Abstract
Transgenic leek (Allium porrum) and garlic (Allium sativum) plants have been recovered by the selective culturing of immature leek and garlic embryos via Agrobacterium-mediated transformation using a method similar to that described by Eady et al. (Plant Cell Rep 19:376–381, 2000) for onion transformation. This method involved the use of a binary vector containing the m-gfp-ER reporter gene and nptII selectable marker, and followed the protocol developed previously for the transformation of onions with only minor modifications pertaining to the post-transformation selection procedure which was simplified to have just a single selection regime. Transgenic cultures were selected for their ability to express the m-gfp-ER reporter gene and grown in the presence of geneticin (20 mg/l). The presence of transgenes in the genome of the plants was confirmed using TAIL-PCR and Southern analysis. This is the first report of leek and ‘true seed’ garlic transformation. It now makes possible the integration of useful agronomic and quality traits into these crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the agronomic benefits of transgenic first-tier crops become more obvious there is an increasing need to develop transformation systems for second tier, niche crops. Leek (Allium porrum) and garlic (A. sativum) are such crops. Because they are closely related to onion (Allium cepa), it is possible that the onion transformation method developed by Eady et al. (2000), which has been shown to be cultivar-independent and effective with different constructs and selective agents (Eady 2002; Eady et al. 2003a, b), might be successfully applied to these crops.
Although there are no reports of leek transformation, somatic hybrid production between leek and onion using protoplast fusion and tissue culture has been reported (Buiteveld et al. 1998). There are three previous reports of garlic transformation. Kondo et al. (2000) used Agrobacterium to transfer the uidA reporter gene and the hygromycin (hyg) selective gene. Park et al. (2002) used biolistics to transfer herbicide resistance to a similar cell culture system. Most recently, Zheng et al. (2004) introduced insect resistance traits into five lines of garlic using Bt constructs. However, the process required lengthy pre-culture periods, and stable transgenic plants were only achieved from one explant of a bulbil and one root explant culture of garlic cv. Printanor. These three systems all required considerable time in tissue culture, raising the possibility of incorporating undesirable somaclonal mutations that cannot easily be eliminated from clonal material (such as garlic). The objective of the work described here was to determine if the immature embryo transformation protocol described by Eady et al. (2000) could be successfully applied to other Allium species (garlic and leek) to produce stable transgenic plants.
Materials and methods
Transformation followed the protocol of Eady et al. (2000) with the following modifications.
Plant material
Field-grown umbels of Allium porrum and A. sativum were used as a source of immature embryos. Immature embryos were isolated following the procedure of Eady and Lister (1998).
Bacterial strain
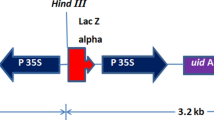
Agrobacterium tumefaciens strain LBA4404 containing the binary vector pBIN m-gfp-ER (Haseloff et al. 1997), which includes the modified gfp (green fluorescent protein) reporter gene that is targeted to the endoplasmic reticulum (ER) (m-gfp-ER) and the nptII (neomycin phosphotransferase II) antibiotic selectable marker gene, was used. Cultures were grown to log phase in LB media containing 50 mg/l of kanamycin and 100 mg/l streptomycin, and then stored at −80°C in 1-ml aliquots containing 15% glycerol. Aliquots were used to inoculate 50-ml overnight cultures. The following morning the cultures were replenished with an equal volume of LB containing the appropriate antibiotics and 100 μM acetosyringone and grown for a further 4 h. Agrobacteria were isolated by a 5-min centrifugation at 4,000 rpm and resuspended in P5 media (consisting of embryogenic induction media from Eady et al. 1998 and 5 mg/l of picloram) (Eady and Lister 1998) containing 200 μM acetosyringone at a density of 0.5–1.0 OD550 nm.
