Abstract
We have developed a high-throughput Agrobacterium-mediated transformation model system using both nptII and the 5-enolpyruvylshikimate-3-phosphate synthase gene from Agrobacterium tumefaciens strain CP4 (cp4) based selections in MicroTom, a miniature rapid-cycling cherry tomato variety. With the NPTII selection system, transformation frequency calculated as independent transgenic events per inoculated explant ranged from 24 to 80% with an average of 56%, in industrial production scale transformation experiments. For CP4, with glyphosate selection, the average transformation frequency was 57%. Stable transformation frequency was positively correlated with transient expression (R=0.85), and variable with the genes of interest. DNA integration and germline transformation were confirmed by biological assay, Southern Blot analysis, and R1 phenotype segregation. Transgene expression was observed in leaf, root, stem, flower, and fruit tissues of the transgenic plants. Ninety-five percent of transgenic events coexpressed two introduced genes based on β-glucuronidase (GUS) and neonmycin phosphotransferase II (NPTII) expression. Seventy-five percent of transgenic events contained one to two copies of the introduced uidA (GUS) gene based on Southern analysis. Transgenic plants from the cotyledon explants to the transgenic plants transferred to soil were produced within about 2–3 months depending on the genes of interest. The utility of this MicroTom model transformation system for functional genomic studies, such as identification of genes related to important agricultural traits and gene function, is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although the global market value of transgenic crops exceeded $4.7 billion in 2004, and the demand for efficient plant transformation methods for functional genomics increases as genome sequences for plant species become rapidly available, the technology of plant transformation is only moderately or marginally successful in many important cultivars of crops, which can be a major limiting factor for the biotechnological exploitation of economically important plant species and it can also impair the wider application of genomic science. Functional genomics in plants involves the use of high-throughput methods for functional analysis of many genes (Jeon et al. 2000; Pereira 2000; Lagudah et al. 2001; Lee et al. 2004; Ostergaard and Yanofsky 2004). Therefore, efficient plant transformation systems are essential for the application of genomic sciences to understand physiological, biochemical, and molecular mechanisms of metabolic pathways (Tyagi and Mohanty 2000; Olhoft et al. 2004; Yang et al. 2004) and agricultural biotechnology.
Tomato (Lycopersicon esculentum) is a major vegetable crop throughout the world, with an annual production of approximately 108 million metric tons in 2002 (Atherton and Rudich 1986; FAS/USDA, 2003). Due to its importance as a human food source and its biological features like relatively small genome and transformability, tomato has long served as a model system for plant genetics, development, pathology, and physiology, resulting in the accumulation of substantial information regarding its biology. Tomato has been used as a good system to isolate mutants using chemical or physical agents and isolate genes by map-based cloning procedures from YAC/BAC libraries (Pereira 2000). Currently a total of 182,765 high-quality BAC end sequences of tomato are available from the International Tomato Sequencing Project (http://www.sgn.cornell.edu/help/about/tomato_sequencing.html, Shibata 2005). In addition, there are other sources of tomato sequence information available in public databases such as approximately 150,000 tomato expressed sequence tag (ESTs) collections (Sol Genomic Network: http://sgn.cornell.edu/); and Japanese tomato EST and full-length cDNA sequences (http://www.jspcmb.jp/pbcontents/pb22-2.html, Takahashi et al. 2005; Tsugane et al. 2005; Yamamoto et al. 2005). High quality tomato microarrays are now commercially available (http://www.affymetrix.com/community/research/consortia.affx). Agrobacterium-mediated transformation has been developed (Koornneef et al. 1986; McCormick et al. 1986) and improved in tomato (Fillatti et al. 1987; Hinchee et al. 1994; Krasnyanski et al. 2001). Agrobacterium-mediated T-DNA insertional mutagenesis for gene identification has been effectively demonstrated in tomato, with targeted tagging yielding mutants at the high rate of about 1/1000 (Jones et al. 1994). However, high-throughput transformation technology, which possesses the five criteria of (1) large scale production capability (high transformation frequency), (2) rapid production cycle, (3) minimal production complexity, (4) inexpensive DNA delivery-method, and (5) high quality (high co-expression and low copy number), in an appropriate genotype of tomato has been lacking for functional genomic applications.
