Abstract
Key message
GmPAP4 , a novel plant PAP gene in soybean, has phytase activity. Over-expressing GmPAP4 can enhance Arabidopsis growth when phytate is the sole P source in culture.
Abstract
Phosphorus (P) is an important macronutrient for plant growth and development. However, most of the total P in soils is fixed into organic phosphate (Po). Purple acid phosphatase (PAP) can hydrolyze Po in the soil to liberate inorganic phosphate and enhance plant P utilization. We isolated a novel PAP gene, GmPAP4, from soybean (Glycine max). It had an open reading frame of 1,329 bp, encoding 442 amino acid residues. Sequence alignment and phylogenetics analysis indicated that GmPAP4 was similar to other plant PAPs with large molecular masses. Quantitative real-time PCR analysis showed that the induced expression of GmPAP4 was greater in P-efficient genotype Zhonghuang15 (ZH15) than in P-inefficient genotype Niumaohuang (NMH) during the periods of flowering (28–35 days post phytate stress; DPP) and pod formation (49–63 DPP). Moreover, peak expression, at 63 DPP, was about 3-fold higher in ‘ZH15’ than in ‘NMH’. Sub-cellular localization showed that GmPAP4 might be on plasma membrane or in cytoplasm. Over-expressing GmPAP4 in Arabidopsis resulted in significant rises in P acquisition and utilization compared with the wild-type (WT). Under phytate condition, transgenic Arabidopsis plants showed increases of approximately 132.7 % in dry weight and 162.6 % in shoot P content compared with the WT. Furthermore, when phytate was added as the sole P source in cultures, the activity of acid phosphatase was significantly higher in transgenic plants. Therefore, GmPAP4 is a novel PAP gene that functions in plant’s utilization of organic phosphate especially under phytate condition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As an essential nutrient, phosphorus (P) is involved in many physiological and biochemical processes in plants, such as the biosynthesis of nucleic acids, ATP production, and membrane stability (Marschner 1995; Hammond et al. 2004). Inorganic phosphate (Pi) is absorbed from the soil by the roots for plant growth and development. Approximately 50–80 % of the total P in soils is fixed into organic phosphate (Po). Of that, however, 60–80 % is inositol hexaphosphate (phytate), a form mostly unavailable to plants (Hayes et al. 2000; George and Richardson 2008). Hence, to improve the P availability is of significance for enhancement of crop yield and quality.
Acid phosphatases (APases) are hydrolytic enzymes that hydrolyze phosphoric acid monoesters and diesters or anhydrides to release Pi with acidic pH optima (Duff et al. 1994; Schenk et al. 2000a). These enzymes are generally divided into two groups, non-specific and specific, based on their particular catalyzing substrate. Purple acid phosphatases (PAPs) are primarily non-specific APases, and their presences are generally defined by the purple color of protein solutions and their tartrate-insensitive activities. According to their molecular masses and protein structures, PAPs are designated as either small (about 35 kDa) or large (about 55 kDa). The PAP active center mainly consists of five conserved motifs and seven invariant metal-ligating amino acid residues (DXG/GDXXY/GNH(D/E)/VXXH/GHXH) (Schenk et al. 1999). These motifs have important roles in coordinating bi-metal nuclear centers, i.e., Fe(III)–Zn(II) or Fe(III)–Mn(II) in plants and Fe(III)–Fe(II) in mammals (Schenk et al. 2000b; Olczak et al. 2003).
To tolerate the stress associated with a low supply of soil P, plants employ several sophisticated mechanisms to enhance P acquisition and utilization. Those steps include modifying root morphology and architecture, enhancing the expression of Pi transporter genes, and improving the activity of PAPs (Devaiah et al. 2007a, b; Zhou et al. 2008). This increase in the activities of internal and secreted PAPs under a P deficiency has been demonstrated in many species, including Arabidopsis (Hurley et al. 2010), white lupin (Gilbert et al. 1999), common bean (Liang et al. 2010), and rice (Zhang et al. 2011). Results from those investigations have suggested that PAPs play an important role in recycling the internal or external Po from old plant leaves and the rhizosphere.
Over-expressing AtPAP15 in soybean can enhance Po efficiency when phytate is supplied as the sole source of P in a culture medium (Wang et al. 2009). Arabidopsis plants transformed with MtPAP1 are better able to cope with Po (Xiao et al. 2006a). PvPAP3, a novel purple acid phosphatase, can enhance extracellular ATP utilization in common bean (Liang et al. 2010). Although 29 PAPs have been identified from Arabidopsis, only AtPAP7–AtPAP13 are induced by P starvation (Li et al. 2002). To date, although some other PAP genes were also studied, it was documented only in some model plants such as Arabidopsis and rice. While in soybean, which is a valuable crop because of its abundant protein and oil contents in the seed, only three PAP genes have been analyzed. Furthermore, there are no obvious evidences to testify that these PAPs can catalyze the phytate in rhizosphere to maintain the plant growth under low P condition. For instance, GmPAP3 is induced by NaCl stress but not by P starvation (Liao et al. 2003). Therefore, it is extremely important to search soybean PAP gene that can efficiently hydrolyze organic phosphate.
The availability of a P-efficient soybean line can help improve P acquisition and utilization (Fageria et al. 2008; Yan et al. 2001). Several soybean varieties show high P efficiency in response to low P soil (Liu 1997). Nutrient-efficient plants are generally defined as those that can produce relatively higher yields or maintain other desirable traits when nutrient levels are reduced (Wang et al. 2010). In present study, by use of two soybean genotypes, Zhonghuang15 (ZH15, P efficient) and Niumaohuang (NMH, P inefficient), with morphological and physiological differences under P-starvation condition (Li et al. 2010; Liu et al. 2012), a novel low P-induced PAP gene, GmPAP4, was isolated from the roots of the soybeans and the functional characteristics were verified of this gene.
