Abstract
Key message
Transgenic plants stably overexpressing ScOPR1 gene enhanced disease resistance by increasing the accumulation of JA, SA, and GST, as well as up-regulating the expression of genes related to signaling pathways.
Abstract
12-Oxo-phytodienoate reductase (OPR) is an oxidoreductase that depends on flavin mononucleotide (FMN) and catalyzes the conversion of 12-oxophytodienoate (12-OPDA) into jasmonic acid (JA). It plays a key role in plant growth and development, and resistance to adverse stresses. In our previous study, we have obtained an OPR gene (ScOPR1, GenBank Accession Number: MG755745) from sugarcane. This gene showed positive responses to methyl jasmonate (MeJA), salicylic acid (SA), abscisic acid (ABA), and Sporisorium scitamineum, suggesting its potential for pathogen resistance. Here, in our study, we observed that Nicotiana benthamiana leaves transiently overexpressing ScOPR1 exhibited weaker disease symptoms, darker 3,3-diaminobenzidine (DAB) staining, higher accumulation of reactive oxygen species (ROS), and higher expression of hypersensitive response (HR) and SA pathway-related genes after inoculation with Ralstonia solanacearum and Fusarium solanacearum var. coeruleum. Furthermore, the transgenic N. benthamiana plants stably overexpressing the ScOPR1 gene showed enhanced resistance to pathogen infection by increasing the accumulation of JA, SA, and glutathione S-transferase (GST), as well as up-regulating genes related to HR, JA, SA, and ROS signaling pathways. Transcriptome analysis revealed that the specific differentially expressed genes (DEGs) in ScOPR1-OE were significantly enriched in hormone transduction signaling and plant–pathogen interaction pathways. Finally, a functional mechanism model of the ScOPR1 gene in response to pathogen infection was depicted. This study provides insights into the molecular mechanism of ScOPR1 and presents compelling evidence supporting its positive involvement in enhancing plant disease resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane (Saccharum spp.) is the main crop for sugar production in China, contributing to over 85% of the total sugar yield (Ruan et al. 2018; Li and Yang 2015; Dotaniya et al. 2016). It is susceptible to a fungal disease called sugarcane smut, caused by Sporisorium scitamineum. The pathogenic mycelium of the smut fungus invades cane shoots and spreads through intercellular filaments, affecting the growing point, resulting in mutations and the production of black whips, which even hinders stem formation in the cane (Rajput et al. 2021; Shamsul et al. 2021; Que et al. 2014). Developing and cultivating sugarcane varieties that are resistant to smut is the primary strategy to combat this disease. Therefore, exploring disease-resistant genes not only provides a genetic resource but also establishes a theoretical foundation for molecular breeding in sugarcane.
Jasmonates (JAs), which include jasmonate acid (JA) and its derivative methyl jasmonate (MeJA), are crucial signaling molecules derived from hydroxyl lipids in plants (Wasternack and Hause 2013; Campos et al. 2014). Generally, the synthesis of JAs occurs in the chloroplast and peroxisome. Within the chloroplast, unsaturated fatty acids are oxygenated by lipoxygenase (LOX) to produce 12-oxo-phytodienoic acid (12-OPDA) through the actions of allene oxide synthase (AOS) and allene oxide cyclase (AOC) (Chini et al. 2018; Mou et al. 2019). In the peroxisome, 12-OPDA is converted into JA by 12-oxo-phytodienoic acid reductase (OPR) and three β-oxidation steps of the carboxylic acid side chain (Chini et al. 2018; Mou et al. 2019). JA is catabolized in the cytoplasm to produce structures like methyl jasmonate (MeJA), jasmonoyl-l-isoleucine (JA-Ile), cis-jasmone (CJ), and 12-hydroxyjasmonic acid (12-OH-JA) (Chini et al. 2018; Mou et al. 2019). Research indicates that OPR, a flavin mononucleotide (FMN)-dependent oxidoreductase, catalyzes OPDA into JA precursor, marking the final step of JA synthesis (Mou et al. 2019; Tani et al. 2008; Breithaupt et al. 2006). OPRs are a multiprotein family with two classes, OPR I and OPR II, based on their substrate preference. Notably, OPR II has the ability to convert (9S, 13S)-OPDA into ( +)-7-epi-JA precursor, while OPR I has different substrate preferences and may aid in substrate (Schaller et al. 1998; Strassner et al. 2002). It was found that after simultaneous mutation of two OPR3 homologous genes by CRISPR/Cas9, the mutant showed complete male sterility and the fertility could be easily restored by exogenous MeJA treatment (Cheng et al. 2023). Besides, a meta-analysis of barley transcriptome datasets revealed that OPR3 was involved in JA biosynthesis (Soltani et al. 2023). Furthermore, OPR3-independent JA biosynthesis pathway is ancient and predates the emergence of the OPR3-independent pathway (Chini et al. 2023). The first plant OPR gene was isolated from Arabidopsis thaliana in 1997, and subsequent research has identified numerous OPR genes (Schaller and Weiler 1997). Currently, there are 3 OPRs in Arabidopsis and Lycopersicon esculentum (Breithaupt et al. 2006; Schaller and Weiler 1997; Biesgen and Weiler 1999), 5 in Citrullus lanatus (Guang et al. 2021), 6 in Pisum sativum (Matsui et al. 2004), 8 in Zea mays (Zhang et al. 2005), 13 in Oryza sativa (Li et al. 2011), and 48 in Triticum aestivum (Mou et al. 2019).
In plants, the OPR gene family is extensively involved in regulating growth and development, resistance to pathogen infection, and tolerance to adversity stress, while the specific function varies among different family members (Ponting et al. 2002; Liu et al. 2020; Tan et al. 2013; Pratiwi et al. 2017; Wang et al. 2016). For example, the Brassica campestris BcOPR3 gene was found to be up-regulated at a higher rate in disease-resistant plants compared to susceptible plants after infection with Hyaloperonospora parasitica (Wen et al. 2017), and its expression could be triggered by the stresses of JA, abscisic acid (ABA), and salicylic acid (SA) (Wen et al. 2017). In Z. mays, ZmOPR1 and ZmOPR2 contributed to defense against several pathogens (Zhang et al. 2005). Moreover, maize opr2 mutants exhibited differing sensitivity to various pathogens (Huang et al. 2023). In Gossypium hirsutum, virus-induced gene silencing (VIGS) revealed that the plants with GhOPR9 knockout were more susceptible to Verticillium dahlia infection (Liu et al. 2020). Similarly, in Solanum lycopersicum, silencing of the SlOPR3 gene resulted in a lower accumulation of OPDA and JA-lle after infection with Botrytis cinerea, making the plants more susceptible to this pathogen (Scalschi et al. 2015). Beyond doubt, these findings strongly support the significant role of OPRs in plant responses to pathogen stress.
