Abstract
Lipid droplets (LDs) have emerged as dynamic and interactive organelles with important roles in lipid metabolism and membrane biogenesis. Here, we report that Saccharomyces cerevisiae Env9 is a novel conserved oxidoreductase involved in LD morphology. Microscopic and biochemical studies confirm localization of tagged Env9 to LDs and implicate its C-terminal hydrophobic domain (aa241–265) in its membrane association and stability. Confocal studies reveal a role for Env9 in LD morphology. Env9 positively affects both formation of large LDs upon overexpression and LD proliferation under poor carbon source. In silico bioinformatic and modeling approaches establish that ENV9 is a widely conserved member of the short-chain dehydrogenase (SDR) superfamily. Bayesian phylogenetic studies strongly support ENV9 as an ortholog of human SDR retinol dehydrogenase 12 (RDH12). Dehydrogenase activity of Env9 was confirmed by in vitro oxidoreductase assays. RDH12 mutations have been linked to Leber Congenital Amaurosis. Similar site-directed point mutations in the predicted Env9 oxidoreductase active site (N146L) or cofactor-binding site (G23–24A) abolished its reductase activity in vitro, consistent with those reported in other retinol dehydrogenases. The same residues were essential for affecting LD size and number in vivo. Taken together, our results implicate oxidoreductase activity of Env9 in its cellular role in LD morphology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipid droplets (LDs) are a structurally unique member of the endomembrane system in eukaryotic cells. For many years, their main role was considered to be as an inert storage depot for neutral lipids. Recently, LDs have garnered intense interest as highly dynamic organelles involved in both biosynthesis and mobilization of neutral lipids in response to nutrient levels (Klug and Daum 2014). LDs are key players in regulation of lipid homeostasis whose defects have been linked to several metabolic diseases. Accumulation of LDs is found in diabetes, obesity, and cancer (Walther and Farese 2012). However, low number of LDs has also been implicated in disease. For example, mutant versions of the LD protein Seipin cause near complete loss of adipose tissue, which leads to hepatic steatosis and insulin resistance (Chen et al. 2014).
Structurally, LDs are composed of a neutral lipid core of triacylglycerol (TG) and steryl ester, surrounded by a phospholipid monolayer with associated LD proteins. The mechanism of LD biogenesis and growth remains poorly characterized. The current accepted models propose that LDs bud from the ER by a process regulated by several gene products including fat storage-inducing transmembrane proteins, FIT (Choudhary et al. 2015), and then grow through expansion and/or homotypic fusion that involves various TG synthesis enzymes (Wang 2015; Thiam and Foret 2016). Additionally, recent data suggest fusion events and communications between LDs and various other organelles including ER, mitochondria, peroxisomes, endosomes, and lysosomes (Dugail 2014; Barbosa et al. 2015; Gao and Goodman 2015; Schrader et al. 2015).
Several dehydrogenases have been reported to be associated with ER and/or LDs. Among those are members of the short-chain dehydrogenase/reductase (SDR) superfamily including retinol dehydrogenase (RDH) 10 and retinol dehydrogenase (RDH) 12 which are involved in redox reactions of the visual cycle in photoreceptor cells. SDRs contain around 250 residues catalyzing oxidation/reduction reactions with substrates ranging from alcohol and sugars to steroids (Kallberg et al. 2002; Kavanagh et al. 2008). RDH10 shows oxidoreductase activity in vitro, while mostly functions in the oxidation reactions in vivo (Belyaeva et al. 2008; Takahashi et al. 2009). It recognizes several retinol substrates and prefers NAD+ as a cofactor (Belyaeva et al. 2008). Subcellular localization studies show that RDH10 is associated with mitochondria/mitochondrial-associated membranes and relocalizes to LD during LD formation (Jiang and Napoli 2013). Mutations in RDH10 disrupt vitamin A metabolism, which leads to embryonic developmental defects (Sandell et al. 2007; Farjo et al. 2011). RDH12 is highly expressed in the retina and involved in retinoid metabolism as well as protection against lipid peroxidation products (Lee et al. 2008). RDH12 is shown to localize to the ER and microsomal fraction in microscopic analysis (Keller and Adamski 2007) and subcellular fractionation (Lee et al. 2007), respectively. While its specific cellular substrate remains unknown, RDH12 displays enzymatic activity on both retinoid and sterol substrates in vitro (Keller and Adamski 2007). Mutations in RDH12 have been genetically linked to several different forms of retinal dystrophies including Stargardt disease (Zolnikova et al. 2016), cone–rod dystrophy (Huang et al. 2016), retinitis pigmentosa (Gong et al. 2015), and Leber congenital amaurosis (LCA), which is the most severe form of childhood retinal dystrophy characterized by early visual loss (Janecke et al. 2004; Thompson et al. 2005; Mackay et al. 2011). In addition, disease-associated mutations in RDH12 cofactor-binding domain correspond to accumulation of bioreactive retinoic acid, which can then induce apoptosis in many cells including photoreceptor cells (Lee et al. 2007).