Materials and methods
Plant materials and growth conditions
Two varieties ‘ZH15’ and ‘NMH’ previously identified as P efficient and P inefficient, respectively, were used from among hundreds of soybean varieties. These two genotypes were with morphological differences under P-starvation condition such as the relative values of shoot dry weight (0.85, 0.52), root dry weight (1.06, 0.80) and whole-plant dry weight (0.89, 0.57), as well as the physiological such as root architecture, P content, P-utilization efficiency and root APase activity (Li et al. 2010; Liu et al. 2012).
For gene cloning experiments, soybean seeds were sown in quartz sand with half-strength Hoagland nutrient solution. Seven days after germination, uniform seedlings were transplanted into full-strength Hoagland solution supplemented with phytate (1 mmol L−1). This nutrient solution was continuously aerated and replaced once a week. After 21 days of culturing under those conditions, roots were harvested and stored at −80 °C for RNA extraction, and the other seedlings were still cultured to observe the growth differences of ‘ZH15’ and ‘NMH’ under phytate condition. For quantitative real-time PCR (qRT-PCR), seeds of each variety were germinated in vermiculite with double distilled water. After 7 days of growth, the seedlings were exposed to one of two phosphate forms (1 mmol L−1 each): KH2PO4 (+Pi, control) or phytate (+Po, treatment). Roots were sampled after 0, 7, 14, 21, 28, 35, 42, 49, 56, 63, and 70 days, respectively.
cDNA and DNA cloning of GmPAP4
Total RNA was extracted from the roots, using TRIzol Total RNA Reagent (Tiangen, China) according to the manufacturer’s instructions. The RNA was treated with DNaseI (Takara, Japan) to prevent contamination with genomic DNA (gDNA). First-strand cDNA was synthesized from 1-μg samples of total RNA with a PrimeScript™ 1st Strand cDNA Synthesis Kit (Takara).
According to the sequence of AtPAP15 (GenBank Accession No. NM_111593), we performed sequence splicing via a Blast search of soybean ESTs. The putative open reading frame (ORF) of novel isolated gene, GmPAP4, was estimated from the NCBI database (http://www.ncbi.nlm.nih.gov/). Two gene-specific primers (F1, 5′-ATGGAACTCAAACAACAAAAACTC-3′; and R1, 5′-TTAGGGCGTCAAAAGTGTACTTCTG-3′) were designed according to the in silico ORF sequence. To verify the putative sequence, we amplified cDNA and gDNA from both ‘ZH15’ and ‘NMH’ with primers F1/R1. Afterward, the target fragments were cloned into the pMD18-T vector (Takara), and 10 positive clones per sample were sequenced by Sangon (China).
Bioinformatics analysis of GmPAP4
The molecular weight and isoelectric point of GmPAP4 were predicted by DNAStar. Multiple sequences were aligned with DNAMAN and a phylogenetic tree was constructed by MEGA5.1. The trans-membrane region and signal peptide were predicted by TMHMM 2.0 and SignalP 4.1 (http://www.cbs.dtu.dk/services), respectively. Sub-cellular localization was estimated with TargetP1.1 (http://www.cbs.dtu.dk/services/) and http://www.predictprotein.org/. Exons, introns and copy numbers of GmPAP4 were analyzed and predicted by the blast results through phytozome genomics program (http://www.phytozome.net).
Quantitative real-time PCR
First-strand cDNA was synthesized with a PrimeScript™ Reagent Kit and the gDNA Eraser (Takara). The qRT-PCR analysis was performed with SYBR Green Mix (Takara) and the Roche LightCycler 2.0 (Roche, Germany). The following primers were used: F2, 5′-CGACCTCTTCCTCGTAAAACC-3′ and R2, 5′-GTGCTTGTCTCCTGCCAAAG-3′ for GmPAP4; and F3, 5′-ATCTTGACTGAGCGTGGTTATTCC-3′ and R3, 5′-GCTGGTCCTGGCTGTCTCC-3′ for the housekeeping gene Actin11 (Wang et al. 2012). The qRT-PCR data, as the means of three replicates, for GmPAP4 were evaluated by the 2−ΔΔCt method (Livak and Schmittgen 2001).
Expression and purification of GmPAP4 in Escherichia coli
A partial ORF of GmPAP4, lacking the predicted signal peptide (21 amino acids of the N terminus) was sub-cloned into pET-32a and transformed into E. coli strain Transetta (DE3). Specific primers for PCR amplification were the following: F4, 5′-GGTACCTCTTACCCTCAACAGGTACACATTTC-3′ (KpnI restriction site) and R4, 5′-CTCGAGTTAGGGCGTCAAAAGTGTACTTCTG-3′ (XhoI restriction site). The transformed E. coli cells were cultured in a Luria–Bertani medium containing 100 μg mL−1 of ampicillin and 50 μg mL−1 of chloromycetin until the density reached OD600 = 0.5 (culture conditions: darkness, 37 °C, 200 rpm). Afterward, 0.8 mmol L−1 of isopropyl β-d-1-thiogalactopyranoside was added to induce gene expression. GmPAP4 was extracted and purified according to the instructions for the His-bind Purification Kit (CWBIO, China). Finally, the quality of the purified GmPAP4 was visualized by SDS-PAGE. The purified GmPAP4-His fusion protein was further verified by Western blotting. Briefly, the primary anti-His-tag monoclonal mouse antibody and the secondary goat anti-mouse IgG, AP conjugated antibody (CWBIO) were diluted at 1:3,000 and 1:5,000, respectively. Nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Amresco, USA) was used as substrate.
Assay for APase and phytase activity in GmPAP4
The activity of APase was analyzed using ρ-nitrophenol phosphate (ρ-NPP) as standard substrate (Sigma, USA). The enzyme detection system entailed 20 μL of purified GmPAP4, which was activated at 37 °C for 30 min in 1,480 μL of NaOAc buffer (200 μmol L−1; pH 5.0) that contained 1 mmol L−1 of ρ-NPP. The reaction was terminated by adding 0.5 mL of NaOH (0.5 mol L−1) for a total volume of 2.0 mL. Afterwards, the reaction product ρ-nitrophenol (ρ-NP) was measured spectrophotometrically at 410 nm (Bradford 1976). Similarly, the pH optimum for GmPAP4 was assessed at pH values of 3.0–8.0, adding different buffers that included NaOAc for pH 3.0 through 6.0 and Tris–HCl for pH 7.0 through 8.0 (Ching et al. 1987; Cashikar et al. 1997). APase activity was defined as ρ-NP released per hour per unit protein at the same condition, and the data were the means of three replicates.
