Abstract
Adult onset Still’s disease (ASD) is a systemic inflammatory disorder of unknown etiology. ASD is characterized by fever with unknown etiology, rash, arthritis, and involvement of several organ systems. FMF and TRAPS are two important autoinflammatory diseases which characterized with recurrent inflammatory attacks. We aimed in this study to investigate the MEFV gene and TNFRSF1A gene variations in ASD. Twenty consecutive Turkish ASD patients (14 female and 6 male; mean age 38.45 ± 14; mean disease duration 3.3 ± 2.3; mean age of the disease onset 35.1 ± 14.4) and 103 healthy controls of Turkish origin were analyzed. All ASD patients were genotyped for the 4 MEFV mutations (M694V, E148Q, V726A, M680I) and TNFRSF1A gene exon 2–3 and exon 4–5 by using sequence analysis. The healthy controls are genotyped using PCR–RFLP method for intron 4 variation. The results of MEFV gene mutations screening show an increase in the MEFV mutation rate in ASD group, but it was not significantly different (p = 0.442, OR 1.64, 95 % CI 0.409–6.589). T–C polymorphism (rs1800692) was the only variation in the intron 4 of TNFRSF1A gene that we observed at the ASD patients. The frequency of TT genotype was 15 %, TC: 45 %, and CC: 40 % in ASD patients and the frequencies were 22, 41, and 37 % in healthy controls, respectively. When we analyzed the allele difference between both groups, there was no difference (p = 0.54, OR 1.24, 0.619–2.496–2.654). The variations in MEFV may have role in ASD pathogenesis. Our findings suggest that there is no significant association between ASD and TNFRSF1A variations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hereditary periodic fever syndromes (HPFS) are a group of diseases including Familial Mediterranean fever (FMF), tumor necrosing factor receptor-associated periodic syndrome (TRAPS), cryopyrin-associated periodic syndromes (CAPSs), pyogenic arthritis, pyoderma gangrenosum and acne (PAPA) syndrome, pediatric granulomatous arthritis, hyperimmunoglobulinemia D with periodic fever syndrome (HIDS), chronic recurrent multifocal osteomyelitis (CRMO), deficiency of the IL-1 receptor antagonist (DIRA), and deficiency of the IL-10 receptor early-onset enterocolitis (IBD) [1]. These monogenic diseases of the innate immune system are characterized with self-limited inflammatory attacks, and they are called autoinflammatory syndromes [2]. Recent years, the definition of autoinflammatory syndromes enlarged and included complex genetic disorders such as ankylosing spondylitis (AS), gout, and diabetes mellitus. Moreover, the clinical spectrum of the classical HPFSs became wider after genomic sequencing of patients with an unnamed autoinflammatory syndrome. Several clinical findings of TRAPS were reported to be associated with different mutations in the TNFRSF1A gene.
FMF (OMIM #249100) is the most common HPF and is caused by the MEFV gene mutations [3]. It was shown that individuals with MEFV mutations are at increased risk for some inflammatory diseases such as AS [4, 5], rheumatoid arthritis (RA) [6, 7], ulcerative colitis [8, 9], juvenile idiopathic arthritis [10, 11], Crohn disease [12], Henoch Schönlein purpura [13, 14], and polyarteritis nodosa [15].
TRAPS (OMIM # 142680) is an autosomal dominantly inherited autoinflammatory syndrome. The mutations in exon 2–3 and 4–5 of the TNFRSF1A gene which is located on chromosome 12p13.2 [16] are the cause of this rare syndrome. TNFRSF1A gene encodes the 55 kDa receptor for tumor necrosis factor [17]. The frequency of TRAPS is 1 per million in the UK [18], and TRAPS is not associated with ethnicity [19]. TRAPS is characterized by irregular recurrent attacks of fever, myalgia, erythematous skin rash, abdominal pain, conjunctivitis, periorbital edema, and amyloidosis. The clinical findings show great variability [20]. Attacks respond dramatically to steroids. The diagnosis is confirmed by sequencing of the TNFRSF1A gene. Because the variability of the clinical findings, genetic confirmation is required for diagnosis. TRAPS patients may present with different clinical manifestations, and several mutations have been found in association with the disease.
Adult onset Still’s disease (ASD) is a systemic inflammatory disorder of unknown etiology. ASD is characterized by fever with unknown origin, erythematous rash, sore throat, arthritis or arthralgias, high glycosylated ferritin levels, and involvement of several organ systems. Steroids are used for treatment of ASD.