LD morphology is variable, and LD size and number vary between cell types and under different growth conditions. For example, white adipocytes have giant LDs, while brown adipocytes and yeast cells have small LDs. LD size can also change in response to diverse signals within the same cell type. The molecular mechanisms that regulate LD size as well as their physiological significance remain poorly understood. Baker’s yeast, however, has served as a seminal model in this respect. Two independent yeast genome-wide screens for defects in LD number and morphology identified Fld1, a homolog of human Seipin, as a regulator of LD dynamics (Szymanski et al. 2007; Fei et al. 2008). Recent studies have reported that Fld1 promotes the initial stage of LD formation (Cartwright et al. 2015) and, along with its interacting protein Ldb16, is involved in the regulation of LD size (Wang et al. 2014). General control nonderepressible (GCN4) transcription factor, which is induced under nutrient starvation and stress conditions, has also been reported to be involved in the regulation of LD size by controlling expression of a TG biosynthetic gene (Yadav and Rajasekharan 2016).
LDs can expand by relocation of TG synthesizing enzymes from the ER to LDs through ER-LD bridges (Wilfling et al. 2013), which involves Coat Protein I (COPI) machinery (Wilfling et al. 2014b). In adipocytes, FSP27 has been implicated in large LD growth by transferring neutral lipids from a smaller LD to a larger LD (Gong et al. 2011; Jambunathan et al. 2011; Grahn et al. 2013; Tan et al. 2014). Additionally, localization mechanisms of LD membrane-associated proteins remain poorly defined.
Previously, we designed a genomic screen that monitored endomembrane trafficking of the vacuolar hydrolase Carboxypeptidase Y (CPY) and uncovered several endosome and vacuole interface (ENV) genes including ENV9 (Ricarte et al. 2011). Deletion of ENV9 leads to internal accumulation of proCPY, aberrant vacuolar morphology and caffeine sensitivity. In this study, we further characterize the ENV9 deletion mutant and the ENV9 encoded product. We establish that Env9 protein is a conserved LD oxidoreductase involved in LD dynamics. The C-terminal hydrophobic stretch (aa241–265) of Env9 is essential for its LD association and stability, while its intact active site and cofactor-binding sites are essential for both its oxidoreductase activity in vitro and its role in LD dynamics in vivo. We also present bioinformatics and phylogenetic data supporting ENV9 as a widely conserved ortholog of mammalian RDH 12, a member of the SDR superfamily and discuss possible involvement of an SDR in the regulation of LD dynamics.
Materials and methods
Yeast strains and media
Yeast strains used in this study are listed in Table 1. Yeast strains were grown at 30 °C in yeast extract-peptone-dextrose (YPD) media (1% yeast extract, 2% peptone, 2% dextrose) or Synthetic Minimal (SM) media (0.67% yeast nitrogen base, 2% glucose, amino acids) from Difco Chemicals (St. Louis, MO). Yeast strains BY4742 (MATα his3Δ leu2Δ ura3Δ lys2Δ) and single-gene deletion mutants were gifts from Dr. Greg Payne (UCLA).
Construction of plasmids and site-directed mutagenesis
Plasmids used in this study are listed in Table 2 and were constructed by homologous recombination as described previously (Oldenburg et al. 1997). HA- and GFP-tagged Env9 were PCR amplified from a genomic DNA using reverse and forward primers (Operon, Huntsville, AL, USA) listed in Table 3. Cells were transformed with PCR-generated inserts and linearized plasmids containing uracil marker and grown on SM-Ura plates to select insert-carrying colonies as described previously (Manandhar et al. 2013). Plasmids were isolated from yeast using Zymoprep Yeast Plasmid Miniprep II (Zymo Research, Irvine, CA, USA) and transformed into E. coli. Deletion or substitution of multiple bases from ENV9 was performed using two PCR-based site-directed mutagenesis as described previously (Mallorqui-Fernandez et al. 2008). Plasmid constructs were confirmed by restriction digestion as well as by DNA sequencing (Macrogen, South Korea). Plasmid expressing Erg6-dsRed as a lipid droplet marker was a gift from Joel Goodman (University of Texas Southwestern Medical Center, Dallas, Texas).