Meanwhile, the activity of phytase in GmPAP4 was analyzed using phytate as substrate (sigma, USA). The molybdenum blue assay was used to measure the phytase activity (Murphy and Riley 1962; Liu et al. 2012, 2011), and the activity was defined as Pi released per hour per unit protein, and the data were the means of three replicates.
In addition, to determine substrate specificity, ρ-NPP and phytate were measured according to the methods described by Ching et al. (1987), and the final concentration of substrate (ρ-NPP and phytate) in 0.1 mol L−1 NaOAc reaction buffer (pH 5.0) was 1 mmol L−1. The released Pi content was determined by the molybdenum blue method (Murphy and Riley 1962), and the data were the means of three replicates.
Sub-cellular localization of GmPAP4-GFP
For sub-cellular location of GmPAP4, the ORF of GmPAP4 (without TAA) was cloned into the pCamE-GFP expression vector using Primers F5 (5′-GTCGACATGGAACTCAAACAACAAAAACTC-3′; SalI restriction site) and R5 (5′-GGTACCGGGCGTCAAAAGTGTACTTC-3′; KpnI restriction site). Subsequently, fusion vector pCamE-GmPAP4-GFP and controlled vector pCamE-GFP were transferred into the epidermal cells of onion (Allium cepa) by a helium-driven accelerator (PDS/1000; Bio-Rad) (Scott et al. 1999). The plasmid DNA and gold particles were prepared as described previously (Scott et al. 1999); bombardment conditions included 5 μL of gold particles, 1,100 psi pressure, and 6-cm bombardment distance. The transformed cells were incubated on MS medium at 28 °C, and sub-cellular localization of GmPAP4-GFP was observed with a fluorescence microscope after 18 h. For plasmolysis, the transformed cells were treated with 30 % sucrose solution.
Arabidopsis transformation and confirmation of transgenic plants
The ORF of GmPAP4 was cloned into the over-expression vector pCamE after PCR amplification with Primers F6 (5′-GTCGACATGGAACTCAAACAACAAAAACTC-3′; SalI restriction site) and R6 (5′-GGTACCTTAGGGCGTCAAAAGTGTACTTCTG-3′; KpnI restriction site). Binary vector pCamE-GmPAP4 was then transformed into Agrobacterium tumefaciens strain GV3101 by a freeze–thaw procedure, and was transferred into Arabidopsis through the floral dipping method (Clough and Bent 1998). The T3 transgenic homozygous lines were screened by hygromycin selection, PCR amplification, and DNA sequencing. To verify that GmPAP4 was being expressed in the transgenics, RT-PCR was performed with cDNA from the transformed and Arabidopsis ecotype Columbia (wild-type, WT) plants.
Bioassays of transgenic Arabidopsis with GmPAP4
To investigate the functions of GmPAP4, transgenic Arabidopsis (GmPAP4) and WT plants were cultured in sand separately. Half-strength Hoagland solution, supplemented with either KH2PO4 or phytate at the same P concentration (1 mmol L−1), was added to each pot in alternate weeks. One month after germination, the Arabidopsis plants were harvested for phenotypic observations, as well as physiological and biochemical determinations. To study the production of primary and lateral roots, we cultured transgenic and WT plants on MS medium containing KH2PO4 or phytate as the sole P source, and recorded lengths and numbers from 14-day-old seedlings. These experiments were repeated three times, with ten plants per replicate.
Total P contents in the shoots were analyzed as described by Ames (1966). Fresh tissues were dried at 80 °C for 24 h and flamed to ashes, which were then incubated in 100 μL of 30 % HCl and 10 % HNO3. Afterwards, 20-μL dissolved samples were mixed with 1.48 mL of distilled deionized water, to which we added 500 μL of a Pi reaction buffer (5 % ammonium molybdate solution and 10 % ascorbic acid; 6:1, v:v). The reaction mixture was incubated at 37 °C for 1 h. Shoot P contents were determined at 820 nm with a spectrophotometer, and were calculated according to a premade standard curve. The experiments were repeated three times, with six plants per replicate.
To quantify chlorophyll, transgenic and WT plants were grown on +Pi or +Po media for 4 weeks. Fresh leaves were harvested and soaked for 1 day under darkness in absolute ethanol to achieve full decolorization. The eluents were evaluated at 649 and 665 nm. Chlorophyll concentrations were computed based on a previous method (Kooten and Snel 1990). Experiments were repeated three times, with six plants per replicate.
For measuring APase activity in the rhizosphere, 20-day-old seedlings of both transgenic and WT lines were transferred to 2-mL Eppendorf tubes containing 1.5 mL of a liquid medium supplemented with 1 mmol L−1 of ρ-NPP. After being held for 1 day at 24 °C, 500 μL of 0.5 mmol L−1 NaOH was added to terminate the reaction. Absorbance was measured at 410 nm, and the situ staining was also completed using ρ-NPP as substrate (Wang et al. 2009; Liu et al. 2012). APase activity was expressed as ρ-NP released per hour per plant. All experiments were repeated three times, with five plants per replicate.
To measure phytase activity in the rhizosphere, we transferred 20-day-old seedlings, both transgenic and WT, to 2-mL Eppendorf tubes containing 1.5 mL of a liquid medium with 1 mmol L−1 phytate. After 2 days at 24 °C, the liquid medium was carbonized with 30 % HCl and 10 % HNO3. We then incubated 1 mL of this carbonized medium plus 500 μL of P reaction buffer with 5 % ammonium molybdate solution and 10 % ascorbic acid (6:1, v:v) at 37 °C for 1 h. The P content in the liquid medium was determined at 820 nm with a spectrophotometer (Murphy and Riley 1962). Phytase activity was defined as Pi released per hour per plant. All experiments were repeated three times, with five plants per replicate.