We aimed to investigate the role of the MEFV and TNFRSF1A gene mutations in the ASD patients, as well as their association with disease phenotype because of the similarity of the clinical findings in ASD, FMF, and TRAPS.
Methods
The study group consisted of 20 ASD patients (14 female and 6 male; mean age 38.45 ± 14; mean disease duration 3.3 ± 2.3; mean age of the disease onset 35.1 ± 14.4) and 103 healthy controls of Turkish origin. All ASD patients fulfilling the classification criteria for ASD [21] were followed up at the Kocaeli Faculty of Medicine, Department of Internal Medicine, Division of Rheumatology. The study protocol was approved by the Local Ethics Committee of the Kocaeli Faculty of Medicine, and a written informed consent was obtained from all patients before inclusion into the study.
All ASD patients were screened for the TNFRSF1A gene variations in exons 2–3 and exons 4–5 by DNA sequencing. Genotyping for the TNFRSF1A gene was carried out with the amplification of genomic DNA for exon 2–3 by using a forward primer of 5′-AGGACTTGAGCCAGGGAAGT-3′ and a reverse primer of 5′-CATAGACAGGCACCCACACA-3′ and for exon 4–5 by using a forward primer of 5′-GGCAGGAAGGTGTGTGTTTT-3′ and a reverse primer of 5′ ATCTGTTGCCCAGCTAATGG-3′. The healthy controls were genotyped for intron 4 polymorphism (rs1800692) using PCR–RFLP method with Mnl1 restriction enzyme.
The four most frequently MEFV mutations (M694V, M680I, V726A, E148Q) were genotyped in all patients and healthy controls with PCR–RFLP method according to our previously study [22].
Allele frequencies of the patients and healthy controls were compared by a Chi Square tests and odds ratios (OR) with 95 % confidence intervals (CI) were calculated by using SPSS16.0.
Results
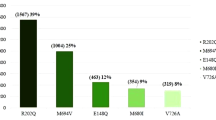
The results of MEFV gene mutations screening were given in the Table 1. Although the mutation rate was increased in ASD group, there was no significant difference between ASD and HC groups (p = 0.442, OR 1.64, 95 % CI 0.409–6.589) (Fig. 1). All ASD patients have been re-examined after the mutation analysis and none of them had FMF-associated symptoms or signs.
All patients were sequenced successfully for the exon 2–5 of the TNFRSF1A gene. No TNFRSF1A gene mutations associated with TRAPS were found in ASD patients. A common polymorphism in intron 4 (T > C, rs1800692) was the only variation found in sequencing; therefore, 103 healthy controls were genotyped by PCR–RFLP for this variation. No association for this variation was found between the ASD group and the HC group. The allele and genotype frequencies are shown in Table 2.
There was no significant difference in distribution of intron 4 polymorphism allele and genotype frequencies between the ASD group and the HC group (p = 0.54). Also, no significant association was observed between the alleles of this polymorphism and clinical manifestations.
Discussion
MEFV gene mutation rate was increased in ASD group, although it was not significantly different (p = 0.442, OR 1.64, 95 % CI 0.409–6.589). We showed only one variation in the TNFRSF1A gene, which was located in the intron 4. We screened this variation in the HC group, but we did not find any difference between both groups (p = 0.54, OR 1.24, 95 % CI 0.619–2.496–2.654). Our findings suggest that there is no significant association between ASD and TNFRSF1A variations. On the other hand, the variations in the MEFV gene may have role in ASD pathogenesis and should be investigated in larger ASD groups.
ASD is an inflammatory disease whose etiology and pathogenesis are unknown. It is characterized with strong inflammatory response to an unknown factor and good response to steroid therapy [23]. No infectious agents have been shown to be causative. ASD could be developed with one attack or non-periodical attacks, which need severe immunosuppressive therapy. Some cases could be treated with anti-TNF or anti-IL-1 therapy, even though it is unknown how these drugs affect to disease [24]. Because of the effectiveness of such therapies, we thought that some innate immune system pathways may play role in the pathogenesis. Genetic and environmental factors such as infectious agents may have role in the pathogenesis [25]. The major genetic association was found in the MHC region, and it was shown that the HLAB17, B18, B35, DR2 are associated with ASD. The other subsequently done studies have shown different HLA loci which are associated with ASD [25]. No link could be demonstrated between ASD and non-MHC gene loci.
