Abstract
A protocol for Agrobacterium-mediated transformation with either kanamycin or mannose selection was developed for leaf explants of the cultivar Prunus dulcis cv. Ne Plus Ultra. Regenerating shoots were selected on medium containing 15 μM kanamycin (negative selection), while in the positive selection strategy, shoots were selected on 2.5 g/l mannose supplemented with 15 g/l sucrose. Transformation efficiencies based on PCR analysis of individual putative transformed shoots from independent lines relative to the initial numbers of leaf explants tested were 5.6% for kanamycin/nptII and 6.8% for mannose/pmi selection, respectively. Southern blot analysis on six randomly chosen PCR-positive shoots confirmed the presence of the nptII transgene in each, and five randomly chosen lines identified to contain the pmi transgene by PCR showed positive hybridisation to a pmi DNA probe. The positive (mannose/pmi) and the negative (kanamycin) selection protocols used in this study have greatly improved transformation efficiency in almond, which were confirmed with PCR and Southern blot. This study also demonstrates that in almond the mannose/pmi selection protocol is appropriate and can result in higher transformation efficiencies over that of kanamycin/nptII selection protocols.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gametophytic self-incompatibility (GSI) is exhibited by many plants belonging to the family Rosaceae (Sedgley 1994) including fruit trees such as apple, pear, cherry and almond, and overcoming self-incompatibility (SI) in almond is a goal of almond-breeding programs. Conventional breeding techniques of woody fruit trees is often difficult and slow because of high levels of heterozygosity and the long generation time between successive crosses (Sriskandarajah et al. 1994). These difficulties necessitate the development of rapid gene transfer methods for the genetic improvement of fruit trees. The pre-requisite for genetic transformation is a regeneration protocol compatible with the gene transfer methods of the targeted species. However, woody fruit trees are very recalcitrant to regeneration and transformation in vitro (Petri and Burgos 2005). Previous success in regenerating transgenic woody plants has been reported in apple (De Bondt et al. 1994; James et al. 1989), pear (Matsuda et al. 2005), apricot (Machado et al. 1992) chestnut (Seabra and Pais 1998) and almond (Miguel and Oliveira 1999).
Regeneration of almond shoots in tissue culture is possible using both immature and mature leaf tissue samples (Ainsley et al. 2000; Mehra and Mehra 1974; Miguel et al. 1996), but robust regeneration of Agrobacterium-mediated transformed almond shoots in culture has been difficult to achieve. Almond callus generated from leaf explants has been successfully transformed (Ainsley et al. 2001a; Archilleti et al. 1995), however, the callus failed to regenerate transformed almond shoots. Using leaf explants derived from germinated almond seeds, Miguel and Oliveira (1999) were successful in regenerating four putative transformed shoots from the almond cultivar Boa Casta, only one of which consistently showed the presence of the transgene (GUS) detected using both PCR and Southern blot analysis. Poor recovery of transformed almond shoots appears to be a consequence of its inadequate growth in culture particularly in the presence of antibiotic (kanamycin) selection pressure. New advances are required to improve upon the inherently low transformation efficiency in almond, which is currently limiting future functional analysis of gene activity and directed genetic enhancement.
Widespread public concern over the use of marker genes conferring antibiotic or herbicide resistance has led to the development of alternative selection systems for transformed tissues (Haldrup et al. 1998; Hansen and Wright 1999; Joersbo et al. 1998). These selection systems operate by inhibiting the growth of the non-transformed tissue through carbohydrate starvation (positive selection). The phosphomannose isomerase structural gene (pmi) from Escherichia coli (Miles and Guest 1984) has been used successfully as a selectable marker in Agrobacterium-mediated herbaceous plant transformations (He et al. 2004; Joersbo et al. 1998; Negrotto et al. 2000; O’Kennedy et al. 2004; Sigareva et al. 2004; Wang et al. 2000; Zhang et al. 2005). Mannose is readily taken up by the plant and converted to mannose-6-phosphate by the action of hexokinase. Accumulation of mannose-6-phosphate in plant cells inhibits phosphogluco isomerase leading to a block in glycolysis (Goldsworthy and Street 1965) which disrupts growth. However, when plants are transformed with pmi, they are able to survive by utilising mannose as a carbohydrate source. Further, unlike standard antibiotics, mannose selection does not result in a direct negative toxic effect as the control callus and shoots continue to grow albeit very slowly and acquire a brown colour (Lindsey and Gallois 1990). In tissue culture, almond can readily utilise sucrose, fructose or glucose as a carbon source but grows poorly on mannose, making the mannose/PMI selection protocol an attractive selection option (Wang et al. 2000).