A Western blotting experiment was performed to detect the secreted GmPAP4-GFP in liquid medium. The seeds of over-expressing GmPAP4-GFP and WT Arabidopsis were sown. Then, 20-day-old seedlings were transferred into liquid medium. After 2 days, the liquid medium was collected, and 1/10 volume 100 % trichloroacetic acid was added, mixed and kept overnight at 4 °C. The mixture was centrifuged at 4 °C 12,000 revolutions per minute for 15 min. Then the supernatant was abandoned and the protein precipitation was washed twice with 1 mL cold acetone (acetone kept at −20 °C). The protein was dried and dissolved with double distilled water for Western blotting. The fusion GmPAP4-GFP protein was determined on SDS-PAGE. Subsequently, Western blotting was performed. Briefly, the primary anti GFP-tag mouse monoclonal antibody and the secondary goat anti-mouse IgG, AP conjugated antibody (CWBIO) were diluted at 1:2,000 and 1:4,000, respectively. Nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Amresco) was used as substrate.
Data analysis
All data were analyzed by one-way ANOVA, using the LSD at P < 0.05 or P < 0.01 to identify the differences between the observations with the statistical program SPSS 17.0 (Kuang et al. 2009).
Results
Molecular cloning and bioinformatics analysis of GmPAP4
By amplifying the cDNA and gDNA sequences of GmPAP4, an ORF of 1,329 bp (GenBank Accession No. HQ162477) was determined, encoding a polypeptide of 442 amino acid residues. Bioinformatics analysis suggested that the molecular mass was approximately 50.3 kDa, with an isoelectric point at 6.14.
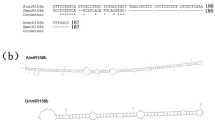
Protein alignments indicated that GmPAP4 shared significant similarity with other plant PAPs in their conserved domain and metal-binding residues (Fig. 1). The protein contained five distinct conserved motifs and seven invariant metal-coordinating amino acid residues. Phylogenetics analysis of these PAPs revealed two distinct groups: high molecular mass (Group I) and low molecular mass (Group II). Here, GmPAP4 was classified into Group I, having high similarity with AtPAP23 (Fig. 2).
Phylogenetic tree constructed with MEGA 5.1 for sequences of GmPAP4, GmPAP1 (AF19820), MtPAP1 (AAX20028), AtPAP10 (NP_179235), SpPAP (BAA92365), GmPAP3 (NP_001236677), AtPAP26 (NP_198334), OsPAP (AAM00197), AtPAP5 (NP_564619), AtPAP11 (NP_179405), AtPAP6 (NP_176033), AtPAP23 (NP_193106), AtPAP15 (NP_187369), GmPhy (AAK49438), StPAP1 (AAT37529), AtPAP3 (NP_172923), AtPAP8 (NP_178298), AtPAP7 (NP_178297), and PvPAP3 (ACO25293). PAP groups were marked I and II. Bootstrap values indicated major branches as percentages
Bioinformatics analysis showed that the 21 amino acids of the N terminus in GmPAP4 were considered as a signal peptide sequence, which indicated that the protein might be on the plasma membrane or be secreted outside of the cells. Meanwhile, the results by blasting against the sequence of soybean genome showed that GmPAP4 contained five exons and four introns, which was identical to the structure of AtPAP15. And there were two copies of GmPAP4 in soybean genome, chromosomes 2 and 14 with one copy each. In addition, the sequence of GmPAP4 in ‘ZH15’ and ‘NMH’ shared 100 % identical via the detections of PCR and DNA sequencing.
Expression patterns for GmPAP4 in soybean
We monitored the temporal expression of GmPAP4 in roots from ‘ZH15’ and ‘NMH’ soybeans under two types of P availability. When phytate was the sole source, the pattern of expression differed between varieties (Fig. 3). For ‘ZH15’, expression was much higher at 28–35 and 49–63 days post phytate stress (DPP), with the peak occurring at 63 DPP. By contrast, expression in ‘NMH’ was relatively consistent over time, with peak levels being only 1.4-fold higher than what was recorded on Day 0 of phytate treatment. When relative expression was compared between ‘ZH15’ and ‘NMH’, levels of induced transcripts were higher in the former from 28 to 35 DPP and from 49 to 63 DPP, with peak transcription being approximately threefold higher in ‘ZH15’ than in ‘NMH’ at 63 DPP.
Expression of GmPAP4 in roots from ‘ZH15’ and ‘NMH’ soybean. Seven-day-old plants were treated with either phytate as P deficiency or KH2PO4 as control for 0, 7, 14, 21, 28, 35, 42, 49, 56, 63, or 70 days. Total RNA isolated from roots was used for qRT-PCR. Data were evaluated by 2−ΔΔCT method and were the means of three replicates. Error bars represented SE for all samples tested. Asterisks indicated that values were significantly different between two varieties at P < 0.05
Prokaryotic expression and GmPAP4 activity
A specific electrophoretic band on the SDS-PAGE showed a molecular mass of about 61.2 kDa (Fig. 4a), which was further confirmed by Western blotting (Fig. 4b). The purified GmPAP4-His fusion protein was nearly consistent with the predicted size.