Localization of Env9
Wild-type cells endogenously expressing ENV9-GFP (Invitrogen) were grown in YPD or SM Complete until mid-log phase and subjected to Nile Red staining as described previously (Greenspan et al. 1985) with following modifications. Cells were washed with 50 mM Tris–HCl, pH 7.5 and stained with Nile Red (1 µg/ml) (Invitrogen, Carlsbad, CA, USA) for 20 min. Cells were then viewed under DIC and an Olympus Fluoview 1000 confocal laser scanning system mounted on an inverted microscope (Olympus IX-81). Images were captured with argon ion (488 nm) laser with Nile Red emission at wavelength range 571–590 nm with 5% laser output power, and GFP emission at wavelength range 501–520 nm with 70% laser output power. Galactose induction of GFP fusion proteins was performed as described previously (Manandhar et al. 2013, 2014). Briefly, env9Δ cells expressing GFP-ENV9 or ENV9-GFP from the inducible promoter were grown in SM-Ura to mid-log phase (OD600 ~ 0.8), washed with sterile H2O and incubated in SM-Ura media containing 0.2% galactose, 1% glycerol, and 1% ethanol for 4–6 h to induce GFP expression. Galactose-induced cells were subjected to Nile Red staining and viewed by DIC and confocal microscopies. Images were captured with argon ion (488 nm) laser with Nile Red emission at wavelength range 571–590 nm with 5% laser output power, and GFP emission at wavelength range 501–520 nm with 25% laser output power. env9Δ cells expressing GFP-ENV9Δ241–265 or ENV9Δ241–265-GFP were also grown under the same conditions and viewed by DIC and confocal microscopies with the same setting as WT GFP-tagged ENV9 expressed from an episomal plasmid. For studies of Env9 localization in the absence of GET complex components, get1Δ or get3Δ cells expressing ENV9-GFP were also grown under the same conditions and viewed by DIC and confocal microscopies with the same setting as GFP-tagged ENV9 expressed from an episomal plasmid in env9Δ cells.
Caffeine sensitivity assay
Wild-type or env9Δ cells expressing ENV9-HA or control plasmid (pWS479) were grown to mid-log phase (OD600 ~ 0.7) in SM-Ura, harvested, and washed with SM media. Washed cells were serially diluted up to 1:1000 and plated 3 μl onto SM-Ura plates with or without 10 mM caffeine. The plates were incubated at 30 °C for up to 10 days and imaged using the Kodak EDAS imager.
Subcellular fractionation
Subcellular fractionation was performed as described previously (Manandhar et al. 2014). env9Δ cells expressing ENV9-HA, ENV9Δ241–265-HA, or control plasmid were grown in SM-Ura until late log phase, treated with 10 mM DTT, and incubated with Zymolyase (Zymo Research, Irvine, CA, USA) for 1 h at 30 °C. The spheroplasts were pelleted and treated with a commonly used lysis agent DEAE-Dextran (0.4 μg/ml) for gentle lysis of the spheroplasts as performed in other studies previously (Manandhar et al. 2013, 2014; Manandhar and Gharakhanian 2014). The lysed spheroplasts were centrifuged at 400×g for 10 min at 4 °C. The resulting supernatant (S0.4) was further centrifuged at 13,000×g for 15 min at 4 °C to obtain P13 and S13 fractions. The supernatant (S13) was subjected to further centrifugation at 100,000×g for 1 h at 4 °C to obtain membrane (P100) and cytosolic fractions (S100). P13 and P100 fractions were solubilized and adjusted to their original volumes with lysis buffer and analyzed by western blotting. For Ficoll gradient centrifugation, fractions were obtained using two subsequent centrifugations as described previously (Lin and Wheeldon 2014) and analyzed by western blotting. For protein stability studies, env9Δ cells harboring either WT ENV9-HA or ENV9Δ241–265-HA plasmid were treated with either dimethyl sulfoxide (DMSO) (control) or a potent proteasomal inhibitor MG-132 (50 μM) in growth media overnight, harvested, and subjected to subcellular fractionation as described previously (Manandhar et al. 2013). Subcellular fractions were assayed for the steady-state levels of HA-tagged protein by western blotting. For membrane association studies, env9Δ cells harboring either WT ENV9-HA or ENV9Δ241–265-HA plasmid were subjected to subcellular fractionation to obtain membrane fraction (P13). P13 fraction was resuspended either in lysis buffer or Triton X100 (1%), NaCl (1 M), and 0.1 M NaCO3 (pH 11.5) and centrifuged at 13,000×g to obtain the soluble (S13) and membrane fractions (P13). P13 fraction was solubilized and adjusted to its original volumes with lysis buffer and analyzed by western blotting. Western blots were performed following semi dry method as described previously (Manandhar et al. 2013) using equal volumes of fractions.