Plant model systems have played an important role in understanding plant biology. The most highly developed plant model system is Arabidopsis because of its small size, rapid life cycle, small genome, and transformability (Meyerowitz and Sommerville 1994; Meinke et al. 1998). MicroTom is a miniature dwarf determinate tomato cultivar, originally bred for home gardening purposes (Scott and Harbaugh 1989). It differs from standard tomato cultivars primarily by two recessive genes, which confer the dwarf phenotype (Meissner et al. 1997). MicroTom shares some features with Arabidopsis that make it successful as model system, such as small size (up to 1,357 plants/m2), short life cycle (70–90 days from sowing to fruit-ripening), and small genome (950 Mb). In addition, MicroTom possesses some unique and useful features. Tomato is susceptible to a wide range of pathogens (Webb et al. 1973). For example, MicroTom is susceptible to Phytophthora infestans, but Arabidopsis is not (Rommens 2000; Dan et al. 2001). Therefore, MicroTom has unique potential as a susceptible model system to study plant disease resistance. MicroTom responds to glyphosate in a similar manner as cotton, particularly with respect to reproductive tolerance, and therefore has been used as a model plant to study glyphosate tolerance (Sidorova et al. 2001). Because of reasonable size of MicroTom's fruits compared with the very tiny size of Arabidopsis seeds, MicroTom has an advantage to be a model system to study yield trait. MicroTom has been used as a model system to dissect tomato molecular genetics through techniques such as mutagenesis, and gene tagging, trapping, and knockouts (Meissner et al. 1997; Mathews et al. 2003). Agrobacterium-mediated transformation methods for MicroTom were described in the same papers, but no details on the transformation experiments were reported by Meissner et al. (1997). Mathews et al. (2003) reported an Agrobacterium-mediated MicroTom transformation method with transformation rates of 40–60% using leaf, stem, hypocotyl, and shoot tip as starting explants and kanamycin selection. However, there were no details on the transformation protocol development, which are important information for advancing transformation technology in Microtom, and no information on the three of the five high-throughput transformation criteria such as (1) production cycle, (2) minimal production complexity, (3) quality of transgenic plants (high co-expression and low copy number), which are critical for functional genomic applications. In addition, there was no determination of the transformation frequency whether it was counted based on the number of independent transgenic events or the number of transgenic plants in both publications. Our transformation frequency was determined as independent transgenic plant events divided by total cotyledons inoculated, then multiplied by 100%. The transformation frequency based on the number of the transgenic plants was approximately two times higher than that based on the number of the independent transgenic plant events in our study.
In this paper, we report systematically a high throughput transformation system development in MicroTom using cotyledon as starting explant and using both kanamycin and glyphosate selection systems. Transformation system using glyphosate selection has not been reported in MicroTom to date, which is essential for studying herbicide tolerance in crops because MicroTom responds similarly to glyphosate as cotton, especially with respect to reproductive tolerance, and therefore has been used as a model plant to study glyphosate tolerance (Sidorova et al. 2001). In addition, we provide the details of experimental data on the cell biology of MicroTom transformation, which has not been reported before. The application of the high throughput transformation system for functional genomic studies is also discussed in this paper.
Materials and methods
Plant materials and transformation vectors
MicroTom seeds (Ball Seed Co., Chicago, Illinois, USA) were surface-sterilized with a two-step process: (1) soaking for 10 min in sterile water, followed by 1 min in 70% alcohol, and then two rinses with sterile water; and (2) soaking in 25% Clorox® with Tween 20 for 10 min, and then rinsing four times with sterile water. The seeds were germinated in Phytatrays® (Sigma Chemical Co., St. Louis, MO, USA) containing a germination medium (Fillatti et al. 1987). Approximately 80 seeds were plated per container. The cultures were placed at 27±1°C in the dark for 5 days, and then grown at 23±1°C under 16 h photoperiod (cool white fluorescent lamps) for 2 days to obtain more uniform germination.
Plasmid pMON15715 was used to develop the transformation system with kanamycin selection. It contains a β-glucuronidase (uidA) gene driven by the FMV promoter (Rogers 1990) and a neomycin phosphotransferase II (nptII) gene, which confers resistance to kanamycin, driven by the nos promoter. Plasmid pMON10125 was used to develop a transformation system with glyphosate selection. Plasmid pMON10125 contains a cp4 gene, the 5-enolpyruvylshikimate-3-phosphate synthase gene (EPSPS) from Agrobacterium strain CP4 (Barry et al. 1992), which confers resistance to glyphosate, driven by the FMV promoter and a uidA gene driven by the cauliflower mosaic virus-enhanced 35S promoter (Kay et al. 1987). Plasmid pMON31892 was used to determine optical density of Agrobacterium for the transformation. It contains a jellyfish gene encoding the green fluorescent protein (GFP) driven by the cauliflower mosaic virus-enhanced 35S promoter and a nptII gene driven by the nos promoter. Plasmid pMON30656 was used to construct all disease resistance gene vectors and as the control vector for transformation experiments. Plasmid pMON30656 contains the nptII gene driven by the nos promoter. All disease resistant gene vectors had the same genetic elements as pMON30656 with the addition of the genes of interest.