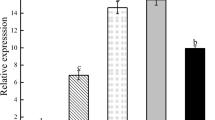
Assays of APase and phytase activity. a Expression of GmPAP4 in E. coli. GmPAP4 sequence was cloned into pET-32a and transferred to BL21 strain. Cells were induced by IPTG, and induced protein was separated via SDS-PAGE. Lanes: M protein marker, 1 control, 2 induced BL21 carrying pET-32a, 3 induced BL21 carrying pET-32a-GmPAP4, 4 purified recombinant GmPAP4. b Western blotting analysis of purified GmPAP4-His fusion protein. c Bacterial cells expressing GmPAP4 had APase activity, which was quantified with synthetic substrate ρ-NPP, expressed as ρ-NP released per hour per unit protein. d GmPAP4 was an APase with pH optimum of 5.0. e Phytate was used as substrate to measure the GmPAP4 phytase activity. Phytase activity in pET-32a and GmPAP4, expressed as Pi released per hour per unit protein. Data were the means of three replicates; error bars represented SE of all samples tested. Asterisks indicated that values were significantly different at P < 0.05
Enzymatic assays demonstrated that the pure GmPAP4 protein had significant APase activity when compared with the control products from pET-32a (P < 0.05; Fig. 4c). Maximum APase activity was noted at pH 5.0 (Fig. 4d). Furthermore, when phytate was used as substrate, GmPAP4 also had a significant phytase activity compared with pET-32a control (P < 0.05; Fig. 4e). The substrate specificity of purified GmPAP4 was examined. APase activity using ρ-NPP as substrate was set as 100 % for comparison with the phytase activity using phytate. Interestingly, GmPAP4 exhibited the relatively high activity with phytate (45 %) (Supplementary Table 1).
Sub-cellular localization of the GmPAP4 protein
Results from the bioinformatics analysis indicated that the 21 amino acid residues in the N terminus of the GmPAP4 could be considered as signal peptide sequences. We had predicted that GmPAP4 was either attached to the plasma membrane or secreted outside of the cell. To test that prediction, we constructed the pCamE-GmPAP4-GFP vector for transient expression in onion epidermal cells. Plasmolysis experiment was used to check whether GmPAP4 was located on plasma membrane or cell wall. The results clearly showed most of pCamE-GmPAP4-GFP fluorescence signals were observed on the plasma membrane or in cytoplasm, while the signals of controlled vector (pCamE-GFP) were detected throughout the intracellular cells (Fig. 5). In addition, we also observed the fluorescence of pCamE-GmPAP4-GFP moved from internal cell to plasma membrane (Supplementary Fig. 1).
Characterization of transgenic Arabidopsis under organic P conditions
Over-expression of GmPAP4 promotes Arabidopsis growth under phytate condition
The T3 transgenic homozygous Arabidopsis plants were confirmed by PCR, RT-PCR (Fig. 6a) and DNA sequencing. When grown with KH2PO4, morphology was very similar between transgenic and WT plants. Under phytate condition, however, the transgenic plants were more robust and had longer petioles (Supplementary Fig. 2) when compared with the WT (Fig. 6b). When plants were grown on +Pi or +Po media, primary root lengths were similar between the transgenic and WT (Fig. 6c). However, the transgenic plants had 35 % more lateral roots, which was a significant increase (P < 0.05) over that of the WT (Fig. 6d). The chlorophyll concentration was 10.8 % lower in the transgenic plants under phytate conditions (Fig. 7a).
Phenotypes of transgenic (GmPAP4) and wild-type (WT) Arabidopsis under different P conditions. a Transcription analysis of GmPAP4 in T3 transgenic lines 1 through 4. Actin11 served as internal control. b The growth of plants after 30 days in sand culture containing either KH2PO4 (+Pi) or phytate (+Po); P concentration was 1 mmol L−1. c Lengths of primary roots from transgenic and WT plants under +Pi or +Po. d Numbers of lateral roots from transgenic (GmPAP4) and WT plants under +Pi or +Po. Data were the means of three replicates (ten plants each). Error bars represented SE of all samples tested. Asterisks indicated that values were significantly different between transgenic plants and WT at P < 0.05
Biochemical measurements of transgenic (GmPAP4) and wild-type (WT) Arabidopsis under different P conditions. a Chlorophyll concentrations in transgenic (GmPAP4) and wild-type (WT) Arabidopsis grown in sand for 30 days under +Pi or +Po. b Dry weights from WT and GmPAP4 plants. c P contents in WT and GmPAP4 plants. For a–c, data were the means of three replicates (six plants each). Error bars represented SE of all samples tested. Asterisks indicated that values were significantly different between transgenic plants and WT at P < 0.01
Biomass and P content are increased in GmPAP4 over-expressing Arabidopsis
Transgenic Arabidopsis plants had markedly enhanced dry weights under phytate conditions, being approximately 132.7 % higher (P < 0.01) than in the WT (Fig. 7b). Furthermore, the transgenics had significantly greater P contents (P < 0.01) that were 162.6 % higher than in the WT (Fig. 7c). However, under +Pi conditions, those traits did not differ between the transgenics and WT.
Over-expressing GmPAP4 improves Arabidopsis root APase and phytase activity
Compared with the WT, secreted APase activity was 22.3 % higher in transgenic plants that over-expressed GmPAP4 (Fig. 8a). Moreover, the culture solution for the transgenics showed more intense yellow coloring (Fig. 8b). Meanwhile, the secreted phytase activity increased by 232.1 % in the transgenic plants when phytate was added to the solution as the sole P source (Fig. 8c). In addition, in transgenic Arabidopsis, GmPAP4-GFP fusion protein secreted into the liquid medium was verified by Western blotting analysis with GFP-tag antibody (Fig. 8d, e). This demonstrated that GmPAP4, a novel purple acid phosphatase gene, functioned in hydrolyzing extracellular phytate and improved the utilization of organic phosphate by Arabidopsis plants.