Protein quantification
Protein concentrations were determined using the Quick Start Bradford protein assay kit (Bio-Rad, Hercules, CA, USA) as described previously (Manandhar et al. 2013).
Lipid droplet morphology studies
For carbon stress studies, cells were grown to stationary phase (72 h) in YPD for control, and a set of YP Galactose, YP Ethanol, and YP Glycerol. For ENV9 overexpression study, cells were grown to stationary phase (72 h) in SM-Ura. Nile Red staining was performed as described in ‘Localization of Env9’ method. Cells were viewed by epifluorescence or confocal microscopy for LD morphology and numbers. Approximately, 150–200 cells were scored randomly in at least two separate experiments.
Reductase assay
Reductase activities were determined spectrophotometrically using HMG-CoA Reductase Assay kit (Sigma, St. Louis, MO, USA). Membrane fractions (P13) or LD-enriched fractions were used as the enzyme sources. Total protein concentrations in aforementioned fractions isolated from env9Δ, WT ENV9-HA, or mutant ENV9-HA overexpressing cells were normalized following protein quantitation by Bradford assays. For P13 reductase assay, P13 fractions (containing 27–30 μg of protein) were washed with enzyme assay buffer and incubated in 1 ml assay mixture containing enzyme buffer, 400 μM NADPH, and 0.3 mg/ml of HMG-CoA or 0.3 mg/ml 4-hydroxynonenal (Cayman Chemical, Ann Arbor, MI, USA) for 30 min at 30 °C. Extended incubations did not result in detection of additional enzyme activity. For LD reductase assay, LD-enriched fractions (containing 12 μg of protein) were incubated with assay mixture as described above for 120 min at 30 °C. Extended incubations of LD-enriched fractions for 120 min result in detected additional enzyme activity. The control reactions contained 2.5–3.5 μg purified HMG-CoA reductase instead of membrane fractions. Three points were plotted for each kinetic curve in at least two separate experiments.
Statistical analysis
Data are shown as average ± standard deviation. Averages were compared with a Chi-square or a t test. All experiments were performed at least twice.
Bioinformatics and phylogenetic analysis
Selection of protein sequences
Potentially orthologous protein sequences were obtained through a Basic Local Alignment Search Tool for proteins (BLASTp) search of non-redundant proteins using Env9 as the query (Altschul et al. 1990). Default parameters were maintained, and the top-scoring sequence results with greater than 70% query coverage and e values of less than 1e-10 were selected for subsequent reciprocal BLASTp searches.
Protein annotation
Domain-, function-, structure- and disease-related annotations were determined using online prediction servers and published data. Details are given in Supplementary Table 1.
Annotations were inputted into the geneious bioinformatics software platform and applied to the protein sequences (Kearse et al. 2012).
Multiple sequence alignment
Protein sequences were aligned using the multiple alignment using Fast Fourier Transform (MAFFT) v6.814b program (Katoh et al. 2002, 2005). The L-INS-I algorithm was used with a BLOSUM62 scoring matrix (Henikoff and Henikoff 1993). Penalties for gaps and offset were 1.53 and 0.123, respectively. Computationally aligned sequences were manually scanned to ensure that no large gaps or regions of misalignment were present. No portions of the proteins sequences were excised for this analysis.
Evolutionary model selection
Multiple sequence alignments were inputted into MEGA v6.06 for evolutionary model selection (Nei and Kumar 2000; Tamura et al. 2013). The model selection process used maximum likelihood methods with a moderate branch swap filter, and only sites that were at least 75% conserved in the alignment were considered.