Agrobacterium preparation
A. tumefaciens strain ABI was grown on Luria-Bertani (LB) medium (Sambrook et al. 1989) supplemented with 50 mg/l each of spectinomycin, streptomycin, and kanamycin, and 25 mg/l chloramphenicol at 28°C for approximately 3 days in the dark. One day before inoculation, a loopful of the bacterial cells from the plate was inoculated into 2 ml of the same LB liquid medium in 17×100 Falcon tube, and the tube was shaken at the speed of approximately 230 rpm and at 28±2°C overnight. The next morning, 0.2 ml of the overnight culture was inoculated into 2 ml of LB plus 0.2 mM acetosyringone in 17×100 mm Falcon tube, and the tube was shaken at the speed of approximately 230 rpm and at 28±2°C for about 4 h. The bacterial culture was diluted to the appropriate concentration using the optical density at 660 nm to determine concentration.
Explanting and Agrobacterium inoculation
Cotyledon explants were isolated from seedlings 7 days after sowing seeds on the germination medium. Cotyledons were harvested into MS liquid medium containing 4 mg/l para-chloro phenoxy acetic acid and 0.5 mg/l kinetin to prevent desiccation and then both ends were trimmed using a scalpel blade. Cotyledons growing in a fairly tight “V” configuration were chosen for use due to higher frequency of transformation based on the frequency of standard tomato transformation (personal communication). The cotyledons were co-cultivated with A. tumefaciens using a modified tobacco feeder layer method (Horsch et al. 1985; Fillatti et al. 1987). The cotyledons were first placed adaxial side down on 100×15 mm “feeder plates” (Fillatti et al. 1987) immediately after isolation and then gently poked using a sharp forceps. Usually six to 10 pokes were made depending on the size of the cotyledons. Three ml of Agrobacterium solution adjusted to the appropriate concentration was then added on top of the cotyledons. The plates were incubated for 10 min at room temperature and the Agrobacterium solution was removed with a sterile pipette. The cotyledons were then co-cultured for 2 days at 23±1°C under 16 h photoperiod (cool white fluorescent lamps). The culture plates were placed in clear plastic boxes and covered by clear plastic bags to maintain humidity, which was preferable for tomato growth.
Transformation and regeneration of transgenic plants
Two days after co-cultivation cotyledons were transferred to various shoot induction media to determine the best medium for shoot organogenesis under different selective pressures. Forty-eight different shoot induction media were screened. These shoot induction media were formulated based on a modified MS medium (Murashige and Skoog 1962), and contained 4.3 g/l MS salts (Sigma Chemical Co., St. Louis, MO, USA), 30 g/l sucrose, B5 vitamins (Gamborg et al. 1968), 10 g/l TC agar (Sigma Chemical Co., St. Louis, MO, USA), pH 5.7; supplemented with various combinations and concentrations of zeatin riboside, IAA, and kanamycin. The standard tomato medium (SI-1) for shoot induction was used as a control, which contains the MS basal medium with 2 mg/l zeatin riboside (Fillatti et al. 1987). After 48 media were examined, the best medium, SI-2, was selected based on the shoot organogenesis response. SI-2 medium contained the MS basal medium supplemented with 2 mg/l zeatin riboside, 0.1 mg/l indole-3-acetic acid (IAA), and 100 mg/l kanamycin. The SI-2 medium was further compared with the control medium SI-1 under 100 mg/l kanamycin selective pressure.
Two to 4 weeks after shoot induction, cotyledons formed buds or shoots, which were separated as putative individual events and transferred to shoot elongation medium. Three shoot elongation media were preliminarily evaluated, and they contained the MS basal medium supplemented with 2, 1, and 0.5 mg/l zeatin riboside, and 100 mg/l kanamycin, respectively.
Three rooting media were first tested using wild-type MicroTom. Three rooting media MR1, MR2, and MR3 were formulated based on MS basal medium supplemented with 0, 1, and 2 mg/l indole-3-butyric acid (IBA), respectively. MR2 medium was selected due to good response to rooting. After shoots elongated to about 2–3 cm in length, they were excised from cotyledons, and transferred to MR2 medium supplemented with 40 mg/l kanamycin.
All plates were placed in clear plastic boxes and covered by clear plastic bags. The cultures were maintained at 23±1°C under a 16 h photoperiod for 3–4, 2–3, and 3–4 weeks for the shoot induction, elongation, and rooting steps, respectively. All the experiments were conducted at least three times.
Plant transformation terminology
All plants regenerated from a single poked area of a cotyledon were considered to be from a single independent transformation event. Only one regenerated plant from a single poke was counted as an independent transgenic event to ensure that each regenerate represented an independent transgenic event. Transformation frequency was measured as independent transgenic plant events divided by total cotyledons inoculated, then multiplied by 100%. The transgenic plants were allowed to self-pollinate by open pollination.
Histochemical β-glucuronidase (GUS) X-gluc assay
Transgenic tissue materials were assayed for histochemical GUS expression using a modified method (half strength) according to Jefferson (1987). The tissues were incubated at 37°C overnight. A cotyledon was considered to have high level of transient expression if more than 30% of the pokes exhibited GUS expression.
