Li X, Valkonen JPT (2005) Viral class 1 RNase III involved in suppression of RNA silencing. J Virol 79(11):7227–7238. doi:10.1128/jvi.79.11.7227-7238.2005
Kumar GBS, Ganapathi TR, Srinivas L, Revathi CJ, Bapat VA (2006) Expression of hepatitis B surface antigen in potato hairy roots. Plant Sci 170(5):918–925
Küster H, Quandt HJ, Broer I, Perlick AM, Pühler A (1995) The promoter of the Vicia faba L. VfENOD-GRP3 gene encoding a glycine-rich early nodulin mediates a predominant gene expression in the interzone II-III region of transgenic Vicia hirsuta root nodules. Plant Mol Biol 29(4):759–772
Larson RL, Wintermantel WM, Hill A, Fortis L, Nunez A (2001) Proteome changes in sugar beet in response to Beet necrotic yellow vein virus. Physiol Mol Plant Pathol 72(1–3):62–72
Lennefors B-L, Savenkov EI, Bensefelt J, Wremerth-Weich E, Roggen P, Tuvesson S, Valkonen JPT, Gielen J (2006) dsRNA-mediated resistance to Beet necrotic yellow vein virus infections in sugar beet (Beta vulgaris L. ssp. vulgaris). Mol Breed 18(4):313–325. doi:10.1007/s11032-006-9030-5
Lennefors BL, van Roggen PM, Yndgaard F, Savenkov EI, Valkonen JP (2008) Efficient dsRNA-mediated transgenic resistance to Beet necrotic yellow vein virus in sugar beets is not affected by other soilborne and aphid-transmitted viruses. Transgenic Res 17(2):219–228. doi:10.1007/s11248-007-9092-0
Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55(399):983–992
Maurel C, Barbier-Brygoo H, Spena A, Tempé J, Guern J (1991) Single rol genes from the Agrobacterium rhizogenes TL-DNA alter some of the cellular responses to auxin in Nicotiana tabacum. Plant Physiol 97(1):212–216
Merz U (1989) Infectivity, inoculum density and germination of Spongospora subterranea resting spores: a solution-culture test system. EPPO Bulletin 19(3):585–592
Mohanpuria P, Kumar V, Ahuja PS, Yadav SK (2011) Agrobacterium-mediated silencing of caffeine synthesis through root transformation in Camellia sinensis L. Mol Biotechnol 48(3):235–243. doi:10.1007/s12033-010-9364-4
Mrosk C, Forner S, Hause G, Küster H, Kopka J, Hause B (2009) Composite Medicago truncatula plants harbouring Agrobacterium rhizogenes-transformed roots reveal normal mycorrhization by Glomus intraradices. J Exp Bot 60(13):3797–3807
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497
Neumann K, Stephan DP, Ziegler K, Hühns M, Broer I, Lockau W, Pistorius EK (2005) Production of cyanophycin, a suitable source for the biodegradable polymer polyaspartate, in transgenic plants. Plant Biotechnol J 3(2):249–258. doi:10.1111/j.1467-7652.2005.00122.x
Ooms G, Hooykaas PJ, Van Veen RJ, Van Beelen P, Regensburg-Tuink TJ, Schilperoort RA (1982) Octopine Ti-plasmid deletion mutants of Agrobacterium tumefaciens with emphasis on the right side of the T-region. Plasmid 7(1):15–29
Palauqui J-C, Elmayan T, Pollien J-M, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16(15):4738–4745
Pieterse CM, van Wees SC, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10(9):1571–1580
Piron F, Nicolai M, Minoia S, Piednoir E, Moretti A, Salgues A, Zamir D, Caranta C, Bendahmane A (2010) An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS One 5(6):e11313. doi:10.1371/journal.pone.0011313
Pistelli L, Giovannini A, Ruffoni B, Bertoli A, Pistelli L (2010) Hairy root cultures for secondary metabolites production. Adv Exp Med Biol 698:167–184
Purcell MK, Hart SA, Kurath G, Winton JR (2006) Strand-specific, real-time RT-PCR assays for quantification of genomic and positive-sense RNAs of the fish rhabdovirus, Infectious hematopoietic necrosis virus. J Virol Methods 132(1–2):18–24. doi:10.1016/j.jviromet.2005.08.017
