Abstract
Increasing evidence indicates that plants, like animals, use basal resistance (BR), a component of the innate immune system, to defend themselves against foreign organisms. Contrary to the hypersensitive reaction (HR)-type cell death, recognition in the case of BR is unspecific, as intruders are recognised based on their common molecular patterns. Induction of BR is not associated with visible symptoms, in contrast to the HR-type cell death. To analyse the early events of BR in tobacco plants we have carried out a subtractive hybridisation between leaves treated with the HR-negative mutant strain Pseudomonas syringae pv. syringae 61 hrcC and non-treated control leaves. Random sequencing from the 304 EBR clones yielded 20 unique EST-s. Real-time PCR has proved that 8 out of 10 clones are activated during BR. Six of these EST-s were further analyzed. Gene expression patterns in a time course showed early peaks of most selected genes at 3–12 h after inoculation (hpi), which coincided with the development-time of BR. Upon treatments with different types of bacteria we found that incompatible pathogens, their hrp mutants, as well as non-pathogens induce high levels of expression while virulent pathogens induce only a limited gene-expression. Plant signal molecules like salicylic acid, methyl jasmonate, ethylene and spermine, known to be involved in plant defense were not able to induce the investigated genes, therefore, an unknown signalling mechanism is expected to operate in BR. In summary, we have identified representative genes associated with BR and have established important features of BR by analysing gene-expression patterns.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants, like animals, face a myriad of potential intruders, both pathogens and non-pathogens. They are well equipped to defend themselves against invading micro-organisms, with preformed barriers and inducible protective mechanisms. Micro-organisms that break through the preformed barriers will face a set of induced defence responses, including the symptomless basal resistance (BR) and hypersensitive cell death (HR).
The HR-type cell death develops following a specific recognition process. In the case of pathogenic bacteria, assembly of the bacterial type-III secretion system enables avirulence (Avr) proteins to be injected into plant cells. Avr proteins are specifically recognised (directly or indirectly) by products of resistance (R) genes, after which the HR develops. During the HR an oxidative burst and rapid death of the infected plant cells are observed along with localisation of the pathogen (Cutt and Klessing 1992; Mehdy 1994; Staskawicz et al. 1995).
The basal resistance response (BR), also mentioned as a form of induced resistance and innate immunity has received less but emerging attention. This type of resistance defends all plant species against most microbes, without any visible symptoms or cell death (Klement et al. 2003). General conserved elicitors of microbes are recognised in a non-specific manner (unlike the R-gene dependent specific HR) (Nurnberger and Brunner 2002; Nurnberger et al. 2004). Flagellin is an example of such a general elicitor: it is found in most bacteria and activates defence responses through MAPK (Mitogen-Activated Protein Kinase) cascades in both plants and animals (Asai et al. 2002; Gómez-Gómez and Boller 2002). General microbial elicitors are often designated PAMPs (Pathogen Associated Molecular Pattern), although they are found not only on pathogens. Other described bacterial PAMPs are lipopolysaccharides (LPS) (Dow et al. 2000; Meyer et al. 2001; Gerber et al. 2004; Keshavarzi et al. 2004); and cold shock proteins (Felix and Boller 2003), etc.
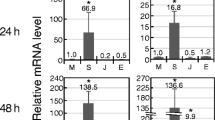
Temporal expression patterns of selected tobacco genes after infiltration with 108 CFU/ml Pseudomonas syringae pv. syringae hrcC mutant. Vertical axis: transcript levels relative to the absolute control. All values were normalised with actin. Error bars indicate SEM n=2. Horizontal axis: time after treatment (h). ▪: Pseudomonas syringae pv. syringae hrcC mutant treatment. ▴: Water control treatment. Note that the values of maximum activation are different for each gene
BR requires active plant metabolism for the induction of defence mechanisms against any non-pathogenic and sometimes opportunistic intruders, as shown by experiments with protein-synthesis inhibitors (Bozsó et al. 1999). The observed mechanisms include cell wall alterations (thickening and lignification, papilla formation), accumulation of phenolics, phytoalexins, and induction of PR (pathogenesis related) genes (Jakobek and Lindgren 1993; Ott et al. 1997; Brown et al. 1998; Dixon 2001; Navarro et al. 2004; Keshavarzi et al. 2004). The basal resistance response is triggered upon infection with heat-killed pathogens, non-pathogens or pathogenicity-deficient mutants (Klement et al. 1999; Burgyán and Klement 1979; Bozsó et al. 1999). The multiplication of bacteria is inhibited and a local resistance response develops at the site of inoculation. This means that an HR will be absent after a second, challenging inoculation with an incompatible pathogen and bacterial growth will be arrested after a challenging inoculation with compatible pathogens (Ott et al. unpublished data). Plants display this kind of resistance not only in response to hrp mutants and nonhost pathogens (P. syringae), but also non-plant pathogens, such as E. coli. Because there are some molecular and pathological differences between the early (0–24 hpi) and later phases of BR (Klement et al. 2003; Burgyán and Klement 1979), we have designated the early phase as EBR and have focused on this part of BR in the present paper. We have used an hrp mutant of Pseudomonas syringae pv. syringae to induce basal resistance. This approach enabled direct comparison to the isogenic, non-mutant strain. The hrp mutant bacteria fail to cause any macroscopically visible symptoms, their multiplication is arrested in the plant followed neither by disease, nor a hypersensitive reaction (Lindgren 1997; Klement et al. 2003).