PCR and ELISA analysis of transgenic plants
Leaf materials from greenhouse grown MicroTom plants were sampled directly into 96-well plates using leaf punches. The samples were mechanically disrupted and then used for DNA isolation or qualitative ELISA. DNA was isolated using the Qiagen DNeasy 96 Plant Kit (Qiagen, Valencia, CA, USA) following the manufacturer's recommendations. Two μl of DNA was used in 50 μl PCR reactions containing 0.25 mM dNTPs, 2 mM MgCl2, 1 U Taq DNA polymerase, and 0.5 μM final concentration of each of the primers NPTIIF (5′TGT GCT CGA CGT TGT CAC TGA A 3′) and NPTIIR (5′CAC CAT GAT ATT CGG CAA GCA G 3′) made against the neomycin phosphotranferase II gene.
ELISA analysis was carried out using the Agdia NPTII ELISA protocol (Agdia, Elkhart, Indiana USA) following the manufacturer's recommendations. Leaf samples were ground in a 1:5 tissue weight:buffer volume and subsequently 100 μl of diluted sample was used per test well. The samples were measured in triplicate using a microplate reader (Biorad Hercules, CA, USA) at 450 nm.
Southern Blot
Genomic DNA from leaves of transgenic MicroTom plants was isolated with a SDS extraction buffer containing 0.1 M Tris, pH 8.0, 0.05 M EDTA, 0.5 M NaCl, and 1% SDS. The DNA was digested with BamHI and transferred from 0.8% agarose gel to blotting membrane (Zeta-Probe GT Genomic Tested Blotting Membranes, BioRad, Catalog 162-0197). The membrane was prehybridized in 15 ml of hybridization buffer, which contained 0.5 M Na phosphate pH 7.2, 7% SDS, tRNA 100 μg/ml, for 1 h at 65°C, and hybridized with P32 labeled GUS probe, which was labeled using Prime It RMT Random Primer Labeling Kit (Stratagene Cat# 300392). The membrane was washed twice at 65° with the solution containing 40 mM Na phosphate pH 7.2, 1% SDS, 0.5×SSC, and exposed with BioMax film (Kodak, catalog 829 4985).
Results and discussion
Effect of Agrobacterium titer on transient GUS expression and stable transformation
Among the optical densities (O.D.) of Agrobacterium from 0.01 to 1.00 tested using the vector of pMON31892, higher Agrobacterium concentrations produced higher transient expression (Table 1). However, the percentage of explants with stable GFP positive shoots was highest at the intermediate O.D. readings of 0.1 and 0.5 (Table 1). When using the O.D. of 1.0, overgrowth of Agrobacterium occurred, which caused the death of explants. When using an O.D. of 0.01, both transient expression and stable transformation were low (Table 1). Since there was no difference between the percentages of explants having stable GFP positive shoots at O.D. readings of 0.1 and 0.5, the O.D. reading of 0.1 was selected for use in subsequent experiments. Others have reported an effect of Agrobacterium titer on the survival and vigor of explants, and overall transformation frequency in L. esculetum (McCormick et al. 1986), while Agrobacterium titer made little or no difference in tobacco and petunia (Horsch et al. 1987).
Effect of explant status on transient expression
Based on the protocol for standard tomato cultivar (Fillatti et al. 1987), cotyledon explants were pre-cultured on “feeder plates” containing a modified MS medium supplemented with 0.1 mg/l kinetin, 0.2 mg/l 2,4-D, 200 μM acetosyringone and 1 mM galacturonic acid for 1 day before inoculation. When MicroTom cotyledons were cultured on the medium for 1 day and then inoculated with Agrobacterium, transient expression of gus gene was poor and some tissue browning occurred around the poking area. Therefore, freshly isolated cotyledons were tested in an attempt to improve Agrobacterium infection. 91.7% of freshly isolated cotyledon explants produced a high level of transient expression compared to only 24.4% for the pre-cultured cotyledons (Fig. 1a; statistically significant difference, p<0.05).