Transformation procedure
Immature embryos from seeds held overnight at 4°C were isolated in batches of 50, cut into approximately 1- to 2-mm sections and used for transformation experiments with no prior pre-culture. Between 14 and 19 batches were used in each of four experiments for both leek and garlic immature embryos. An Agrobacterium culture (400 μl) was added to each batch before vortexing for 30 s. The batches were then placed under vacuum (approx. 20 inch Hg) for 30 min. Embryos were then placed on filter paper for 1 min to remove excess bacteria before being transferred to P5 medium (approx. three batches of embryos per plate). After 6 days of cocultivation, the embryos were transferred to P5 medium containing 12 mg/l geneticin and 200 mg/l timentin. These embryo pieces were cultured in the dark under the conditions described for the production of secondary embryos (Eady and Lister 1998). The cultures were then transferred to fresh medium every 2 weeks for 12 weeks. During this period, the cultures were examined for GFP expression by observation under a fluorescence microscope using 475-nm excitation and 510-nm emission (Haseloff et al. 1997). Any material that was not expressing the transgene was discarded. Actively growing tissue pieces were then transferred to regeneration medium K-7S (Shahin and Kaneko 1986) containing geneticin and timentin at the same concentration as present in the P5 media and placed in a culture room under a 16/8-h (light/dark) photoperiod. This material was transferred to fresh media every 3 weeks and developing shoots were transferred to half-strength MS media (Murashige and Skoog 1962) plus geneticin to induce rooting. Rooted plants were transferred to a perlite, bark and peat mix (1:1:1) containing 400 g/50 l dolomite, 300 g/50 l osmocote, 75 g/50 l superphosphate, 50 g/50 l sulphate of potash, topped with perlite and placed in the glasshouse.
Analyses of transgene expression
Tissue GFP expression was detected as previously described (Eady et al. 2000). Growth and regeneration on media containing geneticin was used as an indication of nptII gene expression.
Initial detection of transgenes
PCR reactions to confirm the presence of the nptII transgene in leek and garlic DNA were performed on DNA isolated using a urea extraction method (Shure et al. 1983). Primers—nptII forward and reverse (Table 1)—were used to amplify a 600-bp fragment. Each 25-μl PCR reaction contained approximately 100 ng DNA, 2.5 μl 10× buffer (Roche, Indianapolis, Ind.), 2.5 μl of 200 nmol of each dNTP, 100 μmol of forward and reverse primers, 0.2 μl Taq (Roche). PCR reactions consisted of one cycle of 5 min at 94°C, followed by 35 cycles at 94°C, 60°C and 72°C for 1 min each, then one cycle of 7°min at 72°C.
TAIL-PCR procedure
Thermal asymmetric interlaced (TAIL)-PCR was performed as described (Liu et al. 1995) with some modifications. Three forward specific nested (SN) primers were designed from sequence data of the right border of pBin19 (from which pBIN m-gfp-ER was derived). These were: RBTAIL1, RBTAIL2 and RBTAIL3. The return primers were the arbitrary degenerate (AD) primers AD1, AD2, AD3 and AD4 (Table 1). Primary TAIL-PCR was performed on approximately 100 ng genomic DNA as a template in 25-μl reactions containing 1× supplied Taq polymerase buffer, 1.5 mM MgCl2, 200 nM of each dNTP, 200 nM of specific nested primer, 2 μM of AD primer and 1 U Taq polymerase (Roche). Primers for the first PCR reaction were the right border SN primer RBTAIL1 and one of the four AD primers as the return primer. The secondary reaction solution was the same as that for the primary reaction except that 1 μl of the primary reaction product diluted 1:50 was used as a template with RBTAIL2 as the forward primer. Likewise, the tertiary reaction solution was the same except that 1 μl of the secondary reaction product diluted 1:50 was used as a template and RBTAIL3 was used as the forward primer. TAIL-PCR reaction conditions are as follows.