To date, there have not been reports of successful transformation and regeneration from a commercial almond cultivar. Here, we report the successful regeneration of transformed shoots from the almond cultivar Ne Plus Ultra via Agrobacterium-mediated transformation using either kanamycin (negative) or mannose/pmi (positive) as selection agents.
Materials and methods
Plant material
Leaf explants from the P. dulcis cv. Ne Plus Ultra were used in the transformation experiments. The explants were taken from clonal shoots propagated in vitro using the protocol of Ainsley et al. (2000).
Bacterial strain and vectors
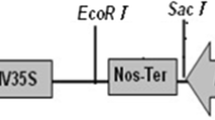
Agrobacterium strain EHA 105 (Hood et al. 1993) transformed with the plasmid pBI121mgfp-5-ER (Haseloff et al. 1997) and AGL1 (Lazo et al. 1991) transformed with the plasmid pNOV2819 manA (Syngenta, NC 27709, USA) were used in the transformation of almond leaf explants. The plasmid pBI121mgfp-5-ER has the nptII gene under the nopaline synthase (nos) promoter, nos terminator and the mgfp-5-ER gene under the control of 35S promoter and the nos terminator. The plasmid pNOV2819 manA contains the pmi gene under the control of CPMS (Cestrium Yellow Leaf Curling Virus Promoter—short version) promoter and nos terminator.
Determination of mannose concentration for selection
To determine the optimum mannose concentration inhibiting shoot formation, almond leaf explants were placed on regeneration medium containing mannose alone (0 and 20 g/l) or in combination with sucrose (0–30 g/l) (Table 1). Ten explants were placed per plate and two plates were used for each mannose–sucrose concentration.
Transformation of in vitro leaf explants
The four youngest fully expanded leaves from in vitro micropropagated clonal shoots were used for transformation. The explants were pre-cultured for 3 days in liquid MS medium (Murashige and Skoog 1962) supplemented with IBA (1.96 mg/l) and BAP (2.5 mg/l) at 23±2°C in dark. The pre-cultured explants were dissected transversely across the midrib into 5 mm sections prior to transformation.
Agrobacterium cultures were grown overnight to turbidity (late log phase) in LB medium supplemented with 0.1% (w/v) glucose and appropriate antibiotics (100 μM kanamycin and 25 μg/ml rifampicin for EHA 105 carrying the plasmid pBI121mgfp-5-ER and 50 μg/ml spectinomycin and 25 μg/ml rifampicin for the AGL1 strain carrying the plasmid pNOV2819 manA). The cultures were centrifuged at 4,000 rpm at 18°C for 5 min and adjusted to OD550 nm of 0.5 with LB medium supplemented with 0.1% (w/v) glucose. Acetosyringone (Sigma) at a final concentration of 100 μM was added to the Agrobacterium cultures and incubated at 28°C with shaking for 2 h. The cultures were centrifuged at 4,000 rpm at 18°C for 5 min and the cells diluted to an OD550 nm of 0.3 with liquid MS medium. The pre-cultured almond leaf explants were co-cultivated with Agrobacterium cultures for 1 h on a rotary shaker at 28°C. The explants were blotted on sterile Whatman filter papers and transferred to plates containing RM1 medium (MS supplemented with 1.96 mg/l IBA, 2.5 mg/l BAP, 30 g/l sucrose, 630 μM cefotaxime) and incubated in the dark at 22°C for 3 days. After co-cultivation the leaf explants were washed twice in liquid MS medium with 1 mM cefotaxime for 10 min each. The explants were blotted and transferred to plates with RM1 medium and incubated in the dark at 25±1°C for 3 weeks, placed in dim light for 2 weeks and subsequently transferred to full light.