A ScOPR1 gene (GenBank Accession Number: MG755745) was identified and characterized in our previous study from the sugarcane cultivar ROC22, and its gene expression was up-regulated by MeJA, SA, and S. scitamineum stresses. Here in our study, transient overexpression of ScOPR1 in Nicotiana benthamiana were performed and three T4 generation stable transgenic lines were selected. The phenotype, vegetative index, SA and JA contents, glutathione S-transferase (GST) enzyme activity, and immune response-associated gene expression were assessed in transgenic plants post-inoculation with two pathogens, Ralstonia solanacearum and Fusarium solanacearum var. coeruleum. Additionally, RNA-Seq in transgenic plants post-inoculation with F. solani var. coeruleum was conducted. The present study aims to establish a theoretical foundation for genetic engineering by ScOPR1 gene for smut resistance improvement in sugarcane breeding.
Materials and methods
Bioinformatics analysis of ScOPR1
The conserved domain prediction of the ScOPR1 protein was conducted using the NCBI (https://www.ncbi.nlm.nih.gov/cdd). The promoter sequence (2000 bp upstream) of two ScOPR1 homologous genes, SsPON.05G0025620-1B and Sh_227A23_contig-1_t000020, were extracted from S. spontaneum (Zhang et al. 2018) and sugarcane cultivar R570 genomes (Garsmeur et al. 2018), respectively. The PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict the cis-regulatory elements (CREs) and the TBtools was used for visualization (Chen et al. 2020).
Transient overexpression of ScOPR1 in Nicotiana benthamiana
Referred to our previous study (Sun et al. 2020), an OPR gene was screened from the sugarcane transcriptome unigene library constructed by our group, and a full-length cDNA sequence, named ScOPR1 (GenBank Accession Number: MG755745), was amplified from ROC22 buds inoculated with smut pathogen for 48 h using RT-PCR. The recombinant vector pEarleyGate 203-ScOPR1 (35S::ScOPR1) and the control vector (35S::00) were produced using the Gateway technique. They were then transiently overexpressed in N. benthamiana leaves via the Agrobacterium-mediated delivery (Choi et al. 2012; Wang et al. 2020). Subsequently, R. solanacearum and F. solani var. coeruleum were inoculated into the leaves of 6-week-old N. benthamiana that transiently overexpressed 35S::ScOPR1 and 35S::00 for 1 d, respectively. Then, the phenotypic changes were tracked and photographed (Dang et al. 2013). Post inoculation with pathogen for 1 d and 6 d, the N. benthamiana leaves were collected for 3,3′-diaminobenzidine tetrahydrochloride (DAB) staining to measure the accumulated hydrogen peroxide (H2O2) content (Choi et al. 2012; Sohn et al. 2007; Wang et al. 2020; Wu et al. 2023). The expression of the ScOPR1 gene in transiently N. benthamiana plants was analyzed through reverse transcription PCR (RT-PCR) with the primers ScOPR1-gate-F/R (Table S1). The expression levels of five immune-related marker genes, consisting of two hypersensitive response (HR) genes (NbHSR201 and NbHSR515) and three SA-related genes (NbPR2, NbPR3, and NbPR1-a/c) (Wang et al. 2023a, b), were analyzed using real-time quantitative PCR (RT-qPCR), respectively (Table S1). Data normalization to the expression level of NbEF-1a (Brogue et al. 1991; Zhang et al. 2019; Wu et al. 2023). All treatments were performed with three biological replicates. The relative expression levels were determined utilizing the 2−ΔΔCT approach (Livak and Schmittgen 2001), and the statistical analysis, including significance (P < 0.05) and standard error, was conducted using DPS 7.05 with Duncan’s new multiple range test.
Generation of transgenic N. benthamiana plants overexpressing ScOPR1 gene and the evaluation of its disease resistance
Agrobacterium tumefaciens GV3101 cells harboring pEarleyGate 203-ScOPR1 were delivered into N. benthamiana utilizing the leaf-disk methodology, followed by the screening of transgenic plant materials in a subculture medium (4.4 g/L MS, 8 g/L agar, pH = 5.8) containing 0.01% basta (Burow et al. 1990). Positive transgenic N. benthamiana lines were screened by RT-PCR using primers ScOPR1-gate-F/R (Table S1). Subsequently, three T4 generation transgenic N. benthamiana plants were generated, referred to as ScOPR1-OE1, ScOPR1-OE2, and ScOPR1-OE3. The pathogens of R. solanacearum and F. solani var. coeruleum were inoculated into the leaves of ScOPR1-OE and wild-type (WT) plants with three biological replicates, respectively. All the subjected materials were grown at 28 °C under a light/dark cycle of 16 h/8 h and 75% relative humidity. The phenotypic changes of the leaves were tracked and observed. Besides, GST activity, as well as SA and JA contents were evaluated using ELISA kits (Shanghai Enzyme-linked Biotechnology, China) at 0 d and 2 d post-inoculation with pathogens, following the manufacturer’s instructions. Furthermore, the expression levels of eight immune-related marker genes, including HR marker genes NbHSR201 and NbHSR515, SA-related genes NbPR2 and NbNPR1, JA-related genes NbLOX1 and NbDEF1 (Torres 2010), and reactive oxygen species (ROS)-related genes NbGST1 and NbAPX (Lai et al. 2013) (Table S1) were analyzed by RT-qPCR using NbEF-1α as an internal reference gene (Brogue et al. 1991; Zhang et al. 2019; Wu et al. 2023).
RNA sequencing and data analysis
The N. benthamiana leaf samples after inoculation with F. solani var. coeruleum at the beginning (0 d, control, CK) and 2 d (treatment, T), resulting in four sample sets (WT-CK, ScOPR1-CK, WT-T, and ScOPR1-T) with three biological replicates, were collected for RNA-Seq. Then, total RNA extraction, cDNA library construction, Illumina sequencing, data analysis, reference N. benthamiana genome mapping, differentially expressed genes (DEGs) identification (fold change ≥ 2 and P-value < 0.05) and DEGs annotation were referred to our previous studies (Wu et al. 2022a, 2022b; Wang et al. 2023a, b; Sun et al. 2023). Seven candidates DEGs including NPR1 (Niben101Scf19043g00002), DELLA (Niben101Scf15437g02006), HDT1 (Niben101Scf09416g05012), CPK28 (Niben101Scf05805g02006), CTL1 (Niben101Scf03036g03023), BKI1 (Niben101Scf03420g01001), and MPK4 (Niben101Scf07241g00013) were randomly screened for RT-qPCR validation.