Bayesian analysis
To generate the phylogenetic tree, the multiple sequence alignment of Env9 and its homologs were inputted into the MrBayes program (Huelsenbeck and Ronquist 2001). The archaeal protein sequence, Haloferax volcanii HVO_2590, was selected as the outgroup, and two heated chains were run for 1,000,000 generations using invariant/gamma rate variation with four gamma categories and a heated chain temperature of 0.25. Subsampling frequency was 1000, and the burn in length was 250,000. The resulting tree was displayed in Geneious and formatted for publication using Adobe Photoshop.
Protein modeling
To obtain a 3D model of Env9, the Env9 protein sequence was submitted to the I-TASSER online protein structure and function prediction server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) (Zhang 2008; Roy et al. 2010). The resulting structure was estimated to be accurate based on a confidence score of −0.50 and a TM score of 0.65 ± 0.13. Confidence scores range from −5.0 to 2.0, with 2.0 indicating the highest confidence. TM scores greater than 0.50 reflect good structural similarity between the query protein and structural templates used in the modeling process. The Env9 protein model was formatted for this study using Molsoft ICM-Browser v3.7-2b.
Results
GFP- and HA-tagged Env9 localize to LDs
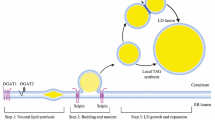
A systematic study reported that GFP-tagged YOR246C, which we have since uncovered as Env9, localized to LDs (Huh et al. 2003). We confirmed localization of endogenously expressed Env9-GFP to lipid droplets (Fig. 1a). We also constructed plasmids expressing C-terminal and N-terminal GFP-tagged Env9 under PGal promoter and studied their localization in env9Δ cells. In line with the native expression results, both GFP-tagged versions of Env9 colocalized with LDs (Fig. 1b). To investigate Env9 membrane localization biochemically, we constructed a plasmid expressing C-terminal HA-tagged Env9 from a constitutive PGK promoter. HA-tagged Env9 was enriched in the P13 vacuolar-enriched fraction in initial fractionations (Fig. 1c, left panels), indicated by colocalization of Env9-HA with full-length alkaline phosphatase (ALP) as vacuolar fraction marker. A common cytoplasmic ALP degradation product was detected in the cytosolic fraction as previously reported by several groups as well as our group (Conibear and Stevens 2000; Kweon et al. 2003; Manandhar et al. 2013, 2014). We further attempted to achieve purification of LDs using consecutive Ficoll high-speed centrifugations and analyzed the obtained fractions. HA-tagged Env9 was enriched in the top layer along with the LD marker Erg6, while vacuolar marker Carboxypeptidase Y (CPY) was enriched in the 8% Ficoll fraction (Fig. 1c, right panels). The low level of CPY, which is a vacuolar lumenal protein, is also present in 0% Ficoll fraction. This may be due to vacuole floatation during Ficoll gradient centrifugation as reported previously (Manandhar et al. 2013). Transformation with the aforementioned plasmid expressing C-terminal HA-tagged Env9 rescued caffeine sensitivity in env9Δ cells, establishing that Env9-HA is biologically active (Fig. 1d). Therefore, we subsequently used Env9-HA for both biochemical localization studies as well as in vitro enzyme assays.
Env9 localizes to lipid droplets. a Chromosomal ENV9-GFP is expressed from its endogenous promoter and localizes to LDs in rich and minimal media. b GFP-tagged Env9 is expressed from an episomal plasmid and localizes to LDs. env9Δ cells expressing GFP-ENV9 (GFP-tag at N-terminal), ENV9-GFP (GFP-tag at C-terminal), or free GFP were induced with galactose for 5 h to express the GFP-tagged proteins, stained with Nile Red and viewed by confocal microscopy. Schematic designs of expression vectors are shown to the right. c HA-tagged Env9 localizes to LDs in subcellular fractionation experiments. Left panels env9Δ cells harboring either control plasmid or ENV9-HA plasmid were spheroplasted and subjected to differential centrifugation to obtain indicated fractions, which were analyzed by western blotting using anti-HA antibody (upper panel). Blots were stripped and reprobed with anti-ALP antibody (lower panel). Asterisk commonly reported cytoplasmic degradation product of ALP. Right panels env9Δ cells expressing ENV9-HA with or without Erg6-dsRed expressing plasmid were spheroplasted, lysed with homogenization in presence of 0.4 μg DEAE-dextran and subjected to Ficoll gradient centrifugation (1st purification). The top layer (0% Ficoll fraction) was