The tissues transformed with the pBI121mgfp-5-ER construct were subcultured onto RM1 medium every 2 weeks and subjected to four different treatments. Three days or twenty-one days after co-cultivation the tissue was transferred to RM1 medium supplemented with or without kanamycin (15 or 20 μM) for 4–6 weeks. The explants with regenerating shoots from the kanamycin-free medium were transferred to RM2 (MS supplemented with 0.1 mg/l IBA, 1.0 mg/l BAP, 30 g/l sucrose, 630 μM cefotaxime) medium and maintained on this medium until the shoots were 1.5–2 cm long (4 weeks). The shoots were then transferred to RM2 medium supplemented with 15 μM kanamycin (70 days after co-cultivation, Table 2) and the shoots that survived the selection were rooted and transferred to soil in pots in the glasshouse.
Putatively transformed tissue from the transformation with the pNOV2819 manA construct was transferred to RM3 medium (MS supplemented with 1.96 mg/l IBA, 2.5 mg/l BAP, 15 g/l sucrose, 2.5 g/l mannose, 630 μM cefotaxime) after 3 weeks in culture and maintained on the selection medium for four passages (8 weeks). The shoots were subsequently transferred to RM1 medium and maintained until they were ready to be rooted and transferred to soil in the containment glasshouse.
Leaf antibiotic resistance assay method
Putative transformed leaves from shoots regenerated from the transformations with the pBI121mgfp-5-ER construct were tested for their susceptibility or resistance to the antibiotic kanamycin. A 1 cm piece of leaf from the growing axis was excised and cut into four pieces and placed on plates with RM2 medium supplemented with kanamycin ranging in concentrations from 5 to 15 μM. The plates were incubated for seven days at 25°C under fluorescent light and scored for resistance/sensitivity to kanamycin. The leaf explants that bleached were scored as sensitive while the leaf explants that did not show bleaching were scored as resistant.
Analysis for transgene insertion in almond
Genomic DNA was extracted from the callus and or shoots regenerated from the transformations with the pBI121mgfp-5-ER and pNOV2819 manA constructs using the DNeasy Plant Mini Kit (QIAGEN) as per manufacturer's instructions. The forward and reverse PCR primers used to amplify the nptII (KanF: GAGGCTATTCGGCTATGACTG, KanR: ATCGGGAGCGGCGATACCGTA) and the pmi gene fragments (PMIF: ACAGCCACTCTCCATTCA, PMIR: GTTTGCCATCACTTCCAG) were designed using the NetPrimer (PREMIER Biosoft International, Palo Alto, CA). All PCR reactions used between 40 and 60 ng of genomic DNA. Aliquots of 5 μl of the PCR products were electrophoresed on a 1.5% (w/v) agarose gel in 0.5×TBE (Tris Borate EDTA buffer). The gels were stained with ethidium bromide (0.5 μg/ml) and visualised under UV light.
For Southern analysis, 5 μg of genomic DNA from PCR-positive shoots were digested with XhoI (transformants containing pBI121mgfp-5-ER) or SalI (transformants containing pNOV2819 manA) to determine the integration of the gene of interest. Following digestion, the DNA fragments were separated on a 1% (w/v) agarose gel and blotted onto Hybond N+ nylon membrane (Amersham). A 700 bp fragment (for nptII gene) or 514 bp fragment (for pmi gene) were generated by PCR with labelled with [32P]-dCTP by random priming using the DECAprime II DNA labelling kit (Ambion) and used as probes. Hybridisation was carried out at 65°C in sodium phosphate buffer [0.5 M sodium phosphate, 1 mM EDTA and 7% (w/v) SDS]. The membranes were washed with 2× SSC + 0.1% (w/v) SDS for 15 min, 1× SSC + 0.1% (w/v) SDS for 15 min and 0.1× SSC + 0.1% (w/v) SDS for 5 min at 65°C. The membranes were exposed to the phosphorimager for 3 days.