Most investigations had been focusing on the HR, therefore our knowledge of the non-specific BR mechanism is limited. More studies are needed to reveal details and relationships between different defence reactions; which was one of the aims of our work. In order to investigate the changes in gene transcription upon infection with non-pathogenic bacteria, we carried out subtractive hybridisation between cDNA populations from Pseudomonas syringae 61 hrcC mutant—injected and untreated control tobacco plants. We obtained a cDNA library containing clones of activated genes including housekeeping genes, genes connected to secondary metabolism and cell wall fortification, signal transduction, detoxification and protection against oxidative stress. Some clones could not be assigned to a function. Focusing on selected genes of cell wall fortification and detoxification, we analysed gene expression patterns by real-time PCR. The application of this rapid and quantitative method allowed us to assess the impact of diverse treatments on the selected set of representative genes and obtain quick and reliable data in order to better understand the nature of BR. This provides a useful basis for future array-based experiments, where a wider set of genes can be investigated, but the possible number of treatments is strongly limited. We have revealed similarities and differences in plant gene expression during the non-pathogen specific BR and the more well-known, pathogen-specific HR, providing insight into the possible molecular mechanism(s) of BR, a less known defence mechanism of plants.
Expression patterns of the selected genes in response to treatments with different bacteria at 6 hpi. Expression values are relative to the absolute control level. All values were normalised with actin values. Error bars indicate SEM n=3. W: water control; C: P. syringae hrcC mutant; P61: P. syringae pv. syringae 61; PF: P. fluorescens TAB KM: kanamycine-inactivated P. tabaci; TAB: P. tabaci; COLI: E. coli; AGR: A. tumefaciens; SM 41: S. meliloti Asterisks (*) denote values determined to be significantly different from the water-treated control (W) at a P<0.05 level
Materials and methods
Plant material and treatment
Tobacco plants (Nicotiana tabacum cv. Samsun nn; Nicotiana tabacum cv. Xanthi NN; Nicotiana tabacum cv. Xanthi NN:NahG) were grown in the greenhouse in soil. NahG-10 were kindly provided by NOVARTIS, Agricultural Biotechnology Research, Research Triangle Park, NC. Before inoculation the 2–2.5-month-old tobacco plants were kept in a growth chamber with 16/8 h light/dark period at 20°C 2 days before and during experiments. Hypodermic syringes fitted with a 25 gauge needle were used for the injection of tobacco leaves as described by Klement (1990). At the appropriate time points, 0.1 g leaf samples were taken, and frozen immediately in liquid nitrogen. In every experiment both non-treated and water-infiltrated control leaves were used. Every experiment was carried out on at least two plants as biological replications and these data were used for evaluation. The experiments were also carried out using other plant generations to confirm the detected trends in gene expression changes. These data were not used in statistical analysis, as the values of fold-changes might differ more between different generations of plants than between ones from the same generation, for example due to slight changes in the greenhouse circumstances during plant growth. This might be observed in the diagrams of Figs. 2 and 3. In the response to C (Pseudomonas syringae pv. syringae hrcC) for example EBR-52 in Fig. 2 had a relative expression level close to 30, while in Fig. 3 it is close to 15.
Bacterial treatments
Bacterium strains and their interactions with plants used in this study are indicated in Table 1. Pseudomonas, Agrobacterium, Escherichia and Sinorhizobium strains were cultured at 28°C on King's medium B (King et al. 1954). Mutants were grown on antibiotic-containing plates (kanamycin, 50 μg/ml). Pseudomonas syringae pv. tabaci was inactivated with 50 μg/ml kanamycin for 10 min, than washed twice in distilled water. Overnight cultures were used for infiltration, suspended in distilled water to 108 CFU/ml, with a densitometer at 560 nm.
Signal molecule treatments
Plant signal molecules or their precursors used in this study were the following. Methyl-jasmonate, 200 μM, salycilic acid, 400 μM, spermine, 100 μM, aminocyclopropane-carboxylic acid (ethylene-precursor), 1 mM (Sigma). All of the molecules were used alone and also as combined with the HR-negative Pseudomonas syringae pv. syringae hrcC (108 CFU/ml).
HR-inhibition test
Detection of EBR was done by an HR-inhibition test as described by Burgyán and Klement (1979). Briefly, intervenials of whole leaf panels were pre-treated with Pseudomonas syringae pv. syringae hrcC (108 CFU/ml) salicylic acid or water. Intervenials were thereafter serially inoculated with Pseudomonas syringae pv. pisi (108 CFU/ml) as challenging bacteria, every half an hour. Total absence of the HR normally caused by P. pisi indicated the presence of EBR.
Construction of a subtracted cDNA library enriched for EBR-related sequences
P. syringae 61 hrcC treated or control leaves (0.1 g pieces) were ground in liquid nitrogen 6 h after inoculation. Total RNA was extracted using the Plant Total RNA Extraction Miniprep System (Viogene). mRNA was obtained from 100 μg total RNA for driver and tester each, using the PolyATtract System (Promega) as recommended by the manufacturer.
cDNA production and subtractive hybridisation were performed using the PCR Select cDNA Subtraction Kit (Clontech). Uninoculated plant material served as “driver” and inoculated plant material as “tester”. Cloning of subtracted fragments was carried out using the TOPO TA Cloning Kit for Sequencing (Invitrogen).
Nucleotide sequencing and data analysis
All of the 304 obtained clones were individually PCR amplified. Sequencing of 20 random clones was carried out using plasmid DNA at the sequencing facility of MWG-Biotech Ag. (Ebersberg, Germany). Vector and adaptor sequences were removed manually. Clone identification was performed using BLAST search (Altschul et al. 1990).
Quantitative RT-PCR analysis of gene expression
Total RNA was extracted using the Plant Total RNA Extraction Miniprep System (Viogene) from 0.1 g treated or control leaf material ground under liquid nitrogen. The concentration of isolated RNA was estimated by measuring its absorbency at 260 nm. Reverse transcription of 2.5 μg total RNA was carried out with the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas) using the oligo(dT) primer, according to the manufacturer's instructions.
For real-time PCR analysis we have designed the primers indicated in Table 2. Specificity of the primers at the applied PCR conditions was initially verified by agarose gel-runs, which yielded a single product at the expected molecular size in the case of each primer pair. Melting curve runs were also performed at the end of each PCR reaction to verify the presence of a single product.