Hereditary periodic fever syndromes constitute a rare disease group, and the most frequently seen disease of this group is FMF. In Turkey, FMF is a very frequent disease and the frequency of MEFV gene mutation was 9.7 % in our study. Because the ASD symptoms resemble the symptoms of FMF attacks, we analyzed in ASD group the common MEFV mutations. In our study, we found that the mutation rate, especially exon 10 mutations rate is increased in ASD group without any symptoms of FMF. However, the difference was not statistically significant. Since our ASD study group was very small, the difference could be greater if the study group papulation had been larger.
MEFV gene mutations cause dysregulation of the inflammasome and increased IL-1β response [26]. This pathway is the major mechanism in FMF pathogenesis. In the populations with a high MEFV mutation carrier rate, it was shown that these mutations are associated with severe disease prognosis in the other inflammatory syndromes [3–15]. The main cause of this effect is the dysregulation of IL-1 β activation [26].
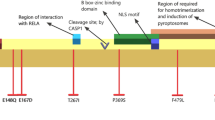
TRAPS is one of the rare hereditary autoinflammatory syndromes. TRAPS is caused by mutations in TNFRSF1A gene. TNFRSF1A, the transmembrane receptor of TNF, play the most important role in TNF-TNFRSF1A binding and therefore in the initiating of inflammation and activation of the NF-KB pathway. In consequence, inflammation is induced and apoptosis is inhibited [18]. This gene has a role in different pathways associated with inflammation, and the mutations in this gene cause in a wide range of clinical findings [27]. They may induce the expression of multiple inflammatory protein encoding genes, receptors, transcription factors, and molecules that have role in the cellular signaling pathway [28]. The clinical findings of TRAPS are variable. Although this may be due to the heterogenous mutations in TNFRSF1A, it was shown that the patients with ‘TRAPS-like’ symptoms are low sTNFRSF1A levels, do not have any mutations in the TNFRSF1A gene [29]. It may be suggested that the other genetic factors that decrease the TNFRSF1A levels may have a role in the pathogenesis of TRAPS. On the other hand, the pathogenesis of the diseases with symptoms similar to TRAPS may be associated with TNFRSF1A gene. It was shown that the patients with early synovitis for less than 6 months had R92Q mutation ratio 2.5 % compared to 1.04 % of HC [30]. A genomewide screening in multiplex rheumatoid arthritis families suggested that the TNFRSF1A gene may play a role in rheumatoid arthritis [31]. The clinical findings of ASD such as fever, erythematous rush, arthritis, response to steroids, and anti-TNF agents are the common characteristics of TRAPS and ASD. There is no research in the literature investigating the association between TRAPS and ASD. A case report from Japan showed that a patient with ASD-like skin manifestations had a novel mutation in the TNFRSF1A gene [32].
It is unknown how the mutations cause the disease. The common mutations are found in exon 2-4 which encode the extracellular domain of TNFRSF1A. More than 80 mutations are described in this gene [33]. The studies have shown that mutations in the extracellular domain of TNFRSF1A resulted to an impaired shedding of TNFRSF1A and so it has been described that the TNFRSF1A mutations caused elevated serum TNF-a levels [34]. The mutations in the extracellular domain of TNFRSF1A gene result the defective shedding of the extracellular part of the TNFRSF1A [35]. The other mechanisms which may have role in the pathogenesis include impaired TNF binding, a defect in the leukocyte apoptosis or a defect of the ER-stress [36–38].
In our study, we found that the intron 4 of the TNFRASF1A gene show wide variation in the Turkish population. We have shown for the first time that this part of the gene locus is variable. But our data showed that this polymorphism was not associated with the disease or any symptoms. Therefore, the variation in intron 4 seems not to be related with ASD.
Conclusion
We herein report that the TNFRSF1A exon 2–5 mutations are not associated with the pathogenesis of ASD and the MEFV gene mutations are increased in ASD group, although the difference was not statistically significant. These results may imply that being an asymptomatic carrier for MEFV mutations may increase susceptibility to ASD. It needs further investigation with larger study populations. The findings of this study suggest that there is no significant association between ASD and TNFRSF1A variations.