A–D Plate showing regeneration of shoots of almond cultivar Ne plus Ultra transformed with pBI121mgfp-5-ER and pNOV2819 manA constructs. A Shoot initiation from tissue transformed with pBI121mgfp-5-ER construct on RM1 [MS + IBA (1.96 mg/l) + BAP (2.5 mg/l)]. B Shoot initiation from tissue transformed with pNOV2819 manA construct on RM1 [MS + IBA (1.96 mg/l) + BAP (2.5 mg/l)]. C Fluorescence due to gfp in the tissue transformed with pBI121mgfp-5-ER construct. D No fluorescence was observed in control tissue
Rooting of shoots
Using a method very similar to that described by Ainsley et al. (2001b), shoots were rooted by brief treatment with 1 mM IBA (no phloroglucinol). Shoot pre-treatment at 4°C was reduced from 4 weeks to either 1 or 2 weeks; water agar was best solidified using 1.5% agar and, after overnight exposure to IBA, shoots in 1/2 strength MS medium were initially kept in the dark for 2 or 3 days. Shoots with roots were potted in a mixture of Nu-Erth Premium Potting Mix:peat moss:vermiculite (3:1:1) and gradually acclimatised in a glasshouse.
Results
The optimum concentration of mannose required for efficient selection against control tissues was determined by observing percent callus growth and regeneration of shoots from inoculated leaf explants on MS medium (Table 1). As the concentration of mannose in the medium was increased relative to sucrose, the percentage of explants producing callus and regenerating shoots decreased. Mannose concentrations above 2.5 g/l severely impacted upon callus growth and shoot regeneration. Subsequent mannose concentrations in the selection medium were set at 2.5 g/l and accompanied by 15 g/l of sucrose.
A PCR analysis of putative transformants (pBI121mgfp-5-ER construct) for the inserted kanamycin gene. M = 100 bp marker. Lanes 1–9, 13, 15, 16 and 17 = putatively transformed shoots. Lanes 10–12 = putatively transformed callus lines. Lane 14 = plasmid DNA. Lane 18 = control. B PCR analysis of putative transformants (pNOV2819 manA construct) for pmi gene. Lane M = 100 bp marker. Lanes 1–5 = putatively transformed shoots. Lane 6 = PCR control (no DNA). Lane 7 = untransformed shoot
Transformation of in vitro leaf explants
The leaf explants transformed with A. tumefaciens (EHA 105) containing the pBI121mgfp-5-ER construct enlarged and started to produce callus 4–5 days after transformation. Shoot initiation was observed after 2 weeks in culture (Fig. 1A). Leaf explants were individually subcultured to RM1 medium with cefotaxime every 2 weeks (4–6 weeks in total) to enable the shoots to grow. Each individual callused leaf explant was then placed on medium with kanamycin 3 days after co-cultivation or following initiation of shoot buds 21 days after co-cultivation. No shoot buds were regenerated when placed on medium with kanamycin 3 days after co-cultivation however, a total of 28 shoot buds were regenerated on medium with 15 μM kanamycin while 10 shoot buds were regenerated on medium with 20 μM kanamycin when selection was delayed for 21 days (Table 2). Regenerated shoot buds showed low vigour, stunted growth, browning of the tissue and eventually stopped growing in subsequent passages on medium with kanamycin. In comparison, the explants placed on kanamycin-free medium regenerated large numbers of shoots and grew vigorously. Individual shoots from each of the independent lines regenerated on kanamycin-free medium were transferred to RM2 medium containing cefotaxime and maintained on this medium until the shoots were 1.5–2 cm in length. A combined total of 197 shoots from all the surviving transformed lines were regenerated on kanamycin-free medium (Table 2). If present, multiple shoots per independent line were separated at this stage and placed on RM2 medium with kanamycin (15 μM) for selection of transformed shoots, 70 days after co-cultivation. When transferred to medium with 15 μM kanamycin, 48 shoots across all the lines survived selection (Table 2). Young leaves from the putatively transformed shoots were also subjected to an in vitro leaf antibiotic resistance assay. The leaves from transformed shoots remained green while those from control shoots bleached showing increasing susceptibility to kanamycin (data not shown).