We used 2.5 μl of a 10-fold dilution of the cDNA stock, in 15 μl reactions. Primer concentrations were 3 μM. PCR was carried out using the iQ SYBR Green 2x Supermix (Biorad), on the DNA Engine Opticon 2 (MJ Research). Cycling parameters were the same for all primers: initial 95°C for 6 min, followed by 40 cycles of 95°C for 30 s, 60°C for 1 min, plate read step, then product melting curve 55°C–95°C. Serial tenfold dilutions (10–108) of the plasmids containing the specific inserts were initially run together with the plant samples to verify that the measurements are within the linear range (correlation coefficient > 0.99).
Relative quantification of gene expression was carried out according to the Applied Biosystems ABI PRISM 7700 Sequence Detection System User Bulletin #2 (1997) using the comparative cycle threshold [C(T)] method for the calculation of ΔC(T) and ΔΔC(T) values. A Microsoft Excel 97 spreadsheet was generated for this. Measured C(T) values were always normalised against actin (GeneBank X69885) as an internal control. Expression ratios of two technical repeats were averaged for each sample.
For the results of the (at least two) biological replications from the same plant generation, standard errors (SEM) were calculated and differences between expression ratios of water-treated controls and other samples were tested for statistical significance using a Students's t test. The absolute controls were given the arbitrary value of 1 i.e. ΔΔC(T) values were calculated using the absolute controls as calibrator.
Results and discussion
Isolation of differentially expressed gene fragments
To map the changes in gene expression during the symptomless EBR of tobacco plants a subtractive hybridisation approach was applied. We were interested in early reactions, so leaf material injected with the non-HR-inducing P. syringae hrcC mutant sampled at 6 hpi was used for mRNA isolation, as well as leaf material from untreated control plants. Double stranded cDNA was reverse-transcribed from both mRNA pools and subsequent rounds of hybridisation and PCR amplification were carried out. The final, amplified pool of cDNA sequences enriched in bacterial treatment-specific gene-fragments was cloned. All of the resulting 304 EBR clones were individually PCR amplified, the insert lengths were between 100 and 700 bp. Sequence information of 20 randomly chosen clones was obtained, six clones were under 200 bp in length, another six were 200–400-bp long and seven were over 400 bp. Similarities and putative functions of the fragments were assigned by using the BLAST tool (Altschul et al. 1990). Six out of 20 sequences corresponded to known genes (similarity>98%), 10 could be assigned some putative function according to BLAST results. Four fragments showed no significant similarity to any known genes. The clones were classified into 5 subgroups: genes with functions related to cell house-keeping; signal transduction; cell protection and detoxification; secondary metabolism and cell wall fortification; and genes with unknown functions (see Table 3). Seven genes (or their most similar homologue) were found in the literature to be induced by biotic stress. However, bacteria were involved only in two cases, the rest of the papers reported induction upon viral or fungal elicititation.
To evaluate the success of subtractive hybridisation and to prove that the isolated genes are induced upon infection with the non-HR causing pathogen, specific primers were designed for 10 of the clones. Real time PCR experiments have proven that eight of the genes are activated during the early (0–24 h) time course of the defence reaction of tobacco against the HR-negative hrcC mutant of P. syringae. Therefore a high percentage of the isolated clones seem to be fragments of activated genes, which demonstrates the efficiency of subtraction.
Two subgroups were selected for further investigation with real-time PCR, the genes related to cell wall fortification and cellular protection. Genes of cell wall fortification are especially interesting, because papilla formation has been suggested as one key feature of the defence response against the nonpathogenic hrcC mutant of Xanthomonas campestris pv. vesicatoria in pepper (Keshavarzi et al. 2004), and also earlier in our laboratory (Ott et al. 1997). Alterations of the plant cell wall are known forms of basal resistance to fungi and to bacteria. Papillae and cell wall appositions might act as a barrier to the transfer of nutrients and water or block the injection of effectors (Keshavarzi et al. 2004). Hauck et al. (2003) have found that a high percentage of genes repressed by type III effectors are associated with plant cell wall functions. We also investigated a subgroup of genes that the plant might use to defend its own cells against reactive oxygen species or toxic metabolites. This ability is also crucial for successful resistance, because plant cells are under heavy (mainly oxidative) stress during defence (Barna et al. 2003).
Homologies and possible functions of the selected EBR genes
Genes of cell wall fortification
EBR-38A (Cinnamic acid 4-hydroxylase)
EBR-38A displayed the highest identity (83%) with C4H (cinnamic acid 4-hydroxylase) of pepper (Capsicum annuum) (AF212318). Cinnamic acid 4-hydroxylase is the second enzyme of the phenyl-propanoid pathway in plants, leading to the synthesis of lignin, pigments, and many defence molecules. However, it is also the rate limiting enzyme directly before the branching of the phenylpropanoid pathway. It catalyses the conversion of cinnamic acid into p-coumarate. C4H belongs to the structural family of P450 heme-thiolate proteins, which catalyse mono-oxygenation of a broad range of substrates within all organisms (Werck-Reichhart and Feyereisen 2000). C4H induction has been reported upon treatment with fungal elicitor in Ammi majus (Hubner et al. 2003), wounding, and chemical treatments (Batard et al. 1997; Anterola et al. 2002).
EBR-43-21 (Ortomethyl transferase a/b)
Among our sequenced clones two were identified as class I ortomethyl transferases (OMTs). These two clones differed only in a few nucleotides from each other. They were identified as OMT I-a (EBR-21, 98%) and OMT I-b (EBR-43, 100%) (Jaeck et al. 1996). In the case of EBR-21 we could not decide if it is in fact identical to OMT I-a, or if we have cloned a new isoform, to be called OMT I-c. During our gene-expression studies however, we have measured cumulative abundance of EBR-21 and -43 transcripts, as it was impossible to design primers that could differentiate between these two, nearly identical genes. OMTs have extensive roles in the later steps of the phenylpropanoid pathway, leading to the formation of coumarins, and different subunits (monolignols) of lignin (Maury et al. 1999). Induction of orthomethyl transferases has been widely studied upon infection with viral and fungal elicitors (Pakusch et al. 1989; Schmitt et al. 1991; Pellegrini et al. 1994). However, data concerning bacterial infections has not been extensive so far.