Results
Sequence characteristics of sugarcane ScOPR1
As depicted in Fig. 1A, the sugarcane ScOPR1 encoded 371 amino acids (AA) and contained a conserved OYE_like_FMN domain from 10 to 349 AA. This gene showed 99.19% and 94.34% similarity with the homologous gene in S. spontaneum (SsPON.05G0025620-1B) and sugarcane cultivar R570 (Sh_227A23_contig-1_t000020) (Fig. S1). Besides, both genes contained cis-regulatory elements related to growth and development, light response, and hormone response, with the unique presence of stress-responsive elements, while Sh_227A23_contig-1_t000020 specifically contained stress-responsive elements (Fig. 1B), suggesting a potential involvement of ScOPR1 gene in various aspects of plant growth and response to environmental stresses. Meanwhile, the expression levels of the ScOPR1 gene were increased under SA, MeJA, and S. scitamineum stresses. Moreover, compared with the control, its expression was up-regulated and reached a peak at 48 h with 2.62-fold compared to the control (0 h) in the smut-resistance variety YC05-179, but down-regulated at 24 h in the susceptible variety ROC22 (Fig. 1C) (Sun et al. 2018). These results suggested a role of the ScOPR1 gene in conferring resistance to S. scitamineum through JA and SA biosynthesis pathways in sugarcane (Fig. 1D).
Characterization of ScOPR1 gene in sugarcane. A Conserved domains of ScOPR1 protein. B Cis-regulatory element (CREs) analysis of the ScOPR1 homologous gene SsPON.05G0025620-1B in S. spontaneum and Sh_227A23_contig-1_t000020 in R570. Different color boxes corresponded to different CREs. C Expression patterns of ScOPR1 in sugarcane under MeJA, SA, and S. scitamineum stresses. Color bars represent the normalized values (log2 Relative exprssion), ranging from blue (low expressionlevel) to red (high expression level). D Jasmonate biosynthetic pathway
Transient overexpression of ScOPR1 led to an enhancement in the disease resistance
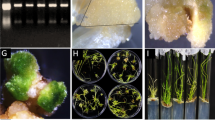
As shown in Fig. 2A, the ScOPR1 gene was successfully transiently overexpressed in N. benthamiana. Following inoculation with R. solanacearum for 1 d, the disease symptoms and DAB staining color had no significant difference between 35S::ScOPR1 and the control (35S::00) (Fig. 2B). However, after 6 d, the symptoms in the leaves of 35S::00 were more severe compared to 35S::ScOPR1. Furthermore, the DAB staining color of 35S::ScOPR1 was darker than the control, indicating a significantly higher H2O2 content in the 35S::ScOPR1 plants (Fig. 2B). Furthermore, the expression levels of genes related to HR and SA pathways were significantly increased in 35S::ScOPR1 plants after infected with R. solanacearum compared to the control. Especially, 6 days after injection with R. solanacearum, the expression levels of NbHSR201, NbHSR515, NbPR-1a/c, and NbPR2 were 4.40-, 9.41-, 46.76-, and 11.56-fold higher than the control, respectively (Fig. 2C). Similarly, there was no significant difference in phenotypes between 35S::ScOPR1 and 35S::00 after inoculation with F. solanacearum var. coeruleum, while a high content of H2O2 accumulated in the 35S::ScOPR1 plants (Fig. 2D). Besides, the expression of HR marker and SA-related genes were significantly up-regulated in 35S::ScOPR1 leaves at 1 d or 6 d, with the NbPR-1a/c gene showed the 10.83-fold higher than 35S::00 (Fig. 2E).
Transient overexpression of the ScOPR1 gene in N. benthamiana. A RT-PCR results of ScOPR1 in N. benthamiana leaves after transient overexpression for 1 d. 35S::ScOPR1, pEarleyGate 203-ScOPR1; 35S::00, the empty vector pEarleyGate 203. B, D Phenotype and DAB staining of N. benthamiana leaves transiently overexpressing 35S::ScOPR1 and 35S::00 after inoculation with R. solanacearum and F. solani var. coeruleum for 1 d and 6 d. C, E The expression levels of HR marker and SA-related genes in N. benthamiana leaves following inoculation with R. solanacearum and F. solani var. coeruleum at 1 d and 6 d. All data points represent means ± standard error (n = 3). The significant differences are represented by different letters
Stable overexpression of ScOPR1 positively regulated the defense response against pathogen infection
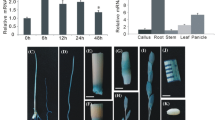
Totally, three T4 lines of ScOPR1 genetically modified N. benthamiana plants were successfully acquired and verified by RT-PCR (Fig. 3A, B). After inoculation with F. solani var. coeruleum 23 d and R. solanacearum 16 d, the WT leaves showed more obvious disease spots and yellowing than that of the transgenic plants (Fig. 3C, F). Compared to the control, the contents of JA and SA, and the activity of GST in ScOPR1-OE2 plants were significantly increased post infection with both two pathogens (Fig. 3D, G). In addition, the expression levels of ROS-, HR-, JA- and SA-related genes were also up-regulated in the transgenic plants after challenging with pathogens (Fig. 3E, H). These results indicated that the stably overexpression of the ScOPR1 gene could enhance the disease resistance of N. benthamiana to pathogen infection by promoting the expression of several genes involved in HR, JA, SA, and ROS signaling pathways.
Disease resistance evaluation of T4 generation of transgenic N. benthamiana overexpressing the ScOPR1 gene. A T4 transgenic N. benthamiana seeds on MS plates with herbicides. WT, wild-type N. benthamiana; OE1–OE3, three ScOPR1 transgenic N. benthamiana lines. B RT-PCR detection of T4 generation transgenic N. benthamiana plants. M, 2000 bp DNA marker; CK, blank control; NC, negative control; PC, positive control. C, F Phenotypes of transgenic N. benthamiana after inoculation with F. solani var. coeruleum 23 d and R. solanacearum 15 d. D, G Determination of SA and JA contents, and GST activity in transgenic N. benthamiana after inoculation with F. solani var. coeruleum and R. solanacearum for 0 d and 2 d. E, H Expression pattern of ROS-, HR-, JA- and SA-related genes in transgenic N. benthamiana after inoculation with R. solanacearum and F. solani var. coeruleum for 0 d and 2 d. All data points were means ± standard error (n = 3). Significant differences are calculated by Duncan’s new multiple range test (P-value < 0.05) and represented by different letters
Transcriptome difference between ScOPR1 overexpressing transgenic lines and WT plants in the process of disease response
Since the WT-CK1 dataset showed a weak correlation with the other biological replicates (Fig. 4A), it was excluded from further analysis. A total of 98.64 GB of high-quality data was obtained, with Q30 above 93% and GC content exceeding 41%, indicating that the sequencing quality of these libraries was excellent and suitable for further analysis (Table S2). Additionally, a total of 2667 (1033 up- and 1634 down-regulated) and 187 DEGs (118 up- and 69 down-regulated) were found in the treatment (ScOPR1-CK_vs_ScOPR1-T) and the control group (WT-CK_vs_WT-T), respectively (Fig. 4B, Tables S3, S4). There were 20 common up-regulated and 29 common down-regulated DEGs in both groups. In addition, the control group had 98 specific up-regulated and 40 specific down-regulated DEGs, while the treatment group had 1013 specific up-regulated and 1605 specific down-regulated DEGs (Fig. 4C). GO enrichment showed that the specific DEGs of ScOPR1-CK_vs_ScOPR1-T were enriched in the JA signaling pathway (GO: 2,000,022), plant-type HR (GO: 0010363), SA metabolic process (GO: 0010337), defense response to fungus (GO: 1,900,150), immune response (GO: 0050776), and response to ABA (GO: 0009737) (Fig. 4D, Table S5). KEGG pathway enrichment indicated that the DEGs specific to treatment group primarily participated in plant–pathogen interaction (ko04626) and several metabolic pathways (ko00860, ko00780, ko00500, ko00520, ko00564, and ko00591) (Fig. 4E, Table S6). These results demonstrated that ScOPR1 transgenic plants could activate more abundant DEGs in biological processes and metabolic pathways in defense against pathogen infection.