The putatively transformed explants were placed under UV light to visualise green fluorescent protein (gfp) 6 days, 2 and 6 weeks after co-cultivation. The transformed explants showed green spots under UV light (Fig. 1C) while the non-transformed explants did not show such fluorescence (Fig. 1D).
The almond leaf explants transformed with A. tumefaciens (AGL1) containing the pmi gene were placed initially on RM1 medium with cefotaxime for callus initiation and growth. Twenty-one days after transformation, the leaf explants were transferred to RM3 medium containing 2.5 g/l of mannose and proliferating tissue was subcultured onto the same medium every 2 weeks. Regeneration of leaf explants with callus tissue could be observed after 3 weeks in culture (Fig. 1B). After four passages on selection medium, the individual explants with regenerating shoots were transferred to RM2 medium for further proliferation and growth. In total 70 shoots were regenerated from these explants (Table 3).
Integration of transgenes
The PCR was used to screen all regenerated shoots from the transformation experiment with the pBI121mgfp-5-ER construct (Table 2). An expected nptII band of 700 bp was amplified in the shoots and four callus lines tested (Fig. 2A). The control tissue showed no amplification of the product (Fig. 2A, lane 18). Overall, 23 independent lines surviving kanamycin selection contained the expected 700 bp PCR-amplified product (Table 2). A preliminary transformation efficiency of 5.6% was calculated as the percentage of single independent kanamycin-positive lines as confirmed by PCR analysis per total number of initial inoculated leaf explants (Table 2).
In the PCR analysis of the shoots transformed with the pmi gene, four out of five shoots amplified a 514 bp pmi product (Fig. 2C). The control shoot did not amplify any product (Fig. 2C, lane 7). Overall, of 70 shoots surviving mannose selection, 27 independent lines tested positive for the pmi gene (Table 3). A preliminary transformation efficiency of 6.8% was calculated as the percentage of single independent pmi positive lines confirmed by PCR analysis per total number of original inoculated leaf explants (Table 3).
To confirm the presence and integration of a transgene into the transformants, six PCR-positive transgenic shoots were used to confirm the insertion of the nptII gene by Southern blot analysis. In each of the six transformants, the nptII probe hybridised to DNA fragments from XhoI digested genomic DNA. The restriction enzyme XhoI cuts the pBI121mgfp-5-ER plasmid once within the T-DNA (Fig. 3A). Examination of the nptII hybridisation pattern indicated approximately one to three integration sites of the transgene per individual line (Fig. 3B). Similarly, for the pmi transformants, Southern blot analysis was used to confirm the presence of the T-DNA in five PCR-positive lines. The restriction enzyme SalI was used to digest genomic DNA isolated from each sample, which is known to cut the pNOV2819 manA T-DNA once (Fig. 3C). The pmi probe hybridised to SalI digested genomic DNA in each of the transformants tested. The analysis revealed at least one to two integration events of the pmi gene into the plant genome (Fig. 3D). The genomic DNA from the control shoots did not hybridise with any of the probes.
A and B Southern analysis of PCR-positive shoots transformed with pBI121mgfp-5-ER construct. Five micrograms of genomic DNA was digested with XhoI (indicated by a line in Fig. 3A) and probed with a 700 bp fragment for the kanamycin gene. Lanes 1–6 = transgenic shoots. Lane 7 = control. C and D Southern analysis of PCR-positive shoots transformed with pNOV2819 manA construct. Five micrograms of genomic DNA was digested with SalI and probed with a 514 bp fragment (indicated by a line in Fig. 3C) for the pmi gene. Lanes 1, 2, 4, 5 and 6 = transgenic shoots. Lane 3 = control
Rooting of shoots
Up to 38% of shoots had rooted after 40 days in 1/2 strength MS medium and 6 transgenic plants were transferred to the containment glasshouse (Fig. 4A). Following establishment they were subjected to natural light and temperature conditions (no artificial light or heating) over winter. These plants subsequently flowered within 18 months of planting (Fig. 4B).