EBR-44 (Glycine-rich protein)
Glycine-rich proteins (GRP-s) are important cell-wall structural proteins beside extensins and proline-rich proteins (Ringli et al. 2001). EBR-44 was 99% identical with NtEIG-E17, a GRP reported to be responsive to fungal elicitors, and incompatible pseudomonads (Takemoto et al. 2003). Several other homologues of EBR-44 have also been reported, with similarity values above 90%. For example, NT16 has been isolated from crown gall tumor tissues induced by Agrobacterium tumefaciens (Yasuda et al. 1997). Other homologues are HR3S and HR4, both isolated from hairy roots induced by Agrobacterium rhizogenes; while the clone GENEBANK/D26454 comes from genetic tumors of tobacco (Fujita et al. 1994). The above genes closely related to EBR-44 seem to have connections to tumors and bacterial infections, consistently with their putative roles in cell-wall formation and fortification. Nodule-specific GRP-s have been described from root nodules of Medicago spp., specifically induced upon bacterial infection (Kevei et al. 2002).
EBR-59 (Extensin)
EBR-59 was similar to the extensin gene 6PExt 1.2 from Nicotiana sylvestris, to a level of 93% (Parmentier et al. 1995). 6Pext was shown to belong to a multigene family of related genes and was induced by wounding and A. tumefaciens infection, so it proved to be a stress-related gene, like EBR-59. Extensins are basic hydroxyproline-rich glycoproteins, which are important structural components of cell walls. Their structure enables them to form intermolecular and intramolecular links, thereby strengthening plant cell walls (Schowalter 1993). They are known to be developmentally regulated and their expression is also induced by different stresses like wounding, or microbial infections and different signal molecules (Hirsinger et al. 1999; Takemoto et al. 2001). In Arabidopsis thaliana the extensin gene is normally expressed in roots and inflorescences, but it is strongly activated in leaves after infection with the compatible pathogen X. campestris pv. campestris (Merkouropoulos and Shirsat 2003), demonstrating the bilateral involvement of extensin genes in tissue development, and self-defence.
Expression patterns of the selected genes in response to treatments with plant signal molecules at 6 hpi. Expression values are relative to the absolute control level and are normalized with actin values. Error bars indicate SEM n=3.W: water control; C: Pseudomonas syringae hrcC mutant; ME: methyl-jasmonate; SA: salicylic acid; SPM: spermine; ACC: amino-cyclopropane-carboxylic acid; ME+C: methyl-jasmonate+PS hrcC; SA+C: salicylic acid + PS hrcC; SPM+C: spermine+PS hrcC; ACC+C: ACC+PS hrcC. Asterisks (*) denote values determined to be significantly different from the water-treated control (W) at a P<0.05 level
Genes of plant cell protection
EBR-52 (Glutathione S-trasferase)
EBR-52 displayed highest identity to the glutathione S-transferase from Hyosciamus muticus (Hmgst-1) (Bilang and Sturm 1995). However, this was only 88%, indicating that we have cloned a distinct, novel member of the GST gene family. The closest relative of EBR-52 from tobacco was the par-B gene (Takahashi and Nagata 1992), where Lipman–Pearson protein alignment revealed a similarity index of 64.1 (Pearson and Lipman 1988). The above genes have a very interesting feature besides GST activity. They are induced by auxin, and are able to specifically bind its molecules. GST-s generally have been associated with detoxification of cytotoxic products, and protection against oxidative damage. In plants, GST-s have been extensively studied in connection with xenobiotic compounds like herbicides (Irzyk and Fuerst 1993). Mauch and Dudler (1993) have described three different GST-s from wheat, two of which seemed to have a specific role in xenobiotic metabolism, and one in defence reactions against pathogens. This was indicated by their differential regulation. We have shown that EBR-52 may play a role in defence reactions against bacterial infections based on transcription data, but no information is available about xenobiotic treatments. However, we have been able to demonstrate pathogen-specificity of EBR-52, as it was strongly activated after bacterial injections, but practically no activation was seen following viral infections (Szatmari et al. unpublished data). The roles of GST-s, a branched family of cell protectant enzymes, remains yet a field to be studied.
EBR-38B (Epoxide hydrolase (EH))
The obtained sequence of EBR-38B displays 98% identity with the Nicotiana tabacum epoxide hydrolase gene EH-1 (Guo et al. 1998) isolated as a gene activated during the resistance response to TMV. Guo et al. (1998) have described tobacco epoxide hydrolases either as a small gene family, or a single gene with several alleles based on genomic DNA hybridisation. Therefore, EBR-38B is likely to be a different allele of the gene, or a closely related member of the small family. Interestingly, the expression of this gene has been reported to be regulated by both SA-independent and SA-dependent pathways, similarly to our result with EBR-38B (see section Signal molecules). Epoxide hydrolases catalyse the conversion of epoxides into diols (Oesch 1973). Some epoxides are highly reactive and toxic, being able to damage biological macromolecules like DNA or proteins. EH-s play a critical role in detoxifying such reactive metabolites in both plants and animals (Murray et al. 1993). EH-s in plants have also been assigned a role in cutin biosynthesis (Pinot et al. 1993). Both of the above putative roles make EH-s promising candidates as genes involved in resistance responses.
Time course
Gene expression levels were investigated in a time course on the first day after inoculation. Figure 1 shows that transcription of the studied genes have followed a general pattern: they were already activated as early as 1–3 h after inoculation (depending on plant material). Activation peaked at 3–12 hpi, and their transcript level was already very low at 24–48 hpi. This pattern is well correlated to the temporal pattern (shown in Fig. 4.) of the HR-inhibiting nature of basal resistance (Klement et al. 1999) therefore these genes seem to be good molecular markers of EBR.