Round-1 PCR
One cycle of 2 min at 95°C; five cycles of 1 min at 94°C; 1 min at 62°C, 2.5 min at 72°C; one cycle of 1 min at 94°C, 6 min at 25°C ramping to 72°C at 0.3°C/sec, 2.5 min at 72°C; fifteen cycles of 30 s at 94°C, 1 min at 62°C, 2.5 min at 72°C, 30 s at 94°C, 1 min at 62°C, 2.5 min at 72°C, 30 s at 94°C, 1 min at 44°C, 2.5 min at 72°C; 5 min at 72°C, then a hold at 4°C.
Round-2 PCR
Fifteen cycles of 30 s at 94°C, 1 min at 62°C, 2.5 min at 72°C, 30 s at 94°C, 1 min at 62°C, 2.5 min at 72°C, 30 s at 94°C, 1 min at 44°C, 2.5 min at 72°C; 5 min at 72°C, then a hold at 4°C.
Round-3 PCR
Thirty cycles of 1 min s at 94°C, 1 min at 44°C, 2.5 min at 72°C; 5 min at 72°C, then a hold at 4°C.
Sequencing of TAIL-PCR products
Amplified TAIL-PCR products were sequenced by reamplifying from agarose plugs of the third-cycle product. This was then run out on a gel to determine purity. Pure single bands were further purified using the Roche High Pure PCR Purification kit. If a single band was not produced, the desired band was separated and purified using the Qiagen Qiaquick Gel Extraction kit (Qiagen, Valencia, Calif.). Cleaned-up PCR product (2 μl) was run next to 1. 5 μl and 3 μl of Low Mass ladder (Invitrogen, Carlsbad, Calif.) to quantify and estimate product size. PCR products were sequenced by standard Big Dye Cycle sequencing with primer RBTAIL3 at the University of Waikato DNA sequencing facility, Hamilton, New Zealand.
Southern analysis
The Southern procedure described by Eady et al. (2000) was used to reconfirm the integrated nature of the transgenes and to determine the copy number.
Results and discussion
Embryo isolation
This research identified several differences between leek, garlic and onion. Leek seeds mature differently in that the seed coat hardens and darkens off later than in onions. This requires the identification of different external signals to gauge the correct immature embryo stages required for transformation. After most of the petals had withered, umbels were checked for the correct seed stage. Leek, despite having considerably smaller seeds, had embryos of a similar size to onion. However, it had less endosperm material. Garlic seed and embryos are smaller than those of onion. The pungency and tearing factor released from garlic seed heads upon disturbance was also noticeably less. Leek flowers produced much more tearing than onion flowers while garlic produced almost none. Finally, the juice released from the ovary wall upon disruption was much more irritating in garlic than in either leek or onion. Gloves were required to prevent skin damage when handling garlic ovary tissue during the isolation process. These observations raised interesting speculations about the temporal and spatial distribution of the various forms of alkyl cysteine sulfoxides and catalytic enzymes such as alliinase and lachrymatory factor synthase within the floral organs of their related Allium species. Such differences between Allium species have been demonstrated (Imai et al. 2002). Unpublished data within our laboratory showing differential temporal expression of alliinase and lachrymatory factor support the above observations.
Leek and garlic seed coats were more sensitive to bleaching than onion. Consequently, seeds were placed in a 30% commercial bleach solution for 10 min instead of the 30 min used for onion sterilisation.
Selection and regeneration
Leek
Immature leek embryos exhibited a quite different response compared to immature onion embryos when placed on P5 media. Cultures were much whiter and more friable than those observed during onion immature embryo selection and culture. When placed on K-7 media the leeks produced mostly root structures with masses of ‘fluffy’ root hairs above the media. When shooting cultures were placed on half-strength MS (Eady et al. 1998) to induce rooting, they grew extremely slowly compared to onion tissue. Ex-flasking these plants was very difficult. The leaves turned a very light-green colour when put in the glasshouse, and many clonal plants died before some were successfully ex-flasked (Fig. 1a). Surviving plants continued to grow slowly with new leaves quickly dying off from the tips, especially under strong light. Shading improved the growth of these plants, but the cause of the poor growth has yet to be determined. It may be due to somaclonal effects arising during in vitro culture, although this has not been observed in non-transgenic leeks regenerated from tissue culture. However, non-transgenic regeneration produces thousands of shoots to choose from so only vigorous ones were likely chosen. With the transgenic line the luxury of choice did not exist. Zheng et al. (2004) had similar problems ex-flasking GFP-positive garlic.