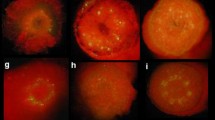
MicroTom transformation. a Effect of freshly cut (FC) and pre-cultured (PC) explants on transient expression 2 days after inoculation, b GUS expression of a transgenic MiroTom plant, c GUS expression of a flower of a transgenic MiroTom plant, d GUS expression of a fruit of a transgenic MiroTom plant, e R1 gus gene segregation of a MicroTom transgenic plant, f Noninoculated cotyledons cultured on SI-2 medium with 100 mg/l kanamycin for 4 weeks, and g Shoot regeneration 4 weeks after cotyledons cultured on SI-2 medium with 100 mg/l kanamycin from transformed cotyledons
Correlation of transient expression and stable transformation frequency
By using freshly isolated cotyledon explants, transient expression was significantly increased. To understand if increasing transient expression can also increase stable transformation frequency, a correlation study between transient expression and stable transformation was conducted. Three random samples of 30–45 cotyledons were examined for GUS transient expression after 1 day of co-culture each from three independent transformation experiments using the vector of pMON15715. Stable transformation frequency was assessed from the same three experiments after a rooting screen for transgenic events. There was a strong positive correlation between transient expression and stable transformation frequency (R=0.85, Fig. 2). The results indicated that optimal initial Agrobacterium infection is critical for high frequency stable transformation with MicroTom when a uidA gene is driven by the FMV promoter and nptII gene is driven by the nos promoter in the cassette of pMON15715. In the contrast with the transient expression using the vector of pMON31892, there was no positive correlation between transient expression and stable transformation frequency (Table 1). This might be due to the effect of the different promoters used for gus and gfp genes in pMON15715 and pMON31892 because both the vectors had the same nos promoter, which drove the nptII gene.
Effect of cytokinin, auxin, and vector on stable transformation frequency
The medium SI-2, which contained 2 mg/l of zeatin riboside, 0.1 mg/l IAA, and 100 mg/l kanamycin, produced the highest percentage of explants responding to bud and/or shoot formation among 48 media tested (data not shown). The medium SI-2 along with the control SI-1 medium was further tested using freshly isolated and pre-cultured cotyledon explants. Freshly isolated explants produced significantly higher transformation frequency (T.F.) of 87.7 and 55.2% than pre-cultured explants for both media tested (19.1 and 11.1%) (Table 2). Transformation frequency from the medium SI-2 was 1.6 and 1.7-fold higher than that from SI-1 for freshly isolated and pre-cultured explants, respectively (Table 2). The best treatment, freshly isolated explants cultured on SI-2 medium, generated a T.F. of 87.7% (Table 2).
The ultimate objective of this study was the development of a high-throughput transformation system to screen a large number of disease resistance genes. In preliminary experiments, we noted a possible interaction between constructs and shoot induction media on T.F. (Fig. 3). Media SI-1 and SI-2 against 33 disease resistance constructs using freshly isolated explants were further screened, and the T.F. of each medium was measured. Among the 33 constructs, 23 produced a higher T.F. on SI-2 medium than on SI-1 medium, 8 generated higher T.F. on SI-1 medium than on SI-2 medium, and 2 had the same T.F. in both SI-2 and SI-1 media (Fig. 4). This indicated that T.F. varied with the genes of interest and a large number of constructs produced higher T.F. when using SI-2 medium. SI-2 medium, which produced higher T.F. than SI-1 medium for the most constructs, contained 2 mg/l zeatin riboside and 0.1 mg/l IAA and SI-1 medium only had 2 mg/l zeatin riboside. Zeatin combined with IAA has also been shown to promote shoot formation in standard tomato regeneration and transformation (Chen and Adachi 1994; Shen et al. 1998).
By testing the standard tomato protocol for MicroTom, shoots elongated first on MS basal medium containing 2 mg/l, then 1 mg/l, and finally 0.5 mg/l zeatin riboside for 1, 2, and 3 weeks, respectively. Shoot elongation took a total of 6 weeks. To reduce shoot elongation time, shoots were directly transferred to MS basal medium supplemented with 2, 1, and 0.5 mg/l zeatin riboside, respectively, instead of using the stepwise approach. Shoot elongation appeared several days faster on MS medium with 0.5 mg/l zeatin riboside than with 2 and 1 mg/l zeatin riboside (data not shown), and therefore 0.5 mg/l zeatin riboside was used for further experiments. Shoots elongated 2–3 weeks faster using this direct 0.5 mg/l zeatin riboside approach than the stepwise approach in the standard tomato protocol.
Of the three rooting media (MR1, MR2, and MR3) tested, rooting media MR2 and MR3 generated higher frequency of shoots producing roots than MR1 medium (Table 3). Roots also formed more rapidly on MR2 and MR3 media than on MR1 medium (Table 3). MR2 medium was selected for all further experiments since there was no difference in the frequency or time of rooting between MR2 and MR3 media.