Within this common pattern some genes could be identified displaying either “quick” or “delayed” induction. Cinnamic acid-4-hydroxilase and epoxide hydrolase were considered to be the earliest induced genes (peak at 1–3 hpi, strongly declining at 6 hpi), while OMT-I and GST were induced with a slight delay (peak at 3 hpi, some decline at 6 hpi). Finally, the two genes encoding for extensin and glycine-rich protein had induction peaks at 12–24 hpi (depending on plant material) and their expression was still relatively high at 24–48 hpi. The results indicated that the general defence response has a well-established course of events. The connection is very clear in the case of cinnamic acid 4-hydroxilase and OMT as they follow each other not only in induction time, but also within the phenyl-propanoid biosynthetic pathway (Maury et al. 1999). Extensin and glycine-rich protein, the latest responders, are both structural proteins of the plant cell-wall, so this kind of cell-wall strengthening might represent a later step of defence events. Revealing sequential patterns of gene expression helps identifying sequential steps of the general defence response, and could provide new insights into its temporal regulation.
Different plant–bacterium interactions
We have examined different plant-bacterial interactions, as illustrated in Fig. 2. We used non-pathogenic as well as pathogenic bacteria, including compatible and incompatible relationships (Table 1). Our general findings were that the EBR-inducing mutant P. syringae hrcC bacterium caused similar gene activation increases to that of the HR-causing isogenic strain (P. syringae 61), the kanamycin-inactivated pathogen (P. tabaci), the saprophytic P. fluorescens and the non-pathogenic E. coli. Only a limited degree of induction was observed in three different cases: the virulent compatible pathogenic P. tabaci, the non-tumorigenic strain of A. tumefaciens and the non-pathogenic (non-symbiont to tobacco) S. meliloti strain.
HR-inhibition test. Demonstration of the ability of salicylic acid to cause a similar HR-inhibition as described in Materials and methods. 4a. Leaf panels pretreated with SA, P. syringae hrcC and water. Challenging bacteria—P. syringae pv. pisi—were injected every 0.5 h. Control leaf was injected with water, and HR was present at all time points. Numbers indicate time of challenging treatment after primary treatment. “x” indicates no challenging infection. Two HR patches in the same intervenial indicate two sites of challenging infection. 4b. Numeric data about the above experiment. Values are means of three independent repeats. 0=No EBR, full HR; 1=HR on cca. 60% of challenged area; 2= HR on cca. 30% of challenged area; 3=Full EBR, no HR
High rate of gene-induction was detected upon infection with HR-causing bacteria. Our results indicate that the genes of EBR are also transcribed intensively during the formation of HR, implying that the processes of these two reactions are overlapping in time. Recently Navarro et al. (2004) found significant overlap between transcriptional changes in array experiments comparing flagellin (general elicitor) treated Arabidopsis and Avr9 treated tobacco (specific, HR-inducing elicitor). Tao et al. (2003) suggested that Arabidopsis responses are quantitative in nature during different interactions with P syringae. They differentiated between two levels: low (compatible interaction) vs. high, (i.e. incompatible and non-host interaction). We have confirmed these two levels in our measurements. However, some independent repeats showed the existence of a 3rd, medium level in the case of EBR-causing bacteria (data not shown). There are some circumstances among which incompatible bacteria induce a stronger induction of EBR-related genes, however, these circumstances remain to be determined in the future. Besides overlaps it is likely that there are qualitative differences between EBR and HR on the expressional level. It should also be subject of future experiments to find selectively EBR-specific and HR-specific genes. One theory that we have considered based on our running experiments (Bozsó, unpublished data) that HR is the combination of EBR plus some complementary mechanisms that lead the process towards plant cell death (Klement et al. 1999).
Some isolated genes showed a limited induction in certain cases. Virulent P. tabaci actively suppresses EBR, as kanamycin-inactivated bacteria showed a “restored” gene-activity inducing feature, indicating that living virulent pathogens in susceptible hosts have special mechanisms to avoid this defence response, as was first suggested by Burgyán and Klement (1979), later by Jakobek et al. (1993), Klement et al. (2003), reviewed by Abramovitch and Martin (2004). Hauck et al. (2003) have recently shown that suppression of cell-wall based defence is carried out by effectors of the type III secretion system as proposed earlier (Brown et al. 1995). A. tumefaciens is also a plant- pathogen, however the strain used in our experiments was a non-tumorigenic one. Still it was able to diminish the induction of the responsive genes, so this species does not elicit or is able to actively suppress plant defence. S. meliloti, a symbiont of alfalfa also caused lower induction of defence genes, although it is not known to be involved in any specific relationship with tobacco.
We found that defence genes are also activated by non-HR inducing mutants, by non-pathogenic bacteria, and killed pathogens. This supports the assumption that the general defence response of plants contributes to the inhibition of the proliferation of a wide range of bacteria that have entered the intercellular spaces, unless those bacteria use special suppressor effectors. Such effectors mainly remain to be identified in the future. Our findings also emphasise the connection between pathogenic and symbiotic lifestyles of bacteria.
Expression patterns of the selected genes in wild type and NahG tobacco plants at 6 hpi. Expression values are relative to the absolute control level and are normalized with actin values. Error bars indicate SEM n=2. W: water control; C: Pseudomonas syringae hrcC mutant;. XA: Nicotiana tabacum cv. Xanthi NN, NahG: Nicotiana tabacum cv. Xanthi NN: NahG. Asterisks (*) denote values determined to be significantly different from the water-treated control (W) at a P<0.05 level
Signal molecules
We used four different signal molecules, which have formerly been associated with processes of pathogenesis and resistance, in order to find out which signal pathway(s) have a role in initiating the BR response. These were salicylic acid (SA), methyl-jasmonate (MeJa), an ethylene precursor (ACC) (aminocyclopropane-carboxylic acid), and spermine (SPM), as indicated by Fig. 3.