Discussion
The development of systems for the successful transformation and culturing of almonds are important milestones in the eventual routine genetic modification of the species. Almonds in general display low transformation efficiencies, which have limited the evaluation of different parameters controlling shoot development, flowering and fruit quality. Agrobacterium strain EHA105 has previously been an effective agent for genetic transformation of recalcitrant fruit tree species (De Bondt et al. 1994; Mourgues et al. 1996; Pena et al. 1995). In this study, in addition to EHA105, we found AGL1 also to be highly effective in transforming almond.
Many plant species including almond show sensitivity to medium supplemented with kanamycin. Alternative selection strategies including ‘delayed selection’ has been successful in obtaining transgenic plants in kanamycin-sensitive plants including apple (Yao et al. 1995; Yepes and Aldwinckle 1994), apricot (Machado et al. 1992) and the almond cultivar Boa Casta (Miguel and Oliveira 1999). In our study, 15 or 20 μM kanamycin was applied at 3, 21 and 70 days after co-cultivation with Agrobacterium in leaf explants transformed with the pBI121mgfp-5-ER construct. Delaying selection by 3 or 21 days after co-cultivation, resulted in the formation of a small number of buds, which developed into small shoots that remained stunted and did not grow further. Kanamycin-free induction medium was subsequently used which resulted in the formation of increased numbers of putatively transformed cell clusters or shoot initials. These shoots were allowed to grow and were subsequently screened on kanamycin selection medium 70 days after transformation. A total of 48 of the 197 shoots from independent lines continued to grow in the presence of kanamycin from which 23 independent lines gave positive results using the PCR to screen for T-DNA insertion. We were able to calculate a preliminary transformation efficiency of 5.6% based on the positive results from the PCR screens relative to the initial number of independent leaf explants used at the beginning of the experiment. This result was encouraging and was further supported by Southern blot analysis on six randomly chosen kanamycin-resistant PCR-positive shoots, which showed integration of the T-DNA into the genomes of each of the almond shoots.
There is growing public concern about the widespread use of antibiotic selection in plant transformation and its perceived risk associated with both human consumption or lateral transfer to other plants or organisms. In recent years, there have been new developments in alternative positive selection systems such as the use of the pmi gene from E. coli. The classical procedure in choosing a selection agent involves identifying concentrations, which limit non-transgenic plants regenerating. In our study, we found 2.5 g/l mannose supplemented with 15 g/l of sucrose to be an effective agent to select and regenerate transgenic almond plants using the pmi system. Combining mannose with sucrose has also previously been shown to give positive results in other pmi transformed plants including sugar beet, maize, rice and cassava (Joersbo et al. 1998; Lucca et al. 2001; Negrotto et al. 2000; Zhang and Puonti-Kaerlas 2000). In the control callus and shoots, mannose selection reduced overall tissue vigour and tissues developed a distinctive brown colour. Interestingly, mannose selection eliminated tissue necrosis (in both transformed and control tissues), which is commonly observed with kanamycin selection in almond (Lindsey and Gallois 1990). However, 5 g/l severely curtailed shoot development in the transgenic shoots (Table 1). Overall, final transformation efficiencies using both mannose and sucrose were calculated to be 6.8% (Table 3) an improvement over the parallel kanamycin-based selection protocol reported in this and other studies (Miguel and Oliveira 1999). Our results obtained using mannose selection are encouraging and show much promise for the continued use of the mannose/pmi transformation and selection method in almond.
Transformed shoots quantified by both PCR and Southern blot analysis for the presence of the T-DNA were rooted in culture and transferred to soil. In less than 18 months, the transgenic almond plants had flowered. This is a significant improvement on the 5–7 years normally required for plants to flower after propagation via traditional breeding methods. It appears that in vitro culture may have overcome the extended vegetative phase common in almond plants raised from seed using traditional breeding methods and offers a promising faster route for further molecular based genetic discovery and manipulation in almond.