Transcriptome variances between ScOPR1-overexpressing transgenic lines and wild-type plants during the process of disease response. A The correlation heat map. WT-CK, WT-T, ScOPR1-CK, and ScOPR1-T represent the wild-type N. benthamiana and transgenic N. benthamiana overexpressing ScOPR1 after inoculation with Fusarium solani var. coeruleum for 0 d (CK) and 2 d (T), respectively. B, C The number of DEGs in WT-CK_vs_WT-T and ScOPR1-CK_vs_ScOPR1-T. D, E GO and KEGG enrichment of specific DEGs in WT-CK_vs_WT-T and ScOPR1-CK_vs_ScOPR1-T
ScOPR1 expression-mediated several signaling pathways in the defense response to pathogen infection
According to the results of KEGG enrichment, three disease resistance pathways including plant hormone signal transduction, MAPK signaling pathway-plant, and plant–pathogen interaction were selected to make a straightforward molecular network. Notably, three WT-CK_vs_WT-T special regulated DEGs, namely PP2CA, EIN2, and MTB1 were up-regulated (Fig. 5A, Table S7). Besides, 11 regulated DEGs (LECRK2, SCL15, MMK2, MMK2, MMK2, PYL4, JAR6, NPR1, PR1, CAT1, and CAT3) specific to ScOPR1-CK_vs_ScOPR1-T were also up-regulated (Fig. 5A, Table S7). While 16 regulated DEGs specific to ScOPR1-CK_vs_ScOPR1-T, including SD31, FLS2, XA21, At3g47570, NLP2, NSP2, CIGR1, SCL23, SCL3, PAT1, TIFY10B, MAKR1, WRKY33, At1g67720, and RBOHA, were down-regulated (Fig. 5A, Table S7). Additionally, six common regulated DEGs (CPK32, CHI14, CTR1, GID1B, LRR1, and At2g23950) and eight common regulated DEGs (CPK32, GID1B, CXE11, SD25, LRK10, and PR5K) were up-regulated in the WT-CK_vs_WT-T and ScOPR1-CK_vs_ScOPR1-T group, respectively. However, there were night common regulated DEGs consist of CPK32, CPK28, CPK1, CTL1, STY46, At1g07650, IRK, CRK33, and LECRK1, were down-regulated in ScOPR1-CK_vs_ScOPR1-T group (Fig. 5A, Table S7). Interestingly, the regulatory mechanisms were different in WT and ScOPR1-OE during the resistance against pathogen infection. Furthermore, seven DEGs (NPR1, DELLA, HDT1, CPK28, CTL1, BKI1, and MPK4) involved in the MAPK signaling, plant–pathogen interaction, and plant hormone signal transduction pathways were randomly selected and verified by RT-qPCR (Fig. 5B, C). It was obvious that the relative expression trend of these seven genes was consistent with (R2 = 0.997) the expression trend of log2 (fold change) in the transcriptome (Fig. 5B, C, and Fig. S2).
Expression patterns of DEGs in disease resistance-related pathways. A Expression patterns of DEGs uniquely or common regulated in the ScOPR1-CK_vs_ScOPR1-T or the WT-CK_vs_WT-T group. B, C Log2 (fold change) values and relative expression levels of seven key genes in WT and ScOPR1 transgenic N. benthamiana inoculated with F. solani var. coeruleum for 2 d
Transcription factors and protein kinases played an important role in disease resistance
As reported, transcription factors (TFs) and protein kinases (PKs) played an important role in plant resistance to the pathogen (Sun et al. 2023). A total of 147 TFs and 126 PKs from the specifically regulated DEGs in ScOPR1-CK_vs_ScOPR1-T, were predicted (Fig. 6A, Tables S8, S9). These 126 PKs (45 up- and 81 down-regulated) were mainly enriched in the CAMK_CDPK, RLK-Pelle_DLSV, RLK-Pelle_LRR-III, RLK-Pelle_LRR-XI-1, RLK-Pelle_RLCK-VIIa-2, and RLK-Pelle_SD-2b families (Fig. 6C, Table S8), with the fact that CAMK_CDPK was mainly acted on regulating plant growth and development through a series of cascading signaling processes (Harmon et al. 2001). Notably, RLK-Pelle was abundant in plants and the RLK-Pelle_DLSV, RLK-Pelle_RLCK-VIIa-2, and RLK-Pelle_SD-2b families were closely related to the plant immune system, involving plant protection from pathogen attack. Interestingly, 147 TFs (57 up- and 90 down-regulated) were closely related to ABA signaling (bZIP and NAC), JA signaling (bHLH), and ethylene (ET) signaling (AP2/ERF) pathways (Fig. 6B, Table S9).
Discussion
Till now, an increasing number of OPR genes have been discovered in various plants due to their significant roles in response to biotic stress (Matsui et al. 2004; Zhang et al. 2005; Nie et al. 2022). According to the results of promoter analysis, the ScOPR1 gene was involved in plant growth and development, as well as response to both biotic and abiotic stresses. Meanwhile, the expression of ScOPR1 gene was not only triggered by the phytohormone signaling molecules MeJA and SA but also could actively respond to S. scitamineum stress (Sun et al. 2018), suggesting that ScOPR1 participated in the response to pathogen invasion in sugarcane. Similarly, two maize OPR genes ZmOPR1 and ZmOPR2, seemed to be involved in defense mechanisms against C. carbonum, C. heterostrophus, and F. verticillioides (Zhang et al. 2005). Likewise, the mutation of ZmOPR2 resulted in decreased resistance to corn smut (Zhang et al. 2005). In the present study, the temporary overexpression of ScOPR1 increased the resistance of N. benthamiana to F. solani var. coeruleum and R. solanacearum (Fig. 2B, D) by up-regulating HR- and SA-related genes (Fig. 2C, E), indicating its positive role in plant disease resistance. Notably, this fact could also be confirmed by the stable overexpression of ScOPR1 in transgenic N. benthamiana (Fig. 3).