Salicylic acid is known to accumulate in plants after infections with necrotising pathogens. SA is able to cause systemic acquired resistance (SAR) and to activate a wide range of pathogenesis related (PR) proteins. Exogenously applied SA is also known to cause the previously mentioned effects (reviewed in Machácková et al. 2004). Jasmonates (JA) are volatile compounds that can modulate resistance to pathogens and insects, fruit ripening, senescence, etc. and are accumulated in plants upon wounding and elicitor treatment (Creelman and Mullet 1997). Ethylene is also a volatile molecule that, apart from developmental functions, has been shown to be involved in disease resistance and also susceptibility. Ethylene was shown not to be required for active defence against avirulent bacteria (Bent et al. 1992). Ethylene and jasmonates have been shown to be in both negative and in positive interaction regarding the regulation of defense-associated genes in some cases. (Lorenzo et al. 2003; Sasaki et al. 2001; Diaz et al. 2002). Spermine is a polyamine, isolated from resistant tobacco plants upon tobacco mosaic virus (TMV) infection. It has been described as a salicylate-independent exogenous inducer for acidic pathogenesis-related proteins and TMV-resistance (Yamakawa et al. 1998). A tobacco peroxidase, tpoxC1 was found not to respond to general defence-related signal compounds (SA, MeJa, ethylene) only to spermine, therefore spermine seems to represent a distinct signalling pathway (Hiraga et al. 2000).
Our genes, however, showed no significant transcriptional alterations in response to MeJa or ACC, nor displayed any significant change in transcription upon spermine treatment at the investigated time point, 6 h after injection.
The tested genes generally did not respond to exogenously applied SA either, but in some repeats a limited activation was observed (see 38B, 44 and 52). So we assayed if there is a BR-like phenomenon caused by SA (Fig. 4.). It was able to inhibit the HR to be caused by challenging incompatible bacteria, although to a lower extent than typical BR. Small necroses and chlorotic patches were still observed. In some repeats the combined application of SA and P. syringae hrcC mutant had a synergistic or additive gene activation effect. (see e.g. 38B and 43). Therefore we assume the existence of a potentiating effect between SA-induced resistance and BR. Shirasu et al. (1997) found a potentiating effect of SA on defence transcript accumulation and hypersensitive cell death in incompatible interactions. They observed the potentiating effect of SA in Avr-involving reactions, which we have now found to exist also in a non-HR reaction. Our results imply that the genes associated with BR are under primary control of an unknown signal that induces their transcriptional activation, but the SA-mediated pathway overlaps with BR events. To find out more about the role of SA, transgenic NahG tobacco plants were applied that metabolise SA into an ineffective compound so SA-dependent mechanisms cannot be induced. In our experiments gene activation was not impaired in NahG plants (Fig. 5.) proving the assumption that SA is not necessary for BR gene activation. Glazebrook et al. (2003) have found a group of genes in bacteria-infected Arabidopsis, that were regulated by SA and an unknown pathway, that was distinct from the group of genes regulated exclusively by SA. This unknown pathway might be related to the primary inducing pathway in our experimental system. DebRoy et al. (2004) have assayed callose deposition in Arabidopsis using hrpA mutants of P. syringae. Their results confirmed the SA-independent nature of cell-wall based immunity elicited by hrp mutants. However they have found another group of bacterium mutants inducing a SA-dependent pathway, approving the existence of two pathways.
In conclusion, none of the signal molecules alone induced significant increases in expression of the BR-responsive genes identified in this study. Based on our results, these signal molecules do not play a primary role in signal transmission during BR. Therefore, we propose the existence of a yet unknown plant defence signalling pathway during BR that might be potentiated by salicylic acid, and should be identified in the future.
Concluding remarks
Innate immunity, local induced resistance and basal resistance are terms in plant-pathology gradually gaining higher interest. We have isolated several genes associated with the early phase of basal resistance (EBR), which is an initial step towards the understanding of defence reactions. In this study we have focused on the expression patterns of genes involved in secondary metabolism, cell-wall fortification and detoxification, plant processes, which might have significant effector functions in plant defence. Our selected genes seem to be under control of a common signalling pathway, as their transcriptional activation patterns were closely similar upon the used treatments. This pathway is not under primary control of the tested signal molecules, however it might be potentiated by salicylic acid. The genes discussed are characteristic markers of the EBR, and could be used in later studies concerning this mechanism. It also remains a task for the future to find out if these marker genes have primary roles in the development of EBR and what their functions are. Real-time PCR assessment of the expression of selected marker genes was an effective and rapid method. Thus we were able to select those treatments, that are worth further studying. In the near future we are planning to perform a high-scale gene-identification and expression-analysis with the help of DNA chip technology, in order to gain as much insight as possible into the basal resistance of plants.