collected and further purified using 8% Ficoll gradient (2nd purification). Fractions were collected and analyzed by western blotting using anti-HA antibody. Blots were stripped and re-probed with anti-CPY antibody (Santa Cruz, California) for detection of CPY as vacuolar marker. Same experimental samples were also subjected to separate SDS-PAGE and western blotting analysis using anti-dsRed antibody (Santa Cruz, California) for detection of Erg6 as LD marker. In addition to Erg6-dsRed, a non-specific higher molecular weight band was detected in anti-dsRed blot. d ENV9-HA is functional and complements caffeine sensitivity of env9Δ cells. env9Δ or WT cells expressing control plasmid or ENV9-HA were plated in serial dilutions on media with or without caffeine
The C-terminal hydrophobic domain of Env9 is necessary for its association with LDs and its stability
Based on Kyle-Doolitle hydropathy plot analysis, we predicted that Env9 associates with the LD phospholipid monolayer membrane through a hydrophobic domain in its C-terminus (amino acids 241–265) (Fig. 2a). Deletion of this putative membrane association domain led to cytoplasmic mislocalization of both N-terminal (GFP-Env9Δ241–265) and C-terminal (Env9Δ241–265-GFP) GFP-tagged Env9 mutants (Fig. 2b). Interestingly, we observed additional mislocalization of C-terminal GFP-tagged Env9Δ241–265 to non-LD punctates as well (Fig. 2b arrows). We also constructed a C-terminal HA-tagged version of Env9Δ241–265. Our biochemical studies of wild-type (WT) and mutant C-terminal HA-tagged Env9 showed Env9Δ241–265-HA in 50:50 ratio in the membrane (P13) and cytosolic fractions (S100) as compared to WT Env9-HA that is highly enriched in the P13 fraction (Fig. 2c). Although soluble partially degraded ALP is prominent relative to mature vacuolar ALP, the levels of both species were relatively the same for both strains. However, steady-state level of Env9Δ241–265-HA was considerably lower than that of WT. To investigate if such low steady-state presence of the mutant protein is due to degradation by proteosomal system, we treated cells either with dimethyl sulfoxide, DMSO (vehicle) or proteosomal inhibitor MG-132 (50μg/ml). Addition of MG-132 stabilized Env9Δ241–265-HA as compared to DMSO-treated cells (Fig. 2d), suggesting that Env9 protein lacking its LD association domain is subject to proteasomal degradation.
A C-terminal membrane association domain anchors Env9 to lipid droplet membranes. a Schematic diagram of WT Env9 and Env9Δ241–265 deleted in the putative membrane association domain (red). b GFP-tagged Env9Δ241–265 mislocalizes to the cytoplasm and non-LD punctates. Left panels env9Δ cells, transformed with indicated plasmids, were grown in SM-Ura media to OD600 = 0.8–1.0, induced with galactose for 5 h to express GFP or GFP fusion proteins, and viewed by confocal microscopy. Right panels Quantification of data presented in left panels. *P < 0.05 compared to WT Env9-GFP. c Env9Δ241–265-HA distribution in membrane and cytosolic fractions. Cells were spheroplasted, lysed with DEAE-Dextran, and subjected to differential centrifugation to obtain indicated fractions. Fractions were collected and analyzed by western blotting using anti-HA (top panel) and anti-ALP (bottom panel, as fractionation marker) antibodies. d Env9Δ241–265-HA is a substrate for proteasomal degradation. Cells were treated with DMSO or proteosomal inhibitor MG-132 (50 μg/ml) and lysed. Fractions were analyzed by Western blotting using anti-HA and anti-ALP antibodies as described in c. e Solubilization of Env9Δ241–265-HA from membranes. Cells were treated with DMSO or MG-132 (50 μg/ml) and lysed. Soluble (S13) and membrane (P13) fractions were resuspended in the indicated solutions (TX-100—Triton X100, pH 11.5–0.1 M NaCO3, pH 11.5) and analyzed by western blotting using anti-HA antibody. f Mislocalization of Env9Δ241–265-HA in proteasome-inhibited cells. Cells were treated with MG-132 (50 μg/ml), spheroplasted, lysed with homogenization in the presence of 0.4 μg DEAE-dextran and subjected to Ficoll gradient centrifugation. Fractions were analyzed by western blotting using anti-HA antibody. g Stabilized and mislocalized Env9Δ241–265-HA is not associated with LD membranes. Cells were subjected to similar treatments as in f followed by sequential Ficoll gradient centrifugation as described in Fig. 1c. Fractions were analyzed by western blotting using anti-HA and anti-dsRed antibodies. In addition to Erg6-dsRed, a non-specific higher molecular weight band was detected in anti-dsRed blot. h Mislocalization of GFP-tagged Env9 in get1Δ and get3Δ. get1Δ or get3Δ cells, transformed with ENV9-GFP plasmid, were grown in SM-Ura media to OD600 = 0.8–1.0, induced with galactose for 5 h to express GFP fusion proteins, and viewed by confocal microscopy. i Quantification of data presented in h. *P < 0.0001 compared to WT