Previous studies have found that it is important to regulate the concentration of ROS at an appropriate level for normal plant growth (Sofo et al. 2015). As an indicator of ROS, H2O2 can rapidly react with DAB under the catalysis of peroxidase to form brown compounds, thereby positioning H2O2 in plant tissues (Mittler et al. 1998). In our study, the DAB staining in the leaves of transgenic tobacco plants overexpressing ScOPR1 was darker compared to the control when they were subjected to pathogen inoculation for 6 days (Fig. 2B, D). Furthermore, we observed an increase in ROS metabolism, including H2O2 accumulation, in ScOPR1-OE2 plants after inoculation with F. solani var. coeruleum for 2 d (Fig. 5A). When plants are attack by pathogens, those genes related to ROS scavenging systems, such as CAT, GST, and APX, play a crucial role in plant disease resistance (Kumar 2014; Boatwright and Pajerowska-Mukhtar 2013; Chan and Lam 2014; Zhang et al. 2016). Likewise, the contents of GST and CAT enzyme of ScOPR1-OE2 were significantly higher after inoculation with pathogens compared to the control (Figs. 3D, G, 5A). It can be reasonably deduced that overexpression of ScOPR1 could activate the ROS signaling pathway during the response of plant to exogenous pathogens. Thordal-Christensen et al. (1997) speculated that ROS was involved in the HR pathway, which is a defense mechanism of plants against pathogen infection in the host-parasite incompatibility relationship. Here in our study, under pathogen stresses, the expression of HR marker genes (NbHSR515 and NbHSR201) was significantly up-regulated in ScOPR1-OE2 plants compared to the control (Fig. 3E, H), indicating that N. benthamiana plants overexpressing ScOPR1 could facilitate the occurrence of HR.
Lipid metabolism is closely related to the synthesis and transport of JA and SA, and OPR3 is a crucial enzyme in JA synthesis (Mou et al. 2019; Tani et al. 2008; Breithaupt et al. 2006). Recent studies demonstrated that plant OPR genes were involved in various defense signaling pathways (Zhang et al. 2005; Sun et al. 2018). In A. thaliana, OPR3 mutants ddel and opr3 both lacked the function of synthesizing JA (Tan et al. 2013). When stimulated by SA, JA, and ET, the expression levels of ClOPR2 and ClOPR4 were notably increased in watermelon (Guang et al. 2021). In cotton, GhOPR9 was identified as a regulator of JA pathway-related gene expression during Verticillium wilt infection, highlighting its crucial role in cotton’s resistance to V. wilt (Liu et al. 2020). The antagonistic relationship between SA and JA in biotrophic and hemibiotrophic pathogen resistance has been extensively documented (Kumar 2014; Boatwright and Pajerowska-Mukhtar 2013). Huang et al. (2023) discovered that SA could counteract JA by utilizing ZmOPR2 to inhibit JA biosynthesis during plant–pathogen interactions in maize. In the present study, the expression levels of SA- and JA-related genes in ScOPR1-OE2 were markedly elevated compared to the control group (Fig. 3E, H). Besides, the enzyme activity assay revealed an increase in the contents of JA and SA (Fig. 3D, G). Overall, the results suggested that transgenic overexpression of ScOPR1 could enhance resistance to external pathogen infection by up-regulating genes associated with the JA and SA pathways. However, it is still unclear why SA and JA do not act antagonistically in pathogen resistance in transgenic ScOPR1-OE2. It is thus hypothesized that the exact in vivo substrates and end products of OPR1 enzyme action are still unknown, warranting further research to elucidate the underlying mechanism. Nonetheless, RNA-seq results showed that DEGs related to SA signaling (NPR1 and PR1) were up-regulated, and the JA pathway was also activated, as evidenced by the down-regulation of JAZ and the up-regulation of both MYC2 and JAR1 (Fig. 5A), suggesting a synergistic relationship between the JA and SA signaling pathways.
The OPR3 gene expression can be triggered by various stimuli, including touch, wind, wounding, UV-light, and brassinosteroids (BRs) (Schaller et al. 2000). Brassinosteroids are a type of steroid hormone that plays a significant role in plant growth, development, and response to stress (Wang et al. 2022). When plants are under stimuli, BRs bind and activate BRI1 and BAK1, and the activated BRI1 can further transmit signals by phosphorylating different substrates (Wang et al. 2022). Similarly, our transcriptome analysis confirmed that BRs participated in disease resistance by activating BRI1 and BAK1 (Fig. 5A). Studies have shown that flg22, a flagellin epitope and PAMP, weakens the hypersensitive cell death, resistance, and biomass reduction induced by Pseudomonas syringae (Pst) AvrRpt2 in Arabidopsis (Wang et al. 2023a, b). It attaches to the receptor-like kinase FLS2, initiating the influx of Ca2+ across the plasma membrane (PM) (Chi et al. 2021). It is widely acknowledged that the FLS2 receptor and ROS burst exhibit sensitivity adaptation upon flg22 stimulation, which is referred to as desensitization and resensitization, to prevent excessive responses to pathogen infection (Chi et al. 2021). In this study, we demonstrated that flg22 bound to FLS2, resulting in the influx of Ca2+ into the PM. CDPK, serving as a Ca2+ receptor, gets activated, leading to the expression of ROS burst and disease-related factors such as PR1, WRKY33, and FPK1, all of which together contribute to plant resistance against pathogen infection (Fig. 5A). Recent study has shown that the ET and JA signaling pathways, along with MPK3/MPK6 signaling pathway, synergistically stimulate camalexin synthesis to enhance plant disease resistance (Zhou et al. 2022). Furthermore, we observed that following inoculation with F. solanacearum var. coeruleum, both ET and JA signals were activated and contributed to disease resistance (Fig. 5A). These results suggested that ScOPR1 functions in enhancing plant resistance against pathogen infection by coordinating the activation of BRs, Ca2+, MAPK, and ET signaling pathways.