Abbreviations
- HR:
-
hypersensitive reaction
- BR:
-
basal resistance
- EBR:
-
early basal resistance
- C4H:
-
cinnamic acid-4-hydroxylase
- OMT:
-
orthomethyl transferase
- GST:
-
glutathione-S-transferase
- EST:
-
expressed sequence tag
- hpi:
-
hours post-inoculation
- SA:
-
salicylic acid
- MeJa:
-
methyl jasmonate
- ACC:
-
aminocyclopropane-carboxylic acid
- SPM:
-
spermine
- C:
-
Pseudomonas syringae pv. syringae 61 hrcC mutant
- PAMP:
-
pathogen-associated molecular pattern
References
Abramovitch RB, Martin GB (2004) Strategies used by bacterial pathogens to suppress plant defenses. Curr Opin Plant Biol 7:356–364
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Anterola AM, Jeon JH, Davin LB, Lewis NG (2002) Transcriptional control of monolignol biosynthesis in Pinus taeda: factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J Biol Chem 277:18272–18280
Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977–983
Barna B, Fodor J, Pogany M, Kiraly Z (2003) Role of reactive oxygen species and antioxidants in plant disease resistance. Pest Manag Sci 59:459–464
Batard Y, Schalk M, Pierrel MA, Zimmerlin A, Durst F, Werck-Reichhart D (1997) Regulation of the cinnamate 4-hydroxylase (cyp73a1) in jerusalem artichoke tubers in response to wounding and chemical treatments. Plant Physiol 113:951–959
Bent AF, Innes RW, Ecker JR and Staskawicz BJ (1992) Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact 5:372–378
Bilang J, Sturm A (1995) Cloning and characterization of a glutathione S-transferase that can be photolabeled with 5-azido-indole-3-acetic acid. Plant Physiol 109:253–260
Bozsó Z, Ott PG, Kecskés ML, Klement Z (1999) Effect of heat and cycloheximide treatment of tobacco on the ability of Pseudomonas syringae pv. syringae 61 hrp/hrmA mutants to cause HR. PhysMol Plant Pathol 55:215–223
Brown I, Mansfield J, Bonas U (1995) Hrp genes in Xanthomonas campestris pv vesicatoria determine ability to suppress papilla deposition in pepper mesophyll cells. Mol Plant Microbe Interact 8:825–836
Brown IR, Trethowan J, Kerry M, Mansfield J, Bolwell GP (1998) Localization of components of the oxidative cross-linking ofglycoproteins and of callose in papillae formed during the interactionsbetween non-pathogenic strains of Xanthomonas campestris and French bean mesophyll cells. Plant J 15:333–343
Burgyán J, Klement Z (1979) Early induced selective inhibition of incompatible bacteria in tobacco plants. Phytopathol Mediterranea 18:153–161
Creelman RA and Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48:355–381
Cutt JR and Klessig DF (1992) Pathogenesis-related proteins. In: Boller T, Meins F (eds) Genes involved in plant defense. Springer-Verlag, Berlin, pp 209–243
DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA 101:9927–9932
Diaz J, ten Have A and van Kan JAL (2002) The role of ethylene and wounding signalling in resistance of tomato to Botrytis cinerea. Plant Physiol 129:1341–1351
Dixon RA (2001) Natural products and plant disease resistance. Nature 411:843–847
Dow M, Newman MA, von Roepenack E (2000) The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu Rev Phytopathol 38:241–261
Felix G and Boller T (2003) Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognised as an elicitor signal in tobacco. J Biol Chem 278:6201–6208
Fujita T, Kouchi H, Ichikawa T, Syono K (1994) Cloning of cDNAs for genes that are specifically or preferentially expressed during the development of tobacco genetic tumors. Plant J 5:645–654
Gerber IB, Zeidler D, Durner J, Dubery IA (2004) Early perception responses of Nicotiana tabacum cells in response to lipopolysaccharides from Burkholderia cepacia. Planta 218:647–657
Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Metraux JP, Katagiri F (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34:217–228
Gómez-Gómez L, Boller T (2002) Flagellin perception: a paradigm for innate immunity. Tends Plant Sci 7:251–256
Guo A, Durner J, Klessig DF (1998) Characterization of a tobacco epoxide hydrolase gene induced during the resistance response to TMV. Plant J 15:647–656
Hauck P, Thilmony R, He SY (2003) A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA 100:8577–8582
Hiraga S, Ito H, Yamakawa H, Ohtsubo N, Seo S, Mitsuhara I, Matsui H, Honma M, Ohashi Y (2000) An HR-induced tobacco peroxidase gene is responsive to spermine, but not to salicylate, methyl jasmonate and ethephon. Mol Plant Microbe Interact 13:210–216
Hirsinger C, Salvà I, Marbach J, Durr A, Fleck J, Jamet E (1999) The tobacco extensin gene Ext 1.4 is expressed in cells submitted to mechanical restraints and in cells proliferating under hormone control. J Exp Bot 50:343–355
Hoekema A, Hirsch PR, Hooykaa PJJ, Schilperoort RA (1983) A binary plant vector strategy based on separation of vir-region and T-region of the Agrobacterium-tumefaciens TI-plasmid. Nature 303:179–180
Huang HC, Schuurink R, Denny TP, Atkinson MM, Baker CJ, Yucel I, Hutcheson SW, Collmer A (1988) Molecular cloning of Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco. J Bacteriol 170:4748–4756
Hubner S, Hehmann M, Schreiner S, Martens S, Lukacin R, Matern U (2003) Functional expression of cinnamate 4-hydroxylase from Ammi majus L. Phytochemistry 64:445–452
Irzyk GP, Fuerst EP (1993) Purification and characterization of a glutathione S-transferase from benoxacor-treated maize (Zea mays). Plant Physiol 102:803–810
Jaeck E, Martz F, Stiefel V, Fritig B, Legrand M (1996) Expression of class I O-methyltransferase in healthy and TMV-infected tobacco. Mol Plant Microbe Interact 9:681–688
Jakobek JL, Lindgren PB (1993) Generalized induction of defense responses in bean is not correlated with the induction of the hypersensitive reaction. Plant Cell 5:49–56
Jakobek JL, Smith JA, Lindgren PB (1993) Suppression of bean defense responses by Pseudomonas syringae. Plant Cell 5:57–63
Keshavarzi M, Soylu S, Brown I, Bonas U, Nicole M, Rossiter J, Mansfield J (2004) Basal defenses induced in pepper by lipopolysaccharides are suppressed by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 17:805–815
Kevei Z, Vinardell JM, Kiss GB, Kondorosi A, Kondorosi E (2002) Glycine-rich proteins encoded by a nodule-specific gene family are implicated in different stages of symbiotic nodule development in Medicago spp. Mol Plant Microbe Interact 15:922–931
King EO, Ward MK, Raney DR (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:301–307
Klement Z (1990) Generally used pathophysiological methods. In: Klement Z, Rudolph K, Sands DC (eds) Methods in phytobacteriology. Akadémiai Kiadó, Budapest, Hungary. pp 96–121