Abbreviations
- AS:
-
Ankylosing spondylitis
- ASD:
-
Adult onset Still’s disease
- CAPS:
-
Cryopyrin-associated periodic syndrome
- CI:
-
Confidence interval
- CRMO:
-
Chronic recurrent multifocal osteomyelitis
- DIRA:
-
Deficiency of the IL-1 receptor antagonist
- ER:
-
Endoplasmic reticulum
- FMF:
-
Familial Mediterranean fever
- HC:
-
Healthy control
- HIDS:
-
Hyperimmunoglobulinemia D with periodic fever syndrome
- HPFS:
-
Hereditary periodic fever syndromes
- IBD:
-
Inflammatory bowel disease
- IL:
-
Interleukin
- MEFV:
-
Mediterranean fever
- NF-kappa B:
-
Nuclear factor-kappa B
- OR:
-
Odds ratio
- PAPA:
-
Pyogenic arthritis, pyoderma gangrenosum, and acne syndrome
- PCR:
-
Polymerase chain reaction
- RA:
-
Rheumatoid arthritis
- RFLP:
-
Restriction fraction length polymorphism
- TNF:
-
Tumor necrosing factor
- TRAPS:
-
TNF receptor-associated periodic syndrome
- TNFRSF1A:
-
Tumor necrosing factor receptor superfamily 1A
References
Henderson C, Goldbach-Mansky R (2010) Monogenic autoinflammatory diseases: new insights into clinical aspects and pathogenesis. Curr Opin Rheumatol 22:567–578
Grateau G, Duruöz MT (2010) Autoinflammatory conditions: when to suspect? How to treat? Best Pract Res Clin Rheumatol 24:401–411
French FMF Consortium (1997) A candidate gene for familial Mediterranean fever. Nat Genet 17:25–31
Cosan F, Ustek D, Oku B, Duymaz-Tozkir J, Cakiris A, Abaci N, Ocal L, Aral O, Gül A (2010) Association of familial Mediterranean fever-related MEFV variations with ankylosing spondylitis. Arthritis Rheum 62:3232–3236
Akkoc N, Sari I, Akar S, Binicier O, Thomas MG, Weale ME, Birlik M, Savran Y, Onen F, Bradman N, Plaster CA (2010) Increased prevalence of M694V in patients with ankylosing spondylitis: additional evidence for a link with familial Mediterranean fever. Arthritis Rheum 62:3059–3063
Koca SS, Etem EO, Isik B, Yuce H, Ozgen M, Dag MS, Isik A (2010) Prevalence and significance of MEFV gene mutations in a cohort of patients with rheumatoid arthritis. Joint Bone Spine 77:32–35
Rabinovich E, Livneh A, Langevitz P, Brezniak N, Shinar E, Pras M, Shinar Y (2005) Severe disease in patients with rheumatoid arthritis carrying a mutation in the Mediterranean fever gene. Ann Rheum Dis 64:1009–1014
Yıldırım B, Tuncer C, Kan D, Tunc B, Demirag MD, Ferda Percın E, Haznedaroglu S, Alagozlu H (2011) MEFV gene mutations and its impact on the clinical course in ulcerative colitis patients. Rheumatol Int 31:859–864
Giaglis S, Mimidis K, Papadopoulos V, Thomopoulos K, Sidiropoulos P, Rafail S, Nikolopoulou V, Fragouli E, Kartalis G, Tzioufas A, Boumpas D, Ritis K (2006) Increased frequency of mutations in the gene responsible for familial Mediterranean fever (MEFV) in a cohort of patients with ulcerative colitis: evidence for a potential disease-modifying effect? Dig Dis Sci 51:687–692
Ayaz NA, Ozen S, Bilginer Y, Ergüven M, Taşkiran E, Yilmaz E, Beşbaş N, Topaloğlu R, Bakkaloğlu A (2009) MEFV mutations in systemic onset juvenile idiopathic arthritis. Rheumatology (Oxford) 48:23–25
Rozenbaum M, Rosner I (2004) Severe outcome of juvenile idiopathic arthritis (JIA) associated with familial Mediterranean fever (FMF). Clin Exp Rheumatol 22:75–78
Villani AC, Lemire M, Louis E, Silverberg MS, Collette C, Fortin G, Nimmo ER, Renaud Y, Brunet S, Libioulle C, Belaiche J, Bitton A, Gaudet D, Cohen A, Langelier D, Rioux JD, Arnott ID, Wild GE, Rutgeerts P, Satsangi J, Vermeire S, Hudson TJ, Franchimont D (2009) Genetic variation in the familial Mediterranean fever gene (MEFV) and risk for Crohn’s disease and ulcerative colitis. PLoS ONE 4:e7154