Selective agents and improvement of transformation system
Transformation systems were developed using both the nptII (kanamycin) and cp4 (glyphosate) selectable markers. With kanamycin selection using the plasmid pMON15715, T.F. ranged from 24 to 80% with an average of 56%, in large production-scale transformation experiments, which generated approximately above 1000 transgenic plants. With glyphosate selection using the plasmid pMON10125, T.F. ranged from 54.5 to 60.8%, with an average of 57%. For the glyphosate selection system, the same media were used at the shoot induction and elongation stages as for the kanamycin selection system, but 0.03 mM glyphosate was included. After a rooting screen with kanamycin selection, 95.6% of plants generated were transgenic based on PCR, ELISA, and GUS expression tests, while 4.4% were either low expressors or escapes. With glyphosate selection, 97.4% of plants generated were transgenic, and 2.6% were either low expressors or escapes. Cotyledons, which were not inoculated by Agrobacterium, did not form any buds under 100 mg/l kanamycin (Fig. 1f) or 0.03 mM glyphosate. Typically shoot regeneration occurred directly from the poked and trimmed area of cotyledons without an obvious callus phase (Fig. 1g). Ninety-five percent of R0 plants generated after the rooting screen were confirmed to be transgenic by GUS expression (Fig. 1b), and 90% of them were transgenic confirmed by PCR and Southern analysis (Fig. 5). This inconsistency of the percentages (95% vs. 90%) from GUS assay, and PCR and Southern analysis (Fig. 6) might be due to experimental deviation because approximately 300 samples were tested for GUS assay, and approximately 50 and 20 samples were examined for PCR and Southern analysis, respectively. Germline transformation was confirmed by R1 GUS phenotype segregation (Fig. 1e).
To date, the majority of tomato transformation frequencies ranging from 0.72 to 25% have been reported (Jiang et al. 1999; Wei and Phillips 2001) although up to 80% of transformation frequencies were reported without the details on the transformation experiments (Meissner et al. 1997). Also, a transformation rate of 40–60% for MicroTom was published without the details on the transformation protocol development (Mathews et al. 2003). In addition there was no determination of the transformation frequency whether it was counted based on the number of independent transgenic events or the number of transgenic plants in both the publications. Our transformation frequency was determined as independent transgenic plant events divided by total cotyledons inoculated, then multiplied by 100%. All plants regenerated from a single poked area of a cotyledon were considered to be from a single independent transformation event. Only one regenerated plant from a single poke was counted as an independent transgenic event to ensure that each regenerate represented an independent transgenic event although there were frequently more than one regenerated plant from a single poke. The transformation frequency based on the number of the transgenic plants obtained was approximately two times higher than that based on the number of the independent transgenic plant events in our study, indicating that our transformation frequency ranged from approximately 48 to 160% if it was counted based on the number of the transgenic plants. Our transformation system had significant improvements over previously published tomato transformation methods in terms of transformation frequency and time frame of transgenic plant production. For example, we compared our new MicroTom transformation protocol with standard tomato transformation protocol in terms of frequency at each stage of the protocol and time frame of the entire transformation cycle. By using freshly isolated cotyledon explants instead of pre-cultured cotyledon explants, the average of percentage of explants with high level of GUS transient expression (Agrobacterium infection) was increased from 24.4 to 88.0% (Table 4). By increasing GUS transient expression (Agrobacterium infection), and determining an optimized shoot induction medium SI-2, the average of frequency of explants forming buds and/or shoots was increased from 44.6 to 89.3% (Table 4), and the time frame for shoot induction was reduced from 4–5 weeks to 3–4 weeks (Table 5). By eliminating the stepwise reduction of zeatin riboside approach in the standard tomato protocol, the time frame for shoot elongation was reduced from 4–6 weeks to 2–3 weeks (Table 5). Therefore, the shoot selection, including both shoot induction and elongation selection steps before rooting, took 5–7 weeks depending on the constructs when using our cotyledon protocol. In addition, there was no sub-culture transfer to fresh medium in each step of our transformation protocol. In contrast with the hypocotyl protocol (Mathews et al. 2003), MicroTom shoot selection took over 8 weeks, and explants were required to be sub-transferred to fresh medium every 2 weeks and the selection level was increased gradually over an 8-week period to 90 mg/l kanamycin. This suggests that our cotyledon protocol is faster, simpler, and more efficient in terms of saving labor and material, and productivity of transgenic plant production than the hypocotyl protocol. Through improvements in shoot induction and rooting, the average of percentage of shoots producing roots was increased from 13.2 to 29.3% (Table 4). This enabled us to achieve an overall increase in T.F. of independent transgenic events from 11 to 57% in MicroTom, and reduce the transformation time frame from approximately 3–4 months to 2–3 months. Subsequently this led us to accomplish an increase in our productivity from 600–700 independent transgenic events to 1500–2000 independent transgenic events per person per year (Table 4).
Southern analysis of the transgenic MicroTom plants from pMON15715. The gus gene was used as a probe. Expected size of the transgene was approximately 3,045 bp (the right border sequences + the promoter + gus gene + the sequences to the first BamHI site in plant). The smaller bands than 3,045 bp might represent truncated gus gene. CK: non-transgenic plant and T1 to T20: transgenic plants
Transgene expression, quality of transgenic plants, and transgene phenotype
Eighty-five percent of R0 transgenic plants expressed the gus gene in whole plants including leaf, stem, and root tissues (Fig. 1b). Eighty-four percent of R0 transgenic plants expressed the gus gene in flower tissue including petal, filament, anther, and ovary, and 91% of them expressed the gus gene in fruit tissue (Fig. 1c and d). Eighty-three percent of R1 seedlings expressed the gus gene (Fig. 1e). Therefore, MicroTom has a potential as a model plant to study gene function such as broader plant diseases including leaf, stem, root, and fruit diseases as well as other important agriculture traits like yield trait because of its ubiquitous GUS expression in MicroTom plant when a constitutive 35S promoter was used.