Klement Z, Bozsó Z, Kecskés ML, Besenyei E, Czelleng A, Ott PG (2003) Local early induced resistance of plants as the first line of defence against bacteria. Pest Manag Sci 59:465–474
Klement Z, Bozsó Z, Ott PG, Kecskés ML, Rudolph K (1999) Symptomless resistant response instead of the hypersensitive reaction in tobacco leaves after infiltration of heterologous pathovars of Pseudomonas Syringae. J Phytopathol 147:467–475
Lindgren PB (1997) The role of hrp genes during plant-bacterial interactions. Annu Rev Phytopathol 35:129–152
Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R (2003) Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15:165–178
Machácková I, Saniewski M, Filek M (2004) Plant hormones in stress signaling. In: Filek M, Biesaga-Koscielniak J, Marcinska I (eds) Analytical methods in plant stress biology. Institute of Plant Physiology, Polish Academy of Sciences, Kraków, Poland
Mauch F, Dudler R (1993) Differential induction of distinct glutathione-S-transferases of wheat by xenobiotics and by pathogen attack. Plant Physiol 102:1193–1201
Maury S, Geoffroy P, Legrand M (1999) Tobacco O-methyltransferases involved in phenylpropanoid metabolism. The different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol 121:215–224
Mehdy MC (1994) Active oxygen species in plant defence against pathogens. Plant Physiol 105:467–472
Merkouropoulos G, Shirsat AH (2003) The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217(3):356–366
Meyer A, Puhler A, Niehaus K (2001) The lipopolysaccharides of the phytopathogen Xanthomonas campestris pv. campestris induce an oxidative burst reaction in cell cultures of Nicotiana tabacum. Planta 213(2):214–222
Murray GI, Paterson PJ, Weaver RJ, Ewen SW, Melvin WT, Burke MD (1993) The expression of cytochrome P-450, epoxide hydrolase, and glutathione S-transferase in hepatocellular carcinoma. Cancer 71:36–43
Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135:1113–1128
Nurnberger T, Brunner F (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr Opin Plant Biol 4:318–324
Nurnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198:249–266
Oesch F (1973) Mammalian epoxide hydrases: inducible enzymes catalysing the inactivation of carcinogenic and cytotoxic metabolites derived from aromatic and olefinic compounds. Xenobiotica 3:305–340
Ott PG, Szabó L, Balázs E, Klement Z (1997) Submicroscopic evidence of bacterially induced resistance in tobacco leaves. Acta Phytopathol Entomol Hung 32:265–280
Pakusch AE, Kneusel RE, Matern U (1989) S-adenosyl-L-methionine:trans-caffeoyl-coenzyme A 3-O-methyltransferase from elicitor-treated parsley cell suspension cultures. Arch Biochem Biophys 271:488–494
Parmentier Y, Durr A, Marbach J, Hirsinger C, Criqui MC, Fleck J, Jamet E (1995) A novel wound-inducible extensin gene is expressed early in newly isolated protoplasts of Nicotiana sylvestris. Plant Mol Biol 29:279–292
Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448
Pellegrini L, Rohfritsch O, Fritig B, Legrand M (1994) Phenylalanine ammonia-lyase in tobacco. Molecular cloning and gene expression during the hypersensitive reaction to tobacco mosaic virus and the response to a fungal elicitor. Plant Physiol 106:877–886
Pinot F, Bosch H, Alayrac C, Mioskowski C, Vendais A, Durst F, Salaun JP (1993) [omega]-hydroxylation of oleic acid in Vicia sativa microsomes (inhibition by substrate analogs and inactivation by terminal acetylenes). Plant Physiol 102:1313–1318
Ringli C, Keller B, Ryser U (2001) Glycine-rich proteins as structural components of plant cell walls. Review. Cell Mol Life Sci 58:1430–1441
Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T, Shimada H, Takamiya X, Ohta H, Tabata S (2001) Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: Self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res 8:153–161
Schmitt D, Pakusch AE, Matern U (1991) Molecular cloning, induction and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. J Biol Chem 266:17416–17423
Schowalter AM (1993) Structure and function of plant cell wall proteins. The Plant Cell 5:9–23
Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9:261–270
Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JDG (1995) Molecular genetics of plant disease resistance. Science 268:661–667
Takahashi Y, Nagata T (1992) ParB: an auxin-regulated gene encoding glutathione S-transferase. Proc Natl Acad Sci USA 89:56–59
Takemoto D, Doke N, Kawakita K (2001) Characterization of elicitor-inducible tobacco genes isolated by differential hybridization. J Gen Plant Pathol 67:89–96
Takemoto D, Yoshioka H, Doke N, Kawakita K (2003) Disease stress-inducible genes of tobacco: expression profile of elicitor-responsive genes isolated by subtractive hybridisation. Physiol Plant 118:545–553
Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. The Plant Cell 15:1–14
Werck-Reichhart, D, Feyereisen, R (2000) Cytochromes P450: a success story. Genome Biol 1(6):reviews3003.1–reviews3003.9
Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW (1989) Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucl Acids Res 17:3469–3478
Yamakawa H, Kamada H Satoh M, Ohashi Y (1998) Spermine is a salicylate- independent endogenous inducer for both tobacco acidic pathogenesis-related proteins and resistance against tobacco mosaic virus infection. Plant Physiol 118:1213–1222
Yasuda E, Ebinuma H, Wabiko H (1997) A novel glycine-rich/hydrophobic 16 kDa polypeptide gene from tobacco: similarity to proline-rich protein genes and its wound-inducible and developmentally regulated expression. Plant Mol Biol 33:667–678
Acknowledgements
We thank Alan Collmer (Cornell University, Ithaca), Steven W. Hutcheson (University of Maryland, College Park) and Eva Kondorosi (Gif-Sur-Yvette, France) for providing different strains of Pseudomonas syringae pv. syringae 61 and Sinorhizobium meliloti. We also thank Lóránt Király for his helpful comments on the manuscript. This research was supported by grants of Hungarian National Science Foundation, OTKA TS-040835 and F037700.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Dudits
Rights and permissions
About this article
Cite this article
Szatmari, A., Ott, P.G., Varga, G.J. et al. Characterisation of basal resistance (BR) by expression patterns of newly isolated representative genes in tobacco. Plant Cell Rep 25, 728–740 (2006). https://doi.org/10.1007/s00299-005-0110-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-005-0110-5