In conclusion, this study demonstrated that the P. dulcis cv. Ne Plus Ultra can be efficiently transformed and cultured to soil using an initial tissue culture selection strategy based on the use of nptII/kanamycin resistance or more favourably through the activity of the pmi gene as a selectable marker with mannose as a selective agent. Previous to this work, success in almond transformation has been limited to in vitro seedlings and the generation of a single transformed line from P. dulcis cv. Boa Casta (Miguel and Oliveira 1999). As also observed in this study, almonds were found to be sensitive to kanamycin and this sensitivity is most likely the source of the reduced transformation efficiencies observed in previous studies (Ainsley et al. 2001a; Archilleti et al. 1995; Miguel and Oliveira 1999). The switch to the positive mannose/pmi section system has enhanced the number of transformed shoots and renders the use of kanamycin markers in almond transformation redundant. It is reasonable to suggest that positive selection may be the preferred option in subsequent almond transformation experiments.
References
Ainsley PJ, Collins GG, Sedgley M (2000) Adventitious shoot regeneration from leaf explants of almond (Prunus dulcis Mill.). In Vitro Cell Dev Biol-Plant 36:470–474
Ainsley PJ, Collins GG, Sedgley M (2001a) Factors affecting Agrobacterium-mediated gene transfer and the selection of transgenic calli in paper shell almond (Prunus dulcis Mill.). J Hort Sci Biotech 76:522–528
Ainsley PJ, Collins GG, Sedgley M (2001b) In vitro rooting of almond (Prunus dulcis Mill.). In Vitro Cell Dev Biol-Plant 33:778–785
Archilleti T, Lauri P, Damiano C (1995) Agrobacterium-mediated transformation of almond pieces. Plant Cell Rep 14:267–272
De Bondt A, Eggermont K, Druart P, De Vil M, Goderis I, Vanderleyden J, Broekaert WF (1994) Agrobacterium-mediated transformation of apple (Malus x domestica): an assessment of factors affecting gene transfer efficiency during early transformation steps. Plant Cell Rep 13:587–593
Goldsworthy A, Street HE (1965) The carbohydrate nutrition of tomato roots VIII. The mechanism of the inhibition by d-mannose of the respiration of excised roots. Ann Bot 29:45–48
Haldrup A, Petersen SG, Okkels FT (1998) The xylose isomerase gene from Thermoanaerobacterium thermosulfurogenes allows effective selection of transgenic plants cells using d-xylose as the selection agent. Plant Mol Biol 37:287–296
Hansen G, Wright MS (1999) Recent advances in the transformation of plants. Trends Plant Sci 4:226–231
Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localisation of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94:2122–2127
He Z, Fu Y, Si H, Hu G, Zhang S, Yu Y, Sun Z (2004) Phosphomannose-isomerase (pmi) gene as a selectable marker for rice transformation via Agrobacterium. Plant Sci 166:17–22
Hood EE, Gelvin SB, Melchers LS, Hoekama A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgen Res 2:208–212
James DJ, Passey AJ, Barbara DJ, Bevan MW (1989) Genetic transformation of apple (Malus pumila Mill.) using a disarmed Ti-binary vector. Plant Cell Rep 7:658–666
Joersbo M, Donaldson I, Krieberg J, Petersen SG, Brunstedt J, Okkels FT (1998) Analysis of mannose selection used for transformation of sugarbeet. Mol Breed 4:111–117
Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio/Technology 9:963–967
Lindsey K, Gallois P (1990) Transformation of sugar beet (Beta vulgaris) by Agrobacterium tumefaciens. J Exp Bot 41:529–536
Lucca P, Ye X, Potrykus I (2001) Effective selection and regeneration of transgenic rice plants with mannose as selective agent. Mol Breed 7:43–49
Machado M, Camara Machado AD, Hanzer V, Weiss H, Regner F, Steinkellner H, Mattanovich D, Plail R, Knapp E, Kalthoff B, Katinger H (1992) Regeneration of transgenic plants of Prunus armenica containing the coat protein gene of plum pox virus. Plant Cell Rep 11:25–29
Matsuda N, Gao M, Isuzugawa K, Takashima T, Nishimura K (2005) Development of an Agrobaterium-mediated transformation method for pear (Pyrus communis L.) with leaf-section and axillary shoot-meristem explants. Plant Cell Rep 24:45–51