Based on gus and nptII gene expression of R0 transgenic events, 95% of transgenic events co-expressed both genes. Seventy-five percent of transgenic events had one to two copies of the introduced gus gene, as determined by Southern analysis. This indicated that high quality as measured of transgenic events was generated using this transformation system. All tested transgenic plants derived from the control vector appeared phenotypically normal and produced fruit and viable seeds. Some genes of interest changed plant morphology such as shape of the plant. Some altered fruit color and fruit shape.
In conclusion, the MicroTom transformation system described in this study possesses five elements of a high throughput transformation system: (1) large-scale production capability with an average transformation frequency of 56% for both kanamycin and glyphosate selections, (2) rapid production cycle, which took 2–3 months from cotyledon explants to transgenic plants which were ready to soil depending on the gene constructs, (3) minimal production complexity, which had a simple protocol with three single steps (shoot induction, shoot elongation, and rooting) and no sub-culture transfer to fresh medium in each step, (4) inexpensive DNA delivery-method, which was Agrobacterium-mediated transformation, and (5) high quality of transgenic plants with high co-expression and low copy number. This high-throughput transformation system has enabled us to use MicroTom in a functional genomics platform to accelerate gene discovery for disease control such as late blight in potato (Dan et al. 2001) and for specific traits such as genes conferring glyphosate tolerance (Sidorova et al. 2001). The same attribute of this system should make it useful for studying many other traits of economic or academic interest as well.
References
Atherton JG, Rudich J (1986) The tomato Crop. Chapman and Hall, London, New York, 661 pp
Barry G, Kishore G, Padgette S, Taylor M, Kolacz K, Weldon M, Re D, Eichholtz D, Fincher D, Hallas L (1992) Inhibitors of amino acid biosynthesis: strategies for imparting glyphosate tolerance to crop plants. In: Sinch BK, Flores HE, Shannon JC (eds) Biosynthesis and molecular regulation of amino acids in plants. American Society of Plant Physiologists, Rochville, MD, pp 139–145
Chen LZ, Adachi T (1994) Plant regeneration via somatic embryogenesis from cotyledon protoplasts of tomato (Lycopersicon esculentum). Breed Sci 44:257–262
Dan YH, Yan H, Munyikwa T, Dong J, Zhang B, Lahman L, Rommens C (2001) MicroTom—a model functional genomics assay. In Vitro Cell Dev Biol Plant 37:22-A
FAS/USDA (2003) Processed tomato products outlook and situation in selected counties. Horticutural and Tropical Products Division
Fillatti J, Kiser J, Rose R, Comai L (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Biotechnology 5:726–730
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirement of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Hinchee MAW, Corbin DR, Armstrong CL, Fry JE, Sato SS, DeBoer DL, Petersen WL, Armstrong TA, Connor-Ward DV, Layton JG, Horsch RB (1994) Plant transformation. In: Vasil IK, Thorpe TA (eds) Plant cell and tissue culture. Academic, Dordrecht, The Netherlands, pp 231–270
Horsch RB, Fry J, Hoffmann N, Wallroth M, Eichholtz D, Rogers S, Fraley R (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Horsch R, Fraley R, Rogers S, Fry J, Klee H, Shah D, McCormick S, Niedermeyer J, Hoffmann N (1987) Agrobacterium-mediated transformation of plants. In: Green CE, Somers DA, Hackett WP, Biesboer DD (eds) Plant tissue and cell culture. Geo J Ball, West Chicago, pp 317–329
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Lee S, Yang K, Nam J, An K, Han MJ, Sung RJ, Choi HS, Yu JH, Choi JH, Cho SY, Cha SS, Kim SI, An G (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22:561–570
Jiang GY, Jin DM, Weng ML, Guo BT, Wang B (1999) Transformation and expression of trichosanthin gene in tomato. Acta Bot Sin 41:334–336
Jones DA, Thomas CM, Hammond-Kossack KE, Balint-Kurti PJ, Jones JDG (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266:789–793
Kay R, Chan A, Daly M, McPherson J (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 230:1299–1302
Koornneef M, Hanhart C, Jongsma M, Toma I, Weide R, Zabel P, Hille J (1986) Breeding of a tomato genotype readily accessible to genetic manipulation. Plant Sci 45:201–208
Krasnyanski S, Sandhu J, Domier LL, Buetow DE, Korban SS (2001) Effect of enhanced CAMV 35S promoter and a fruit-specific promoter on uida gene expression in transgenic tomato plants. In Vitro Cell Dev Biol Plant 37:427–433