Qu F, Ye X, Hou G, Sato S, Clemente TE, Morris TJ (2005) RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol 79(24):15209–15217. doi:10.1128/JVI.79.24.15209-15217.2005
Quandt HJ (1994) Entwicklung eines Transformationssystems zur Analyse der Aktivität von Nodulinpromotoren in transgenen Wurzelknöllchen von Pflanzen der Gattung Vicia—analyse der expression des chimären pVfgLb3-gusA-int Gens. Dissertation (Dr. rer. nat.), Universität Bielefeld; Fakultät für Biologie
Quandt HJ, Pühler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta : a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6(6):699–706
Rhodes MJC, Parr AJ, Giulietti A, Aird ELH (1994) Influence of exogenous hormones on the growth and secondary metabolite formation in transformed root cultures. Plant Cell Tissue Organ Cult 38(2):143–151
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Bioinformatics methods and protocols: methods in molecular biology, vol 132. Humana Press, Totowa
Santala J, Samuilova O, Hannukkala A, Latvala S, Kortemaa H, Beuch U, Kvarnheden A, Persson P, Topp K, Ørstad K, Spetz C, Nielsen SL, Kirk HG, Budziszewska M, Wieczorek P, Obrępalska-Stęplowska A, Pospieszny H, Kryszczuk A, Sztangret-Wiśniewska J, Yin Z, Chrzanowska M, Zimnoch-Guzowska E, Jackeviciene E, Taluntytė L, Pūpola N, Mihailova J, Lielmane I, Järvekülg L, Kotkas K, Rogozina E, Sozonov A, Tikhonovich I, Horn P, Broer I, Kuusiene S, Staniulis J, Uth JG, Adam G, Valkonen JPT (2010) Detection, distribution and control of Potato mop-top virus, a soil-borne virus, in northern Europe. Ann Appl Biol 157(2):163–178
Sasaya T, Fujii H, Ishikawa K, Koganezawa H (2008) Further evidence of Mirafiori lettuce big-vein virus but not of Lettuce big-vein associated virus with big-vein disease in lettuce. Phytopathology 98(4):464–468. doi:10.1094/PHYTO-98-4-0464
Sauerwein M, Wink M, Shimomura K (1992) Influence of light and phytohormones on alkaloid production in transformed root cultures of Hyoscyamus albus. J Plant Physiol 140(2):147–152. doi:10.1016/S0176-1617(11)80925-9
Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18(5):1121–1133. doi:10.1105/tpc.105.039834
Shen WH, Petit A, Guern J, Tempé J (1988) Hairy roots are more sensitive to auxin than normal roots. Proc Natl Acad Sci USA 85(10):3417–3421
Shen WH, Davioud E, David C, Barbierbrygoo H, Tempé J, Guern J (1990) High sensitivity to auxin is a common feature of hairy root. Plant Physiol 94(2):554–560
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol 1(9):784–791
Simón-Mateo C, García JA (2011) Antiviral strategies in plants based on RNA silencing. Biochim Biophys Acta 1809(11–12):722–731. doi:10.1016/j.bbagrm.2011.05.011
Slightom JL, Durand-Tardif M, Jouanin L, Tepfer D (1986) Nucleotide sequence analysis of TL-DNA of Agrobacterium rhizogenes agropine type plasmid—identification of open reading frames. J Biol Chem 261(1):108–121
Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Gene expression: total silencing by intron-spliced hairpin RNAs. Nature 407(6802):319–320. doi:10.1038/35030305
Sonoda S, Nishiguchi M (2000) Graft transmission of post-transcriptional gene silencing: target specificity for RNA degradation is transmissible between silenced and non-silenced plants, but not between silenced plants. Plant J 21(1):1–8
Spanò L, Mariotti D, Cardarelli M, Branca C, Costantino P (1988) Morphogenesis and auxin sensitivity of transgenic tobacco with different complements of Ri T-DNA. Plant Physiol 87(2):479–483
Spena A (1993) Transgenic plants altered in phytohormone metabolism. Acta Bot Gallica 140(6):693–700
Stiller J, Martirani L, Tuppale S, Chian RJ, Chiurazzi M, Gresshoff PM (1997) High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J Exp Bot 48(312):1357–1365