Assays of enzyme activity in transgenic Arabidopsis. a APase activity in wild-type (WT) and transgenic Arabidopsis (GmPAP4), expressed as ρ-NP released per hour per plant. b Secretion of APase from roots of WT and GmPAP4, indicated by yellow color. c Phytase activity in WT and GmPAP4, expressed as amount of Pi released per hour per plant. Data were the means of three replicates (five plants each). Error bars represented SE of all samples tested. d Detection of total protein in root exudates by SDS-PAGE analysis. M protein marker, 1 wild-type Arabidopsis (WT), 2 transgenic Arabidopsis (GmPAP4). e Detection of GmPAP4-GFP in root exudates by Western blotting. 3 wild-type Arabidopsis (WT), 4 transgenic Arabidopsis (GmPAP4) (color figure online)
Discussion
Although some APase genes have been cloned and shown to be functional in some model plant species (Gilbert et al. 1999; Hegeman and Grabau 2001; Xiao et al. 2006b; Hurley et al. 2010; Liang et al. 2010; Zhang et al. 2011), only a few such genes have been characterized in soybean. More importantly, there were no direct evidences to testify that these PAPs could catalyze the phytate in culture medium to maintain the plant normal growth and development under low P condition. For example, GmPAP1 and GmPAP2 were only confirmed to be induced by low P stress, and GmPAP3 was approved to associate with the adaptation of soybean to NaCl stress, rather than to low P starvation (Liao et al. 2003). Thus, to obtain a soybean PAP gene related to phytate utilization in rhizosphere, we performed RT-PCR to clone and sequence GmPAP4 from soybean roots. The Blast search indicated that GmPAP4 was a purple acid phosphatase. Multiple alignment and phylogenetics analysis revealed high similarities between GmPAP4 and other PAPs in the high molecular mass Group I. Its conserved domain had five distinct conserved motifs and seven invariant metal-coordinating residues that were typical of PAPs. Furthermore, many obviously experimental evidences testified that GmPAP4 enhanced extracellular phytate utilization in Arabidopsis and soybean subsequently.
As species have evolved, plants have developed a series of adaptive strategies to improve P acquisition and utilization from the soil, such as altering root morphology and architecture or enhancing APase activity (Chiou and Lin 2011). The diverse mechanisms that operate in response to low phosphorus are controlled by a coordinated array of genes. The polyploidy soybean has a large and complex genome, and most of its genes have multiple copies, thereby leading to different patterns of expression (Schmutz et al. 2010). The exon–intron structure of its genes also influences transcription via alternative splicing (Fedorova and Fedorov 2003; He et al. 2007, 2009). We found that the gDNA sequence of GmPAP4 had five exons and four introns, which was consistent with our bioinformatics predictions. Because two varieties did not differ in either their cDNA or gDNA sequences, it may be concluded that GmPAP4 was conserved, similar to that noted with the deduced sequences for the whole genome of ‘Williams 82’ soybean (Supplementary Fig. 3; Schmutz et al. 2010).
Analysis of temporal expression by GmPAP4 showed that, after phytate treatment, the induced accumulation of transcripts was significantly greater at 28–35 DPP and 49–63 DPP in ‘ZH15’, with levels always being higher than in ‘NMH’, where expression remained low and relatively stable over time. The two spans parallel the stages of flowering (28–35 DPP) and pod formation (49–63 DPP) are critical times in soybean growth and development when more phosphorus is required. Thus, it can be inferred from these qRT-PCR data that GmPAP4 had a vital function in hydrolyzing and utilizing soil-available organic phosphate by soybean plants. Our laboratory has previously confirmed that ‘ZH15’ and ‘NMH’ differed significantly in their phenotypic and biochemical characteristics under P-deficient conditions (Supplementary Fig. 4; Li et al. 2010; Liu et al. 2012). Those differences might be explained GmPAP4 with different patterns of expression in these two varieties.
Arabidopsis genome-wide transcriptional analysis has shown that phosphate starvation-induced genes are directly regulated by transcription factors such as PHR1 (Schachtman and Shin 2007; Chiou and Lin 2011). Bustos et al. (2010) have reported that PHR1 is a central integrator of transcription regulation, based on observations from mutants in which transcriptional activation and repression are influenced in response to Pi starvation. In rice, one or two OsPHR2 binding elements have been found in the promoter region of 10 PAP genes, implying that those genes are directly regulated by OsPHR2 (Zhou et al. 2008). Wang et al. (2011) have demonstrated that the promoter of AtPAP10 is affected by Pi deficiency, but not by K or N deficiencies. Using dose–response experiments with common bean, Liang et al. (2010) have shown that, at the same level of P, induction of PvPAP3 transcripts is stronger in the leaves and roots of ‘G19833’ than those of ‘DOR364’. All of these reports support our findings that differences exist in the transcriptional levels of GmPAP4 from ‘ZH15’ and ‘NMH’ soybeans. Thus, the above results provided new focus in the regulation of transcription factors or cis-acting elements to GmPAP4.
PAP genes play an important downstream role in the signaling network for plant P acquisition and utilization (Schachtman and Shin 2007; Chiou and Lin 2011). Various researchers have investigated whether PAP can release Pi from organic phosphate and recycle internal phosphorus under a P deficiency. Testing the theory that PAPs in plants generally function under acidic conditions, Kuang et al. (2009) have proposed that AtPAP15 shows maximum APase activity at pH 4.5, and also found that AtPAP15 has a phytase activity, which is about 4 % relative to its APase activity. In addition, recombinant AtPAP10 protein has 29 % phytase activity compared with its APase activity (Wang et al. 2011). The result of in vitro enzyme assay showed that purified GmPAP4 possessed the features of APase, with maximum activity at pH 5.0. More importantly, GmPAP4 also significantly possessed a relative higher phytase activity in vitro assay (45 %) compared with its APase activity, which indicated that GmPAP4 in eukaryotic cells might have a relative higher capability to catalyze phytate in plant rhizosphere.
To verify whether GmPAP4 in plant has a relative higher activity of APase or phytase, the GmPAP4 was over-expressed in Arabidopsis. The activity of APase in transgenic Arabidopsis showed that over-express GmPAP4 had relatively higher APase activity compared with the WT. The results from in situ staining using ρ-NPP as substrate for the activity of APase in the whole-plant roots also provided direct evidence that GmPAP4 could hydrolyze the organic P in the growth media and release Pi for plant growth. Meanwhile, bioinformatics analysis showed that GmPAP4 might be on the plasma membrane or secreted outside of the cell, and the subsequent sub-cellular localization of GmPAP4-GFP in onion epidermal cells showed that part of GmPAP4-GFP signals was detected on the plasma membrane. Furthermore, the Western blotting results of GmPAP4-GFP fusion protein extracted from GmPAP4 over-expression plants liquid culture medium demonstrated that GmPAP4 had the ability to secrete outside of plant cells. Taken together, the novel GmPAP4 could secrete outside of plant cells, and had a higher APase activity to catalyze Po in growth medium.