We further explored the nature of membrane association of Env9Δ241–265-HA. We treated P13 fraction of WT and mutant Env9 with high pH (11.5), high salt (1 M), or ionic detergent Triton X-100 (1%) and analyzed both soluble and insoluble fractions by western blotting using anti-HA antibody. Env9-HA remained in the pellets with each treatment, suggesting its tight association with LD membranes. However, Env9Δ241–265-HA was prone to solubilization especially upon high pH and detergent treatment (Fig. 2e), indicating either its aberrant association with membranes or aggregate formation as reported for other proteins in response to different cellular signals (Saarikangas and Barral 2016). Ficoll gradient centrifugation of MG-132 treated cells revealed that Env9-HA is mostly present in the LD-enriched top layer fraction (0% Ficoll), whereas Env9Δ241–265-HA is in the pellet (Fig. 2f). To investigate if Env9Δ241–265-HA leads to changes in LD properties that shift LDs from top layer (0% Ficoll) to pellet, we examined the localization of LD marker Erg6 using separate sequential Ficoll gradient centrifugation of MG-132 treated cells. While in both samples Erg6 was mostly detected in the top layer fraction (0% Ficoll) where LDs are expected, Env9Δ241–265-HA was consistently present in the pellet (Fig. 2g). Thus, stabilized Env9Δ241–265-HA is not associated with LD membranes and forms aggregates and/or aberrant associations, which may render it prone to proteasomal degradation. This result is in line with our microscopic observation, which shows mislocalization of C-terminal GFP-tagged Env9Δ241–265 to the cytoplasm as well as non-LD punctates, whereas N-terminal GFP-tagged Env9Δ241–265 mislocalizes to cytoplasm only. These suggest that HA- or GFP-tagging at C-terminus of Env9Δ241–265, where the Env9 membrane association domain is normally located, may enhance aberrant aggregation or association of Env9 mutant protein.
Based on the absence of a signal sequence in Env9 and the cytoplasmic mislocalization of Env9 upon deletion of its single C-terminal hydrophobic domain, we hypothesized that Env9 membrane association is via a tail-anchored (TA) insertion-like mechanism following its synthesis in the cytoplasm. Therefore, we studied the localization of Env9 in the absence of Get1 or Get3 components of the guided entry of TA proteins (GET) complex, which is known to anchor proteins with C-terminal TA region into the ER membrane (Schuldiner et al. 2008; Jonikas et al. 2009; Wang et al. 2011; Johnson et al. 2013). Single deletion of GET complex components, Get1 or Get3, leads to both LD and diffused cytoplasmic localizations of C-terminal GFP-tagged Env9 in get1Δ and get3Δ strains (Fig. 2h and i), suggesting GET-dependent membrane association for Env9. Taken together, these results establish that C-terminal hydrophobic domain of Env9 is essential to its wild-type association with LD membranes, which may involve a GET-dependent mechanism.
Env9 is involved in LD morphology
We uncovered ENV9 through a genome-wide immunodetection screen for endomembrane trafficking defects and found that ENV9 deletion mutant has defects in vacuolar CPY maturation. Thus, our initial functional characterizations remained focused on vacuolar events. Using metabolic labeling and immunoprecipitation, we confirmed a CPY maturation delay in env9∆ cells (Supplementary Fig. 1). The maturation delay extended to p1CPY → p2CPY conversion that occurs at ER to Golgi stage of trafficking, consistent with early endomembrane trafficking defects. We also explored vacuolar function and morphology under various stress conditions in env9∆ cells. Our results show that env9∆ cells were not defective in autophagy (Supplementary Figs. 2–6), in maintaining vacuolar fragmentation during hyperosmotic stress (Supplementary Figs. 7, 8), or in vacuolar acidification (Supplementary Fig. 9). env9∆ cells also show normal vacuolar morphology under different carbon stress conditions (Supplementary Figs. 10–14) and under high level of ENV9 overexpression from a constitutive PGK promoter (Supplementary Figs. 15, 16). Thus, ENV9 deletion affects early endomembrane events and singularly is not essential for the range of tested vacuolar events.