Stougaard J, Abildsten D, Marcker KA (1987) Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol Gen Genet 207(2):251–255
Sun JH, Cardoza V, Mitchell DM, Bright L, Oldroyd G, Harris JM (2006) Crosstalk between jasmonic acid, ethylene and nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J 46(6):961–970
Thimmaraju R, Venkatachalam L, Bhagyalakshmi N (2008) Morphometric and biochemical characterization of red beet (Beta vulgaris L.) hairy roots obtained after single and double transformations. Plant Cell Rep 27(6):1039–1052
Vancanneyt G, Schmidt R, O’Connor-Sanchez A, Willmitzer L, Rocha-Sosa M (1990) Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet 220(2):245–250
Vanhala L, Eeva M, Lapinjoki S, Hiltunen R, Oksman-Caldentey K-M (1998) Effect of growth regulators on transformed root cultures of Hyoscyamus muticus. J Plant Physiol 153(3–4):475–481. doi:10.1016/S0176-1617(98)80177-6
Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16(1):69–79. doi:10.1016/j.molcel.2004.09.028
Veena V, Taylor CG (2007) Agrobacterium rhizogenes : recent developments and promising applications. In Vitro Cell Dev Biol Plant 43(5):383–403
Vervliet G, Holsters M, Teuchy H, Vanmontagu M, Schell J (1975) Characterization of different plaque-forming and defective temperate Phages in Agrobacterium strains. J Gen Virol 26:33–48
Vieweg MF, Frühling M, Quandt HJ, Heim U, Bäumlein H, Pühler A, Küster H, Andreas MP (2004) The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Mol Plant Microbe Interact 17(1):62–69
Vijn I, Christiansen H, Lauridsen P, Kardailsky I, Quandt HJ, Broer I, Drenth J, Jensen EO, vanKammen A, Bisseling T (1995) A 200 bp region of the Pea ENOD12 promoter is sufficient for nodule-specific and nod factor induced expression. Plant Mol Biol 28(6):1103–1110
Vuorinen AL, Gammelgård E, Auvinen P, Somervuo P, Dere S, Valkonen JPT (2010) Factors underpinning the responsiveness and higher levels of virus resistance realised in potato genotypes carrying virus-specific R genes. Ann Appl Biol 157(2):229–241. doi:10.1111/j.1744-7348.2010.00424.x
Weathers PJ, Bunk G, McCoy MC (2005) The effect of phytohormones on growth and artemisinin production in Artemisia annua hairy roots. In Vitro Cell Dev Biol Plant 41(1):47–53. doi:10.1079/ivp2004604
White FF, Taylor BH, Huffman GA, Gordon MP, Nester EW (1985) Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. J Bacteriol 164(1):33–44
Yamada T (1993) The role of auxin in plant-disease development. Annu Rev Phytopathol 31(1):253–273. doi:10.1146/annurev.py.31.090193.001345
Yang YK, Lee SY, Park WT, Park NI, Park SU (2010) Exogenous auxins and polyamines enhance growth and rosmarinic acid production in hairy root cultures of Nepeta cataria L. Plant OMICS 3(6):190–193
Acknowledgments
This project was carried out as part of the Baltic Sea Region MOP-TOP project supported by the Nordic Joint Committee for Agricultural Research (Project NKJ-122). Dr. Stuart Wale at the Scottish Agricultural College is thanked for providing potato tubers with powdery scab. Financial support from the Ministry of Agriculture and Forestry (Grant 1386/39/2005) and the Academy of Finland (Grants 1134579 and 1253126) to J.P.T.V., and from The Viikki Doctoral Programme of Molecular Biosiences to J.S. is gratefully acknowledged.
Conflict of interest
There are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Eugenio Benvenuto.
Rights and permissions
About this article
Cite this article
Horn, P., Santala, J., Nielsen, S.L. et al. Composite potato plants with transgenic roots on non-transgenic shoots: a model system for studying gene silencing in roots. Plant Cell Rep 33, 1977–1992 (2014). https://doi.org/10.1007/s00299-014-1672-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-014-1672-x