Conclusions
By integrating phenotypic observations, DAB staining, physiological and biochemical changes, immune-related gene expression, and RNA-seq analysis, our study revealed that the ScOPR1 overexpression in N. benthamiana plants post-pathogen infection facilitated the interaction between pathogen-associated molecular proteins (PAMPs) and RLK proteins, which activated the MAPK cascade signaling pathway. This activation then induced the expression of AP2/ERF-ERF, bHLH, NAC, C2H2, MYB, bZIP, and WRKY transcription factors and DEF1, LOX1, PR2, NPR1, and GST1 defense-related genes involved in JA, SA, ET, and ABA pathways, thereby increasing the disease resistance of tobacco to pathogens. At the same time, the binding of PAMPs to RLK triggered a release of Ca2+ and activation of CDPKs calcium receptor proteins. Furthermore, pathogen infection resulted in the production of ROS, which to some extent induced an immune response known as HR in the plant itself, ultimately leading to increased resistance. Finally, a functional mechanism model of ScOPR1 overexpression-mediated defense response of transgenic plants to pathogen infection was depicted (Fig. 7). This study offered valuable insights into the role of the ScOPR1 gene in conferring pathogen resistance and highlighted its molecular mechanisms in sugarcane.
A functional model of ScOPR1 overexpression-mediated defense response of transgenic plants to pathogen infection. PAPMs pathogen-associated molecular proteins; CDPKs calcium-dependent protein kinases; RKLs receptor-like kinases; MAPK mitogen-activated protein kinase; JA jasmonic acid; ABA abscisic acid; ROS reactive oxygen species; ET ethylene; TFs transcription factors; ScOPR1 overexpressing transgenic lines, respectively
Data availability
Data will be made available on request.
References
Biesgen C, Weiler EW (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208(2):155–165. https://doi.org/10.1007/s004250050545
Boatwright JL, Pajerowska-Mukhtar KM (2013) Salicylic acid: an old hormone up to new tricks. Mol Plant Pathol 14(6):623–634. https://doi.org/10.1111/mpp.12035
Breithaupt C, Kurzbauer R, Lilie H, Schaller A, Strassner J, Huber R, Macheroux P, Clausen T (2006) Crystal structure of 12-oxophytodienoate reductase 3 from tomato: self-inhibition by dimerization. Proc Natl Acad Sci 103(39):14337–14342. https://doi.org/10.1073/pnas.0606603103
Brogue K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S, Broglie R (1991) Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254(5035):1194–1197. https://doi.org/10.1126/science.254.5035.1194
Burow MD, Chlan CA, Sen P, Lisca A, Murai N (1990) High-frequency generation of transgenic tobacco plants after modified leaf disk cocultivation with Agrobacterium tumefaciens. Plant Mol Biol Rep 8:124–139. https://doi.org/10.1007/BF02669766
Campos ML, Kang JH, Howe GA (2014) Jasmonate-triggered plant immunity. J Chem Ecol 40(7):657–675. https://doi.org/10.1007/s10886-014-0468-3
Chan C, Lam HM (2014) A putative lambda class glutathione S-transferase enhances plant survival under salinity stress. Plant Cell Physiol 55(3):570–579. https://doi.org/10.1007/s10886-014-0468-3
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Cheng HT, Hao MY, Sang SF, Wen YF, Cai YT, Wang H, Wang WX, Mei DS, Hu Q (2023) Establishment of new convenient two-line system for hybrid production by targeting mutation of OPR3 in allopolyploid Brassica napus. Hortic Res 10(12):uhad218. https://doi.org/10.1093/hr/uhad218
Chi Y, Wang C, Wang M, Wan D, Huang F, Jiang Z, Pei ZM (2021) Flg22-induced Ca2+ increases undergo desensitization and resensitization. Plant Cell Environ 44(12):3563–3575. https://doi.org/10.1111/pce.14186
Chini A, Monte I, Zamarreño AM, Hamberg M, Lassueur S, Reymond P, Weiss S, Stintzi A, Schaller A, Porzel A, García-Mina JM, Solano R (2018) An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat Chem Biol 14(2):171–178. https://doi.org/10.1038/nchembio.2540
Chini A, Monte I, Zamarreño AM, García-Mina JM, Solano R (2023) Evolution of the jasmonate ligands and their biosynthetic pathways. New Phytol 238(5):2236–2246. https://doi.org/10.1111/nph.18891
Choi DS, Hwang IS, Hwang BK (2012) Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24(4):1675–1690. https://doi.org/10.1105/tpc.112.095869
Dang FF, Wang Y, Yu L, Eulgem T, Lai Y, Liu ZQ, He SL (2013) CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ 36(4):757–774. https://doi.org/10.1111/pce.12011
Dotaniya ML, Datta SC, Biswas DR, Dotaniya CK, Meena BL, Rajendiran S, Regar KL, Lata M (2016) Use of sugarcane industrial by-products for improving sugarcane productivity and soil health. Int J Recycl Org Waste Agric 5:185–194. https://doi.org/10.1007/s40093-016-0132-8
Garsmeur O, Droc G, Antonise R, Grimwood J, Potier B, Aitken K (2018) A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat Commun 9(1):2638. https://doi.org/10.1038/s41467-018-05051-5
Guang Y, Luo S, Ahammed GJ, Xiao X, Li J, Zhou Y, Yang Y (2021) The OPR gene family in watermelon: genome-wide identification and expression profiling under hormone treatments and root-knot nematode infection. Plant Biol 23:80–88. https://doi.org/10.1111/plb.13225
Harmon AC, Gribskov M, Gubrium E, Harper JF (2001) The CDPK superfamily of protein kinases. New Phytol 151:175–183. https://doi.org/10.1046/j.1469-8137.2001.00171.x
Huang PC, Tate M, Berg-Falloure KM, Christensen SA, Zhang J, Schirawski J, Meeley R, Kolomiets MV (2023) A non-JA producing oxophytodienoate reductase functions in salicylic acid-mediated antagonism with jasmonic acid during pathogen attack. Mol Plant Pathol 24(7):725–741. https://doi.org/10.1111/mpp.13299
Kumar D (2014) Salicylic acid signaling in disease resistance. Plant Sci 228:127–134. https://doi.org/10.1016/j.plantsci.2014.04.014
Lai Y, Dang FF, Lin J, Yu L, Shi Y, Xiao YH, Huang MK, Lin JH, Chen CC, Qi AH, Liu ZQ, Guan DY, Mou SL, Qiu AL, He SL (2013) Overexpression of a Chinese cabbage BrERF11 transcription factor enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Physiol Biochem 62:70–78. https://doi.org/10.1016/j.plaphy.2012.10.010