Garlic
Garlic immature embryos responded to selection and culture in a fashion more similar to that of onions than leek. The major difference in garlic was that a high proportion of the immature embryos (approx. 50% in some experiments) simply produced elongated cotyledons (similar to etiolated seedlings) in response to the P5 media. These embryos failed to produce embryogenic cultures. Cultures that were transgenic responded to regeneration and rooted, although like leek they took much longer to regenerate than onion. Fortunately, like onion they were relatively easy to ex-flask and once in the glasshouse grew into mature garlic plants, producing cloves and topsets (Fig. 1a).
Transformation results
Leek
In three experiments approximately 2,250 embryos were isolated from field-grown Allium porrum umbels and transformed with the binary vector pBIN m-gfp-ER (Haseloff et al. 1997) (Table 2). A transformation frequency of about 0.09% was obtained (two events from 2,250 embryos). However, there are many explanations for this low frequency. The first experiment used leeks from a commercial source that had been in transit at ambient temperature for 4 days. The embryos consequently did not arrive in a healthy state nor were they at the optimum stage of development. In experiments 0119 and 0120, embryos were isolated from a small population of locally grown Musselburgh cultivar and may have contained many selfed individuals. Finally, umbels in experiment 0120 had been refrigerated for 3 days prior to embryo isolation. This appears to be detrimental to transformation success (Eady, unpublished data).
Initial transgenic tissue was selected using GFP expression and growth on geneticin (Fig. 1d). Several clonal plants of two independent transgenic events were regenerated and ex-flasked (Fig. 1b), and these have been confirmed as transgenic by molecular diagnostics.
Garlic
In four experiments (experiments 0122–0125) approximately 3,200 embryos were isolated from field-grown Allium sativum umbels and transformed with the binary vector pBIN m-gfp-ER (Haseloff et al. 1997) (Table 2). A transformation frequency of approximately 0.06% was obtained (two events from 3,200 embryos). Both of these plants were the result of a steady improvement in transient gene expression achieved during progression from experiments 0122 to 0125 (such as skill in isolating the embryos and choosing larger, apparently healthy embryos). In experiment 0122, embryos were very small (average approx. 2 mm), and most succumbed to Agrobacterium overgrowth. Developing skills in isolating embryos markedly improved the success of transient gene expression. In experiment 0125, a multicellular GFP sector development (Fig. 1c) of 1% was achieved, and the transformation efficiency was 0.1%
Molecular analysis
Detection of integrated transgenes
TAIL-PCR (Liu et al. 1995) from right borders of T-DNA inserts was used to determine unique integration events, as product size and banding pattern of TAIL-PCR are dependent on the sequence composition of flanking genomic DNA. As outlined by Eady (2002), this process has been adopted to detect and confirm transformants in Allium species as it can be applied to small amounts of tissue and is relatively rapid, providing a screen of putative clonal material for escapes and different transformation events.
Leek
PCR analysis of the plants using the nptII primers indicated the presence of the nptII gene in the transformants tested (Fig. 2). TAIL-PCR of these plants has confirmed different integrated independent transgenic events (Fig. 3).