Ozçakar ZB, Yalçinkaya F, Cakar N, Acar B, Kasapçopur O, Ugüten D, Soy D, Kara N, Uncu N, Arisoy N, Ekim M (2008) MEFV mutations modify the clinical presentation of Henoch-Schönlein purpura. J Rheumatol 35:2427–2429
Bayram C, Demircin G, Erdoğan O, Bülbül M, Caltik A, Akyüz SG (2011) Prevalence of MEFV gene mutations and their clinical correlations in Turkish children with Henoch-Schönlein purpura. Acta Paediatr 100:745–749
Yalçinkaya F, Ozçakar ZB, Kasapçopur O, Oztürk A, Akar N, Bakkaloğlu A, Arisoy N, Ekim M, Ozen S (2007) Prevalence of the MEFV gene mutations in childhood polyarteritis nodosa. J Pediatr 151:675–678
Derré J, Kemper O, Cherif D, Nophar Y, Berger R, Wallach D (1991) The gene for the type 1 tumor necrosis factor receptor (TNF-R1) is localized on band 12p13. Hum Genet 87:231–233
McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, Gadina M, Karenko L, Pettersson T, McCarthy J, Frucht DM, Aringer M, Torosyan Y, Teppo AM, Wilson M, Karaarslan HM, Wan Y, Todd I, Wood G, Schlimgen R, Kumarajeewa TR, Cooper SM, Vella JP, Amos CI, Mulley J, Quane KA, Molloy MG, Ranki A, Powell RJ, Hitman GA, O’Shea JJ, Kastner DL (1999) Germline mutations in the extracellular domains of the 55 kDa TNF receptor (TNFR1) define a family of dominantly inherited auto-inflammatory syndromes. Cell 97:133–144
Lachmann HJ, Hawkins PN (2009) Developments in the scientific and clinical understanding of autoinflammatory disorders. Arthritis Res Therapy 11:212
Padeh S, Berkun Y (2007) Auto-inflammatory fever syndromes. Rheum Dis Clin North Am 33:585–623
Hull KM, Drewe E, Aksentijevich I, Singh HK, Wong K, McDermott EM, Dean J, Powell RJ, Kastner DL (2002) The TNF receptor-associated periodic syndrome (TRAPS): emerging concepts of an autoinflammatory disorder. Medicine (Baltimore) 81:349–368
Nowlan ML, Drewe E, Bulsara H, Esposito N, Robins RA, Tighe PJ, Powell RJ, Todd I (1992) Preliminary criteria for classification of adult Still’s disease. J Rheumatol 19:424–430
Ustek D, Ekmekci CG, Selçukbiricik F, Cakiris A, Oku B, Vural B, Yanar H, Taviloglu K, Ozbek U, Gül A (2007) Association between reduced levels of MEFV messenger RNA in peripheral blood leukocytes and acute inflammation. Arthritis Rheum 56:345–350
Bagnari V, Colina M, Ciancio G, Govoni M, Trotta F (2010) Adult-onset Still’s disease. Rheumatol Int 30:855–862
Riera E, Olivé A, Narváez J, Holgado S, Santo P, Mateo L, Bianchi MM, Nolla JM (2011) Adult onset Still’s disease: review of 41 cases. Clin Exp Rheumatol 29:331–336
Efthimiou P, Georgy S (2006) Pathogenesis and management of adult-onset Still’s disease. Semin Arthritis Rheum 36:144–152
Dinarello CA (2011) A clinical perspective of IL-1β as the gatekeeper of inflammation. Eur J Immunol 41:1203–1217
Masters SL, Lobito AA, Chae J, Kastner DL (2006) Recent advances in the molecular pathogenesis of hereditary recurrent fevers. Curr Opin Allergy Clin Immunol 6:428–433
Rebelo SL, Amel-Kashipaz MR, Radford PM, Bainbridge SE, Fiets R, Fang J, McDermott EM, Powell RJ, Todd I, Tighe PJ (2009) Novel markers of inflammation identified in tumor necrosis factor receptor–associated periodic syndrome (TRAPS) by transcriptomic analysis of effects of TRAPS-associated tumor necrosis factor receptor type I mutations in an endothelial cell line. Arthritis Rheumatism 60:269–280