Li YR, Yang LT (2015) Sugarcane agriculture and sugar industry in China. Sugar Tech 17:1–8. https://doi.org/10.1007/s12355-014-0342-1
Li WY, Zhou F, Liu B, Feng DR, He YM, Qi KB, Wang HB, Wang JF (2011) Comparative characterization, expression pattern and function analysis of the 12-oxo-phytodienoic acid reductase gene family in rice. Plant Cell Rep 30(6):981–995. https://doi.org/10.1007/s00299-011-1002-5
Liu SC, Sun RB, Zhang XJ, Feng ZL, Wei F, Zhao LH, Zhang YL, Zhu LF, Feng HJ, Zhu HQ (2020) Genome-wide analysis of OPR family genes in cotton identified a role for GhOPR9 in Verticillium dahliae resistance. Genes 11(10):1134. https://doi.org/10.3390/genes11101134
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Matsui H, Nakamura G, Ishiga Y, Toshima H, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y (2004) Structure and expression of 12-oxophytodienoate reductase (subgroup I) genes in pea, and characterization of the oxidoreductase activities of their recombinant products. Mol Genet Genomics 271:1–10. https://doi.org/10.1007/s00438-003-0948-6
Mittler R, Feng X, Cohen M (1998) Post-transcriptional suppression of cytosolic ascorbate peroxidase expression during pathogen-induced programmed cell death in tobacco. Plant Cell 10(3):461–473. https://doi.org/10.1105/tpc.10.3.461
Mou YF, Liu YY, Tian SJ, Guo QP, Wang CS, Wen SS (2019) Genome-wide identification and characterization of the OPR gene family in wheat (Triticum aestivum L.). Int J Mol Sci 20(8):1914. https://doi.org/10.3390/ijms20081914
Nie WF, Chen Y, Tao J, Li Y, Liu J, Zhou Y, Yang Y (2022) Identification of the 12-oxophytodienoic acid reductase (OPR) gene family in pepper (Capsicum annuum L.) and functional characterization of CaOPR6 in pepper fruit development and stress response. Genome 65(11):537–545. https://doi.org/10.1139/gen-2022-0037
Ponting CP, Ito T, Moscat J, Diaz-Meco MT, Inagaki F, Sumimoto H (2002) OPR, PC and AID: all in the PB1 family. Trends Biochem Sci 27(1):10. https://doi.org/10.1016/s0968-0004(01)02006-0
Pratiwi P, Tanaka G, Takahashi T, Xie XN, Yoneyama K, Matsuura H, Takahashi K (2017) Identification of jasmonic acid and jasmonoyl-isoleucine, and characterization of AOS, AOC, OPR, and JAR1 in the model lycophyte Selaginella moellendorffii. Plant Cell Physiol 58(4):789–801. https://doi.org/10.1093/pcp/pcx031
Que YX, Xu LP, Wu QB, Liu YF, Ling H, Liu YH, Zhang YY, Guo JL, Su YC, Chen JB, Wang SS, Zhang CG (2014) Genome sequencing of Sporisorium scitamineum provides insights into the pathogenic mechanisms of sugarcane smut. BMC Genomics 15(1):996. https://doi.org/10.1186/1471-2164-15-996
Rajput ML, Rajput AN, Syed RN, Lodhi AM, Que YX (2021) Sugarcane smut: current knowledge and the way forward for management. J Fungi 7(12):1095. https://doi.org/10.3390/jof7121095
Ruan HY, Feng PY, Wang B, Xing HT, Leary GJ, Huang ZG, Guo H, Liu DL (2018) Future climate change projects positive impacts on sugarcane productivity in southern China. Eur J Agron 96:108–119. https://doi.org/10.1016/j.eja.2018.03.007
Scalschi L, Sanmartín M, Camañes G, Troncho P, Sánchez-Serrano JJ, García-Agustín P, Vicedo B (2015) Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J 81(2):304–315. https://doi.org/10.1111/tpj.12728
Schaller F, Weiler EW (1997) Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana: structural and functional relationship to yeast old yellow enzyme. J Biol Chem 272(44):28066–28072. https://doi.org/10.1074/jbc.272.44.28066
Schaller F, Hennig P, Weiler EW (1998) 12-Oxophytodienoate-10, 11-reductase: occurrence of two isoenzymes of different specificity against stereoisomers of 12-oxophytodienoic acid. Plant Physiol 118(4):1345–1351. https://doi.org/10.1104/pp.118.4.1345
Schaller F, Biesgen C, Müssig C, Altmann T, Weiler EW (2000) 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210(6):979–984. https://doi.org/10.1007/s004250050706
Shamsul AB, Robert CM, Meredith DM, Karen SA (2021) Sugarcane smut, caused by Sporisorium scitamineum, a major disease of sugarcane: a contemporary review. Phytopathology 111(11):1905–1917. https://doi.org/10.1094/PHYTO-05-21-0221-RVW
Sofo A, Scopa A, Nuzzaci M, Vitti A (2015) Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int J Mol Sci 16(6):13561–13578. https://doi.org/10.3390/ijms160613561
Sohn SI, Kim YH, Kim BR, Lee SY, Lim CK, Hur JH, Lee JY (2007) Transgenic tobacco expressing the hrpNEP gene from Erwinia pyrifoliae triggers defense responses against Botrytis cinerea. Mol Cells 24(2):2–239. https://doi.org/10.1016/s1016-8478(23)07333-8
Soltani Z, Moghadam A, Tahmasebi A, Niazi A (2023) Integrative systems biology analysis of barley transcriptome-hormonal signaling against biotic stress. PLoS ONE 18(4):e0281470. https://doi.org/10.1371/journal.pone.0281470
Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32(4):585–601. https://doi.org/10.1046/j.1365-313x.2002.01449.x
Sun TT, Wang DJ, Liu F, Wang L, Li Z, Dai MJ, Que YX, Xu LP, Sun YC (2018) Molecular cloning, subcellular localization, and expression analysis of jasmonic acid synthesis gene ScOPR1 from sugarcane. J Appl Environ Biol 24:1365–1374. https://doi.org/10.19675/j.cnki.1006-687x.2018.01005
Sun T, Cen G, You C, Lou W, Wang Z, Su W, Wang W, Li D, Que Y, Su Y (2020) ScAOC1, an allene oxide cyclase gene, confers defense response to biotic and abiotic stresses in sugarcane. Plant Cell Rep 39(12):1785–1801. https://doi.org/10.1007/s00299-020-02606-z
Sun TT, Chen Y, Feng AY, Zou WH, Wang DJ, Lin PX, Chen YL, You CH, Que YX (2023) The allene oxide synthase gene family in sugarcane and its involvement in disease resistance. Ind Crops Prod 192:116136. https://doi.org/10.1016/j.indcrop.2022.116136
Tan CT, Carver BF, Chen MS, Gu YQ, Yan LL (2013) Genetic association of OPR genes with resistance to Hessian fly in hexaploid wheat. BMC Genomics 14:369. https://doi.org/10.1186/1471-2164-14-369