Two individual TAIL-PCR products were amplified from leek transformants. One line, from experiment 0119, showed a single TAIL-PCR step down, which indicated integration of the T-DNA right border into the leek genome. Sequence information (Fig. 4) from this step-down band confirmed this. The other TAIL-PCR step-down suggested a more complex integration pattern as two step-downs were apparent. This was confirmed by Southern analysis, which confirmed that a line from experiment 0120 contained multiple integration events. (Fig. 5). PCR primers designed from the TAIL sequence of a transgenic leek line from experiment 0119 amplified the expected fragment size and failed to amplify non-transgenic DNA or transgenic leek from experiment 0120, confirming the unique nature of the integration site.
TAIL-PCR sequence across the right-border integration junction showing the T-DNA right border sequence (black) and the leek genomic DNA (grey). The lowercase c at position 91 is different to the Genbank consensus, which is a T. The leek DNA has no sequence similarity with any sequence in the Genbank
Plants are presently being grown to maturity to obtain seed, which will allow the inheritance and stability of the transgene in leeks to be tested. However, this is proving difficult as containment glasshouse facilities do not reflect natural conditions and getting good growth for floral induction is an issue. To date, clonal regeneration through bulbs has been used to maintain the material (Fig. 6)
Garlic
PCR analysis of the plants using nptII primers indicated the presence of the nptII gene in the transformants tested (Fig. 2). An individual TAIL-PCR product was amplified from a garlic transformant line from experiment 0124. (Fig. 3.) Sequence from the third-cycle TAIL amplification was used to confirm integration and showed that the clonal material was indeed clonal. Southern analysis (Fig. 5) of clones showed that a line from experiment 0125 contained two transgenic loci, with possibly one locus containing two copies of the transgene. Different migration distances of the transgene fragments reflected differences in the running distances of the digested DNA (not shown), and this needs to be considered before deciding upon whether integration sites are different or the same. Also,it was noted that the lower band is more intense in lane 8 than in lane 7, this could be attributed to a chimeric transformant and is similar to that noted by Domínguez et al. (2004). However, Southern blot analysis of large genomes is often complicated by the large amount of DNA that has to be loaded, and such a phenomenon could just as likely be attributed to different impurities in the preparations or differences in degradation status of the DNA in the sample. Analysis of topsets from these plants and TAIL sequence can help confirm this situation. Kondo et al. (2000) produced Southern data for two lines from 1,000 callus pieces (0.2% transformation frequency). While this is higher than reported here (0.1% in the last experiment), their process required a pre-culture period of at least 2 months, a selective callus culture period of 2 months, and 4–5 months on shoot selection medium before the shoots were induced. A lengthy culture period of 14 months was also required by Park et al. (2002) to produce a few chlorosulfuron-resistant garlic lines via bombardment. The technique reported here can process up to 1,500 embryos per experiment and two experiments per week. Shoots can be regenerated within just 4 months so although transformation frequency is low, efficiency is relatively high.
Zheng et al. (2004) claimed rapid (6 months) and high-frequency transformation (1.47% from one experiment). However, after their exhaustive transformation experiments on several different cultivars and sources, only two lines survived in the glasshouse and Southern data was produced for just one of these. This result is thus comparable with that reported here following much less exhaustive experimentation.
Plants were grown to maturity to obtain topsets and cloves (Fig. 6). These are being regenerated to determine the stability of the transgene through successive generations. It is unlikely that glasshouse conditions can replicate conditions required for floral induction, so sexual inheritance studies are not feasible unless the plants can be field-grown in the correct environment.
Conclusion
The repeatable transformation system for onion developed by Eady et al. (2000) has been extended and applied to other members in the Allium genus. Further optimisation of conditions is necessary to improve the response of leek and garlic to the transformation process, particularly aspects of media composition, co-cultivation conditions and immature embryo health. A higher than expected frequency of multiple copy inserts has been reported by Zheng et al. (2004) when transformating garlic and onion. This was not observed previously by Eady et al. (2000) but could not be determined in this report due to small sample size. Copy number may reflect the quality of the material being transformed and its ability to readily regenerate under the given culture conditions rather than any innate difference amongst Alliums to integrate T-DNA.