Aganna E, Hammond L, Hawkins PN, Aldea A, McKee SA, van Amstel HK, Mischung C, Kusuhara K, Saulsbury FT, Lachmann HJ, Bybee A, McDermott EM, La Regina M, Arostegui JI, Campistol JM, Worthington S, High KP, Molloy MG, Baker N, Bidwell JL, Castañer JL, Whiteford ML, Janssens-Korpola PL, Manna R, Powell RJ, Woo P, Solis P, Minden K, Frenkel J, Yagüe J, Mirakian RM, Hitman GA, McDermott MF (2003) Heterogeneity among patients with tumor necrosis factor receptor–associated periodic syndrome phenotypes. Arthritis Rheum 48:2632–2644
Aksentijevich I, Galon J, Soares M, Mansfield E, Hull K, Oh HH, Goldbach-Mansky R, Dean J, Athreya B, Reginato AJ, Henrickson M, Pons-Estel B, O’Shea JJ, Kastner DL (2001) The tumor-necrosis-factor receptor-associated periodic syndrome: new mutations in TNFRSF1A, ancestral origins, genotype-phenotype studies, and evidence for further genetic heterogeneity of periodic fevers. Am J Hum Genet 69:301–314
Seldin MF, Amos CI, Chen WV, Shigeta R, Monteiro J, Kern M, Criswell LA, Albani S, Nelson JL, Clegg DO, Pope R, Schroeder HW Jr, Bridges SL Jr, Pisetsky DS, Ward R, Kastner DL, Wilder RL, Pincus T, Callahan LF, Flemming D, Wener MH, Gregersen PK (2001) A genomewide screen in multiplex rheumatoid arthritis families suggests genetic overlap with other autoimmune diseases. Am J Hum Genet 68:927–936
Nakamura M, Kobayashi M, Tokura Y (2009) A novel missense mutation in tumour necrosis factor receptor superfamily 1A (TNFRSF1A) gene found in tumour necrosis factor receptor-associated periodic syndrome (TRAPS) manifesting adult-onset Still disease-like skin eruptions: report of a case and review of the Japanese patients. Br J Dermatol 161:968–970
Jesus AA, Oliveira JB, Aksentijevich I, Fujihira E, Carneiro-Sampaio MM, Duarte AJ, Silva CA (2008) TNF receptor-associated periodic syndrome (TRAPS): description of a novel TNFRSF1A mutation and response to etanercept. Eur J Pediatr 167:1421–1425
Huggins ML, Radford PM, McIntosh RS, Bainbridge SE, Dickinson P, Draper-Morgan KA, Tighe PJ, Powell RJ, Todd I (2004) Shedding of mutant tumor necrosis factor receptor superfamily 1A associated with tumor necrosis factor receptor–associated periodic syndrome: differences between cell types. Arthritis Rheum 50:2651–2659
Todd I, Radford PM, Draper-Morgan KA, McIntosh R, Bainbridge S, Dickinson P, Jamhawi L, Sansaridis M, Huggins ML, Tighe PJ, Powell RJ (2004) Mutant forms of tumour necrosis factor receptorI that occur in TNF-receptor-associated periodic syndrome retain signaling functions but show abnormal behaviour. Immunology 113:65–79
Siebert S, Amos N, Fielding CA, Wang EC, Aksentijevich I, Williams BD, Brennan P (2005) Reduced tumor necrosis factor signaling in primary human fibroblasts containing a tumor necrosis factor receptor superfamily 1A mutant. Arthritis Rheum 52:1287–1292
Lobito AA, Kimberley FC, Muppidi JR, Komarow H, Jackson AJ, Hull KM, Kastner DL, Screaton GR, Siegel RM (2006) Abnormal disulfide-linked oligomerization results in ER retention and altered signaling by TNFR1 mutants in TNFR1-associated periodic fever syndrome (TRAPS). Blood 108:1320–1327
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cosan, F., Emrence, Z., Erbag, G. et al. The association of TNFRSF1A gene and MEFV gene mutations with adult onset Still’s disease. Rheumatol Int 33, 1675–1680 (2013). https://doi.org/10.1007/s00296-012-2609-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-012-2609-8