Tani T, Sobajima H, Okada K, Chujo T, Arimura S, Tsutsumi N, Nishimura M, Seto H, Nojiri H, Yamane H (2008) Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 227(3):517–526. https://doi.org/10.1007/s00425-007-0635-7
Thordal-Christensen H, Zhang ZG, Wei YD, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11:1187–1194. https://doi.org/10.1046/j.1365-313x.1997.11061187.x
Torres MA (2010) ROS in biotic interactions. Physiol Plant 138(4):414–429. https://doi.org/10.1111/j.1399-3054.2009.01326.x
Wang YK, Yuan GL, Yuan SH, Duan WJ, Wang P, Bai JF, Zhang FT, Gao SQ, Zhang LP, Zhao CP (2016) TaOPR2 encodes a 12-oxo-phytodienoic acid reductase involved in the biosynthesis of jasmonic acid in wheat (Triticum aestivum L.). Biochem Biophys Res Commun 470(1):233–238. https://doi.org/10.13345/j.cjb.210236
Wang DJ, Wang L, Su WH, Ren YJ, You CH, Zhang C, Que YX, Su YC (2020) A class III WRKY transcription factor in sugarcane was involved in biotic and abiotic stress responses. Sci Rep 10(1):20964. https://doi.org/10.1038/s41598-020-78007-9
Wang L, Yang R, Sun J (2022) Regulation of crop agronomic traits and abiotic stress responses by brassinosteroids: a review. Chin J Biotechnol 38(1):34–49. https://doi.org/10.13345/j.cjb.210236
Wang D, Wei L, Liu T, Ma J, Huang K, Guo H, Huang Y, Zhang L, Zhao J, Tsuda K, Wang Y (2023a) Suppression of ETI by PTI priming to balance plant growth and defense through an MPK3/MPK6-WRKYs-PP2Cs module. Mol Plant 16(5):903–918. https://doi.org/10.1016/j.molp.2023.04.004
Wang DJ, Qin LQ, Wu MX, Zou WH, Zang SJ, Zhao ZN, Lin PX, Guo JL, Wang HB, Que YX (2023b) Identification and characterization of WAK gene family in Saccharum and the negative roles of ScWAK1 under the pathogen stress. Int J Biol Macromol 224:1–19. https://doi.org/10.1016/j.ijbiomac.2022.11.300
Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann Bot 111(6):1021–1058. https://doi.org/10.1016/j.bbrc.2016.01.043
Wen K, Wang Y, Hu D, Yuan JP, Hou XL, Li Y (2017) Cloning and expression analysis of BcOPR3 gene in non-heading Chinese cabbage. J Nanjing Agric Univ 40:804–811. https://doi.org/10.7685/jnau.201612018
Wu QB, Pan YB, Su YC, Zou WH, Xu F, Sun T, Grisham MP, Yang SL, Xu LP, Que YX (2022a) WGCNA identifies a comprehensive and dynamic gene co-expression network that associates with smut resistance in sugarcane. Int J Mol Sci 23(18):0770. https://doi.org/10.3390/ijms231810770
Wu QB, Su YC, Pan YB, Xu F, Zou WH, Que BB, Lin PX, Sun TT, Grisham MP, Xu LP, Que YX (2022b) Genetic identification of SNP markers and candidate genes associated with sugarcane smut resistance using BSR-Seq. Front Plant Sci 13:1035266. https://doi.org/10.3389/fpls.2022.1035266
Wu QB, Chen YL, Zou WH, Pan YB, Lin P, Xu LP, Grisham MP, Ding QG, Su YC, Que YX (2023) Genome-wide characterization of sugarcane catalase gene family identifies a ScCAT1 gene associated with disease resistance. Int J Biol Macromol 232:123398. https://doi.org/10.1016/j.ijbiomac.2023.123398
Zhang JL, Simmons C, Yalpani N, Crane V, Wilkinson H, Kolomiets M (2005) Genomic analysis of the 12-oxo-phytodienoic acid reductase gene family of Zea mays. Plant Mol Biol 59(2):323–343. https://doi.org/10.1007/s11103-005-8883-z
Zhang JX, Wang XL, Vikash V, Ye Q, Wu DD, Liu YL, Dong WG (2016) ROS and ROS-mediated cellular signaling. Oxid Med Cell Longev 2016:4350965. https://doi.org/10.3390/ijms231810770
Zhang JS, Zhang XT, Tang HB, Zhang Q, Hua XT, Ma XK, Zhu F, Jones T, Zhu XG, Bowers J (2018) Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet 50(12):1565–1573. https://doi.org/10.1038/s41588-018-0237-2
Zhang C, Chen H, Zhuang RR, Chen YT, Deng Y, Cai TC, Wang SY, Liu QZ, Tang RH, Shan SH, Pan RL, Chen LS, Zhuang WJ (2019) Overexpression of the peanut CLAVATA1-like leucine-rich repeat receptor-like kinase AhRLK1 confers increased resistance to bacterial wilt in tobacco. J Exp Bot 70(19):5407–5421. https://doi.org/10.1093/jxb/erz274
Zhou J, Mu Q, Wang X, Zhang J, Yu H, Huang T, Meng X (2022) Multilayered synergistic regulation of phytoalexin biosynthesis by ethylene, jasmonate, and MAPK signaling pathways in Arabidopsis. Plant Cell 34(8):3066–3087. https://doi.org/10.1093/plcell/koac139
Funding
This study was supported by the Special Projects for the Central-guided Local Science and Technology Development (2022L3086), National Key Research and Development Program of China (2022YFD2301100), China Agriculture Research System of MOF and MARA (CARS-17), Project of National Key Laboratory for Tropical Crop Breeding (1630052024003 and NKLTCB-YAAS-2024-S01), Director’s Research Fund of National Engineering Research Center for sugarcane, Fujian Agriculture and Forestry University (NERD202212), and Natural Science Foundation of Fujian Province, China (2021J01137 and 2020J01591).
Author information
Authors and Affiliations
Contributions
Wenhui Zou: data curation, investigation, methodology, software, validation, visualization, writing—original draft; Tingting Sun: data curation, funding acquisition, investigation, methodology, software, validation, visualization, writing—original draft; Yao Chen: data curation, investigation, software, validation, visualization; Dongjiao Wang: data curation, investigation, validation, visualization; Chuihuai You: data curation, investigation, software; Shoujian Zang: data curation, validation, visualization; Peixia Lin: data curation, investigation, software; Qibin Wu: conceptualization, project administration, supervision, writing—review and editing; Yachun Su: conceptualization, funding acquisition, supervision, writing—review and editing; Youxiong Que: conceptualization, funding acquisition, project administration, supervision, writing—review and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Leandro Peña.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zou, W., Sun, T., Chen, Y. et al. Sugarcane ScOPR1 gene enhances plant disease resistance through the modulation of hormonal signaling pathways. Plant Cell Rep 43, 158 (2024). https://doi.org/10.1007/s00299-024-03241-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00299-024-03241-8