The development of garlic production systems capable of producing commercial quality ‘true seed’ and the short in vitro culture periods of the transformation method described here would make immature embryo transformation an appealing option for garlic. It is also the first demonstration of stable transformation in leek, opening the way for transgenics to be used to introduce beneficial traits into this species.
References
Buiteveld J, Suo Y, Campagne MMV, Creemersmolenaar J (1998) Production and characterization of somatic hybrid plants between leek (Allium ampeloprasum L.) and onion (Allium cepa L.). Theor Appl Genet 96:765–775
Domínguez A, Cervera M, Pérez RM, Romero J, Fagoaga C, Cubero J, López MM, Juárez JA, Navarro L, Peña L (2004) Characterisation of regenerants obtained under selective conditions after Agrobacterium-mediated transformation of citrus explants reveals production of silenced and chimeric plants at unexpected high frequencies. Mol Breed 14:171–183
Eady C (2002) Genetic transformation of onions. In: Rabinowitch MD, Currah L (eds) Allium crop science: recent advances. CABI Publ, Oxon, pp 234–301
Eady CC, Lister CE (1998) A comparison of four selective agents for use with Allium cepa L. immature embryos and immature embryo derived cultures. Plant Cell Rep 18:117–121
Eady CC, Butler RC, Suo Y (1998) Somatic embryogenesis and plant regeneration from immature embryo cultures of onion (Allium cepa L.). Plant Cell Rep 18:111–116
Eady C, Weld R, Lister C (2000) Agrobacterium tumefaciens-mediated transformation and regeneration of onion (Allium cepa L.). Plant Cell Rep 19:376–381
Eady C, Davis S, Farrant J, Reader J, Kenel F (2003a) Agrobacterium tumefaciens-mediated transformation and regeneration of herbicide resistant onion (Allium cepa) plants. Ann Appl Bio 142:213–217
Eady C, Reader J, Davis S, Dale T (2003b) Inheritance and expression of introduced DNA in transgenic onion plants (Allium cepa L.). Ann Appl Biol 142:219–224
Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94:2122–2127
Imai S, Tsuge N, Tomotake M, Nagatome Y, Sawada H, Nagata T, Kumagai H (2002) Plant biochemistry: an onion enzyme that makes the eyes water. Nature 419:685
Kondo T, Hasegawa H, Suzuki M (2000) genetic transformation and hybridization: Transformation and regeneration of garlic (Allium sativum L.) by Agrobacterium-mediated gene transfer. Plant Cell Rep 19:989–993
Liu YG, Mitsukawa NTO, Wittier R (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8:457–463
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Park M, Yi N, Lee H, Kim S, Kim M, Park J, Kim J, Lee J, Cheong J, Choi Y (2002) Generation of chlorosulfuron-resistant transgenic garlic plants (Allium sativum L.) by particle bombardment. Mol Breed 9:171–181
Shahin E, Kaneko K (1986) Somatic embryogenesis and plant regeneration from callus cultures of nonbulbing onions. Hortscience 21:294–295
Shure M, Wessler S, Fedoroff N (1983) Molecular identification and isolation of the Waxy locus in maize. Cell 35:225–233
Zheng SJ, Henken B, Ahn Y, Krens F, Kik C (2004) The development of a reproducible Agrobacterium tumefaciens transformation system for garlic (Allium Sativum L.) and the production of transgenic garlic resistant to beet armyworm (Spodoptera exigua Hubner). Mol Breed 14: 293–307
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Peña
Rights and permissions
About this article
Cite this article
Eady, C., Davis, S., Catanach, A. et al. Agrobacterium tumefaciens-mediated transformation of leek (Allium porrum) and garlic (Allium sativum). Plant Cell Rep 24, 209–215 (2005). https://doi.org/10.1007/s00299-005-0926-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-005-0926-z