Mehra A, Mehra PN (1974) Organogenesis and plantlet formation in vitro in almond. Bot Gaz 135:61–73
Miguel CM, Druart P, Oliveira MM (1996) Shoot regeneration from adventitious buds induced on juvenile and adult almond (Prunus dulcis Mill.) explants. In Vitro Cell Dev Biol-Plant 32:148–153
Miguel CM, Oliveira MM (1999) Transgenic almond (Prunus dulcis Mill.) plants obtained by Agrobacterium-mediated transformation of leaf explants. Plant Cell Rep 18:387–393
Miles JS, Guest JR (1984) Nucleotide sequence and transcriptional start point of the phosphomannose isomerase gene (manA) of Escherichia coli. Gene 32:41–48
Mourgues F, Chevreau E, Lambert C, De Bondt A (1996) Efficient Agrobacterium-mediated transformation and recovery of transgenic plants from pear (Pyrus communis L.). Plant Cell Rep 16:245–249
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Negrotto D, Jolley M, Beer S, Wenck AR, Hansen G (2000) The use of phosphomannose-isomerase as a selectable marker to recover transgenic maize plants (Zea mays L.) via Agrobacterium transformation. Plant Cell Rep 19:798–803
O’Kennedy MM, Burger JT, Botha FC (2004) Pearl millet transformation system using the positive selectable marker gene phosphomannose isomerase. Plant Cell Rep 22:684–690
Pena L, Cerevera M, Juarez J, Ortega C, Pina JA, Duran-Vila N, Navarro L (1995) High efficiency Agrobacterium-mediated transformation and regeneration of citrus. Plant Sci 104:183–191
Petri C, Burgos L (2005) Transformation of fruit trees. Useful breeding tool or continued future prospect? Transgen Res 14:15
Seabra R, Pais MS (1998) Genetic transformation of European chestnut. Plant Cell Rep 17:177–182
Sedgley M (1994) Self-incompatibility in woody horticultural species. In: Williams EG, Clarke AE, Knox RB (eds) Genetic control of self-incompatibility and reproductive development in flowering plants, Kluwer Academic Publishers, London, pp 141–163
Sigareva M, Spivey R, Willits MG, Kraimer CM, Chang Y-F (2004) An efficient mannose selection protocol for tomato that has no adverse effect on the ploidy level of transgenic plants. Plant Cell Rep 23:236–245
Sriskandarajah S, Goodwin PB, Speirs P (1994) Genetic transformation of the apple scion cultivar Delicious via Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult 36:317–329
Wang AS, Evans RA, Altendorf PR, Hanten JA, Doyle MC, Rosichan JL (2000) A mannose selection system for production of fertile transgenic maize plants from protoplasts. Plant Cell Rep 19:654–660
Yao JL, Cohen D, Atkinson R, Richardson K, Morris B (1995) Regeneration of transgenic plants from the commercial apple cultivar Royal Gala. Plant Cell Rep 14:407–412
Yepes LM, Aldwinckle HS (1994) Factors that affect leaf regeneration efficiency in apple and effect of antibiotics in morphogenesis. Plant Cell Tissue Organ Cult 37:257–269
Zhang P, Puonti-Kaerlas J (2000) PIG-mediated cassava transformation using positive and negative selection. Plant Cell Rep 19:1041–1048
Zhang S, Zhu L-H, Li X-Y, Ahlman A, Welander M (2005) Infection by Agrobacterium tumefaciens increased the resistance of leaf explants to selective agents in carnation (Dianthus caryophyllus L. and D. chinensis). Plant Sci 168:137–144
Acknowledgements
This work was supported by an ARC (Australian Research Council) linkage grant and the Almond Board of Australia. We thank Dr. Ursula Langridge for care of the almond plants in the PC2 containment glasshouse. We thank SYNGENTA for providing the pNOV2819 manA plasmid
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Peña
Rights and permissions
About this article
Cite this article
Ramesh, S.A., Kaiser, B.N., Franks, T. et al. Improved methods in Agrobacterium–mediated transformation of almond using positive (mannose/pmi) or negative (kanamycin resistance) selection-based protocols. Plant Cell Rep 25, 821–828 (2006). https://doi.org/10.1007/s00299-006-0139-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-006-0139-0