Worldwide, P is the most important nutrient that can limit crop production (Marschner 1995). In soils, only the inorganic form is readily available for uptake by roots. However, abundant total P is fixed and exists as Po, with approximately 60–80 % being phytate. A few PAPs play roles in hydrolyzing phytate. For example, Xiao et al. (2006a) have found that transgenic Arabidopsis plants grow better on a medium when phytate is added as the sole P source, and Wang et al. (2011) have used enzyme assays to show that AtPAP10 has moderate phytase activity. Wang et al. (2009) have reported that dry weights are significantly increased in soybean plants over-expressed AtPAP15, and that their P contents are 18.2–90.1 % higher compared to the WT while both are grown on sand cultures containing phytate. In soybean, as we know, there are no clear evidences that the existing PAPs can promote plant growth under phytate condition to date (Liao et al. 2003). Here, GmPAP4 was a phytate utilization-related PAP gene confirmed by some accurately experimental evidences. On media supplemented with phytate, GmPAP4-over-expressing Arabidopsis plants were more robust, and had longer petioles compared with the WT. The reduced concentration of chlorophyll in transgenic leaves implied that those plants had greater photosynthetic efficiency in a P-deficient environment. The significant increases in dry weight and total P content in transgenic shoots also demonstrated that GmPAP4 improved the P efficiency of Arabidopsis when phytate was added as the sole source of phosphorus.
An interesting phenomenon recorded in this work was the plant root architecture altered. It had been generally known that increased number and length of lateral roots could help plants maintain Pi level under Pi starvation (Williamson et al. 2001). One reference documented that when transgenic plants over-expressing AtPAP10 were grown under different phosphorus level conditions (0, 10, 50, 150 μM as low P level; normal P as control), the number and length of lateral roots at 150 μM P level were higher than the other P levels (Wang et al. 2011). It seemed to infer that lateral roots could grow more and better at the moderate low P stress level than the serious Pi deficiency or Pi sufficiency. In our study, transgenic plants over-expressing GmPAP4 on phytate medium had significantly more and longer lateral roots than WT plants, which indicated that GmPAP4 over-expressing made Arabidopsis hydrolyze phytate to supply a certain amount of Pi. However, whether GmPAP4 can directly or indirectly affect lateral root growth needs to be further investigated.
Abbreviations
- APase:
-
Acid phosphatase
- DPP:
-
Days post phytate stress
- GFP:
-
Green fluorescent protein
- ZH15:
-
‘Zhonghuang15’ soybean
- NMH:
-
‘Niumaohuang’ soybean
- ORF:
-
Open reading frame
- P:
-
Phosphorus
- PAP:
-
Purple acid phosphatase
- Phytate:
-
Inositol hexaphosphate
- Pi:
-
Inorganic phosphate
- Po:
-
Organic phosphate
- qRT-PCR:
-
Quantitative real-time PCR
- RT-PCR:
-
Reverse-transcription PCR
References
Ames BN (1966) Assay of inorganic phosphate, total phosphate and phosphatase. Methods Enzymol 8:115–118
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1):248–254
Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6(9):e1001102
Cashikar AG, Kumaresan R, Rao NM (1997) Biochemical characterization and subcellular localization of the red kidney bean purple acid phosphatase. Plant Physiol 114(3):907–915
Ching TM, Lin TP, Metzger RJ (1987) Purification and properties of acid phosphatase from plump and shriveled seeds of triticale. Plant Physiol 84:789–795
Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62:185–206
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Devaiah BN, Karthikeyan AS, Raghothama KG (2007a) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143(4):1789–1801
Devaiah BN, Nagarajan VK, Raghothama KG (2007b) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145(1):147–159
Duff SMG, Sarath G, Plaxton WC (1994) The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant 90(4):791–800
Fageria N, Baligar VC, Li YC (2008) The role of nutrient efficient plants in improving crop yields in the twenty first century. J Plant Nutr 31(6):1121–1157
Fedorova L, Fedorov A (2003) Introns in gene evolution. Genetica 118:123–131
George TS, Richardson AE (2008) Potential and limitations to improving crops for enhanced phosphorus utilization. Plant Ecophysiol 7:247–270
Gilbert GA, Knight JD, Vance CP, Allan DL (1999) Acid phosphatase activity in phosphorus-deficient white lupin roots. Plant Cell Environ 22(7):801–810
Hammond JP, Broadley MR, White PJ (2004) Genetic responses to phosphorus deficiency. Ann Bot 94(3):323–332
Hayes JE, Richardson AE, Simpson RJ (2000) Components of organic phosphorus in soil extracts that are hydrolysed by phytase and acid phosphatase. Biol Fertil Soils 32:279–286
He XY, He ZH, Zhang LP, Sun DJ, Morris CF, Fuerst EP, Xia XC (2007) Allelic variation of polyphenol oxidase (PPO) genes located on chromosomes 2A and 2D and development of functional markers for the PPO genes in common wheat. Theor Appl Genet 115(1):47–58
He XY, He ZH, Ma W, Appels R, Xia XC (2009) Allelic variants of phytoene synthase 1 (Psy1) genes in Chinese and CIMMYT wheat cultivars and development of functional markers for flour colour. Mol Breed 23(4):553–563
Hegeman CE, Grabau EA (2001) A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiol 126(4):1598–1608
Hurley BA, Tran HT, Marty NJ, Park J, Snedden WA, Mullen RT, Plaxton WC (2010) The dual- targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiol 153(3):1112–1122
Kooten O, Snel JF (1990) The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth Res 25(3):147–150
Kuang R, Chan KH, Yeung E, Lim BL (2009) Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol 151(1):199–209