Lagudah ES, Dubcovsky J, Powell W (2001) Wheat genomics. Plant Physiol Biochem 39:335–344
Lee MK, Kim HS, Kim JS, Kim SH, Park YD (2004) Agrobacterium-mediated transformation system for large-scale production of transgenic Chinese cabbage (Brassica rapa L. ssp pekinensis) plants for insertional mutagenesis. J Plant Biol 47:300–306
Mathews H, Clendennen KS, Caldwell GC, Liu XL, Connors K, Matheis N, Schuster KD, Menasco DJ, Wagoner W, Lightner J, Wagner DR (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15:1689–1703
McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R (1986) Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep 5:81–84
Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M (1998) Arabidopsis thaliana: a model plant for genome analysis. Science 282:679–682
Meissner R, Jacobson Y, Melame S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A (1997) A new model system for tomato genetics. Plant J 12:1465–1472
Meyerowitz E, Sommerville CR (1994) Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Olhoft PM, Flagel LE, Somers DA (2004) T-DNA locus structure in a large population of soybean plants transformed using the Agrobacterium-mediated cotyledonary-node method. Plant Biotechnol J 2:289–300
Ostergaard L, Yanofsky MF (2004) Establishing gene function by mutagenesis in Arabidopsis thaliana. Plant J 39:682–696
Pereira A (2000) A transgenic perspective on plant functional genomics. Transgenic Res 9:245–260
Rogers SG (1990) Promoter for transgenic plants. US Patent No. 05378619
Rommens C (2000) Discovery of new signaling genes associated with late blight resistance in patato. Cambridge Healthtech Institute's Third Annual Impact of Molecular Biology on Crop Production and Crop Protection (abstract)
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Scott JW, Harbaugh BK (1989) MicroTom—a miniature dwarf tomato. Fla Agr Exp Sta Circ 370:1–6
Shen L, Sheng JP, Luo YB (1998) Tomato transgenic system with Ti plasmid of Agrobacterium tumefaciens. J China Agric Univ 3:101–110
Shibata D (2005) Genome sequencing and functional genomics approaches in tomato. J Gen Plant Pathol 71:1–7
Sidorova N, Layton JG, Armstrong CL (2001) Comparison of model transformation systems. In Vitro Cell Dev Biol Plant 37:20-A
Takahashi H, Shimizu A, Arie T, Rosmalawati S, Fukushima S, Kikuchi M, Hikichi Y, Kanda A, Takahashi A, Kiba A, Ohnishi K, Ichinose Y, Taguchi F, Yasuda C, Kodama M, Egusa M, Masuta C, Sawada H, Shibata D, Hori K, Watanabe Y (2005) Catalog of Micro-Tom tomato responses to common fungal, bacterial, and viral pathogens. J Gen Plant Pathol 71:8–22
Tsugane T, Watanabe M, Yano K, Sakurai N, Suzuki H, Shibata D (2005) Expressed sequence tags of full-length cDNA clones from the miniature tomato (Lycopersicon esculentum) cultivar Micro-Tom. Plant Biotechnol 22:161–165
Tyagi AK, Mohanty A (2000) Rice transformation for crop improvement and functional genomics. Plant Sci 158:1–18
Webb RE, Stoner AK, Gentile AG (1973) Tomatoes. In: Nelson RR (ed) Plants for disease resistance breeding. Pennsylvania State University Press, University Park, pp 344–360
Wei H, Phillips GC (2001) A combination of overgrowth-control antibiotics improves Agrobacterium tumefaciens-mediated transformation efficiency for cultivated tomato (L. esculentum). In Vitro Cell Dev Biol Plant 37:12–18
Yamamoto N, Tsugane T, Watanabe M, Yano K, Maeda F, Kuwata C, Torki M, Ban Y, Nishimura S, Shibata D (2005) Expressed sequence tags from the laboratory-grown miniature tomato (Lycopersicon esculentum) cultivar Micro-Tom and mining for single nucleotide polymorphisms and insertions/deletions in tomato cultivars. Gene 356:127–134
Yang YZ, Peng H, Huang HM, Wu JX, Ha SR, Huang DF, Lu TG (2004) Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci 167:281–288
Acknowledgements
The authors would like to thank David Duncan for critically reviewing the manuscript and for helpful suggestions. The authors are grateful to Jeanne Layton and Pamela Delaquil for providing us technical information on standard tomato transformation at Monsanto. The authors would like to thank Linda Lahman, Salim Hakimi, Glenda Debrecht, and Bei Zhang for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. C. Jordan
Rights and permissions
About this article
Cite this article
Dan, Y., Yan, H., Munyikwa, T. et al. MicroTom—a high-throughput model transformation system for functional genomics. Plant Cell Rep 25, 432–441 (2006). https://doi.org/10.1007/s00299-005-0084-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-005-0084-3