Li DP, Zhu HF, Liu KF, Liu X, Leggewie G, Udvardi M, Wang DW (2002) Purple acid phosphatases of Arabidopsis thaliana comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277(31):27772–27781
Li XH, Gai JY, Chang WS, Zhang CY (2010) Identification of phosphorus starvation tolerant soybean (Glycine max) germplasms. Front Agric China 4(3):272–279
Liang CY, Tian J, Lam HM, Lim BL, Yan XL, Liao H (2010) Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol 152(2):854–865
Liao H, Wong FL, Phang TH, Cheung MY, Li WYF, Shao G, Yan X, Lam HM (2003) GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 318:103–111
Liu K (1997) Soybeans: chemistry, technology, and utilization. Aspen Publishers Inc., Press, Gaithersburg
Liu Y, Li XH, Sun X, Zhang CY (2012) Variation of acid phosphatase activity and analysis of genotypic difference in P efficiency of soybean under phosphorus stress. J Plant Genetic Resour 13(4):521–528
Liu JF, Zhao CY, Ma J, Zhang GY, Li MG, Yan GJ, Wang XF, Ma ZY (2011) Agrobacterium-mediated transformation of cotton (Gossypium hirsutum L.) with a fungal phytase gene improves phosphorus acquisition. Euphytica 181:31–40
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Marschner H (1995) Mineral nutrition of higher plants. Academic Press Harcourt Brace, London
Murphy J, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36
Olczak M, Morawiecka B, Watorek W (2003) Plant purple acid phosphatases-genes, structures and biological function. Acta Biochim Pol 50(4):1245–1256
Schachtman DP, Shin R (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58:47–69
Schenk G, Ge YB, Carrington LE, Wynne CJ, Searle IR, Carroll BJ, Hamilton S, de Jersey J (1999) Binuclear metal centers in plant purple acid phosphatases: Fe–Mn in sweet potato and Fe–Zn in soybean. Arch Biochem Biophys 370(2):183–189
Schenk G, Guddat LW, Ge Y, Carrington LE, Hume DA, Hamilton S, de Jersey J (2000a) Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 250(1):117–125
Schenk G, Korsinczky MLJ, Hume DA, Hamilton S, de Jersey J (2000b) Purple acid phosphatases from bacteria: similarities to mammalian and plant enzymes. Gene 255(2):419–424
Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26(6):1125–1128
Wang XR, Wang YX, Tian J, Lim BL, Yan XL, Liao H (2009) Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol 151(1):233–240
Wang XR, Yan XL, Liao H (2010) Genetic improvement for phosphorus efficiency in soybean: a radical approach. Ann Bot 106:215–222
Wang LS, Li Z, Qian WQ, Guo WL, Gao X, Huang LL, Wang H, Zhu HF, Wu JW, Wang DW, Liu D (2011) The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157(3):1283–1299
Wang F, Duan YX, Chen LJ, Wang YY, Zhu XF, Li W (2012) Glutathione peroxidase cDNA cloning and expression in soybean roots under heterodera glycines infection. J Agric Sci 4(7):163
Williamson LC, Ribrioux SP, Fitter AH, Leyser HO (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126(2):875–882
Xiao K, Katagi H, Harrison M, Wang ZY (2006a) Improved phosphorus acquisition and biomass production in Arabidopsis by transgenic expression of a purple acid phosphatase gene from M. truncatula. Plant Sci 170(2):191–202
Xiao K, Harrison M, Wang ZY (2006b) Cloning and characterization of a novel purple acid phosphatase gene (MtPAP1) from Medicago truncatula Barrel Medic. J Integr Plant Biol 48(2):204–211
Yan XL, Liao H, Trull MC, Beebe SE, Lynch JP (2001) Induction of a major leaf acid phosphatase does not confer adaptation to low phosphorus availability in common bean. Plant Physiol 125:1901–1911
Zhang Q, Wang C, Tian J, Li K, Shou H (2011) Identification of rice purple acid phosphatases related to phosphate starvation signaling. Plant Biol 13(1):7–15
Zhou J, Jiao FC, Wu ZC, Li YY, Wang XM, He XW, Zhong WQ, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146(4):1673–1686
Acknowledgments
This research was funded by the Natural Science Foundation of China (No. 31071441), the National Project from the Ministry of Agriculture of China (No. 2014ZX08004-004B), and the Natural Science Foundation of Hebei Province (No. C2010000749). The authors are grateful to Priscilla Licht and Professor Zhiying Ma for their critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Chong.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 Sub-cellular localization of GmPAP4. (a-c) 15 h post-treatment, with most fluorescence signals for pCamE-GmPAP4-GFP occurring in cytosol. (d-f) 18 h post-treatment, with most fluorescence signals for pCamE-GmPAP4-GFP occurring on plasma membrane. (g-i) the fluorescence signals of empty vector pCamE-GFP.
Supplementary Fig. 2 Lengths of petioles from transgenic (GmPAP4) and wild-type (WT) Arabidopsis under +Pi or +Po. Data were the means of 3 replicates (10 plants each). Error bars represented SE of all samples tested. Asterisks indicated that values were significantly different between transgenic plants and WT at P < 0.05.
Supplementary Fig. 3 Amino acid sequences alignment of GmPAP4 (HQ162477) and Gm02g45950 deduced sequence in ‘Williams 82’ soybean.
Supplementary Fig. 4 The different growth of ZH15 and NMH under phytate condition.
Supplementary Table 1 Substrate specificity of GmPAP4. Enzymatic activities were assayed at 37 °C for 1 h in 0.1 mol L−1 NaOAc buffer (pH5.0) containing ρ-NPP or phytate (final concentration, 1.0 mmol L−1). Each value represented the means of three experiments and was expressed as a percentage relative to the measurement using ρ-NPP as substrate.
Rights and permissions
About this article
Cite this article
Kong, Y., Li, X., Ma, J. et al. GmPAP4, a novel purple acid phosphatase gene isolated from soybean (Glycine max), enhanced extracellular phytate utilization in Arabidopsis thaliana . Plant Cell Rep 33, 655–667 (2014). https://doi.org/10.1007/s00299-014-1588-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-014-1588-5