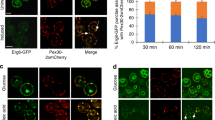
Due to its localization to LD membranes, we hypothesized that Env9 is involved in LD morphology, mainly LD size and number in cells. The current models propose that LDs bud from the ER and can grow through expansion, permeation, and homotypic fusion (Walther and Farese 2012; Wilfling et al. 2014a). As LDs are involved in lipid metabolism and TG storage, LD morphology was compared under various nutrient conditions such as variations of carbon source and lipid-loading states. Under normal (YPD 2%) and low glucose levels (YPD 1 and 0.5%), env9Δ cells did not exhibit significantly different LD morphology from WT (Supplementary Fig. 17). Exogenous loading of monounsaturated fatty acids (FAs) such as oleic acid can promote TG synthesis and induce LD proliferation, presumably, to protect both mammalian and yeast cells from lipotoxicity (Listenberger et al. 2003; Connerth et al. 2010). Studies suggest that oleic acid may be a preferred substrate for TG synthesis enzyme and may also increase the stability of LDs (Listenberger et al. 2003; Fujimoto et al. 2006). To determine whether ENV9 is involved in oleic acid-induced LD proliferation, cells were treated with 0.1% oleic acid, and LD morphology was analyzed by confocal microscopy. Throughout this study, we analyze LD morphology and numbers using live cell epifluorescence or confocal microscopy as performed previously. We scored LD morphology following a conventional scoring that classifies cells with LD number ranging from 3 to 7 as having normal LDs, cells with fewer than three LDs as having few LDs, and cells with more than seven LDs as having many LDs (Fei et al. 2008). In the presence of oleic acid as sole carbon source, env9Δ cells form many and large LDs analogous to WT cells (Supplementary Figs. 18 and 19), indicating that ENV9 is not essential in promoting oleic acid-induced LD morphology. To explore ENV9 role in LD morphology under poor carbon source, we analyzed LD morphology of env9Δ cells in glycerol-containing media and observed that WT cells form many LDs while env9Δ cells do not (Fig. 3a, b). Although glycerol is a representative non-fermentable carbon source, yeast can also utilize and convert glycerol into dihydroxyacetone phosphate (DHAP), which enters glycolytic or gluconeogenic pathway through phosphorylation and oxidation processes (Gancedo et al. 1968; Sprague and Cronan 1977; Schüller 2003). Thus, glycerol can also be used as a substrate for TG biosynthesis, and yeast lipidome studies have shown that TG levels are increased in glycerol-grown cells (Klose et al. 2012). So, glycerol loading may induce LD proliferation as simply a non-fermentable poor carbon source leading to carbon stress or, alternatively, due to an increased pool for TG biosynthesis.
Env9 is involved in LD morphology. a ENV9 promotes glycerol-induced LD proliferation. WT and env9Δ cells were grown until stationary phase in media containing 2% dextrose or 3% glycerol, stained with Nile Red, and viewed by confocal microscopy for lipid droplet morphology. b Quantification of data presented in panel a. Approximately, 200–300 cells were scored for number of lipid droplets and categorized into ‘few lipid droplets’ (<3), ‘normal lipid droplets’ (3–7), and ‘many lipid droplets’ (>7). *P < 0.005 compared to WT under the same conditions. c ENV9 plays role in carbon stress-induced LD proliferation. WT and env9Δ cells were grown until stationary phase in media containing 2% dextrose, 2% galactose, or 3% ethanol, stained with Nile Red, and viewed by confocal microscopy for lipid droplet morphology. d Quantification of data presented in c. *P < 0.005 compared to WT under the same conditions. e Moderate overexpression of ENV9 does not affect LD morphology. WT (expressing control plasmid) or ENV9 overexpressing cells were grown until stationary phase in SM-Ura media, stained with Nile Red, and viewed by epifluorescence microscopy. f Quantification of data presented in e. g High-level overexpression of ENV9 leads to fewer and larger LDs. WT (expressing control plasmid) or ENV9-HA overexpressing cells were grown until stationary phase in SM-Ura media, stained with Nile Red, and viewed by confocal microscopy. h Quantification of data presented in g. *P < 0.0001 compared to WT
Therefore, we further investigated whether other poor carbon sources also lead to the same phenotype. We analyzed LD morphology of WT and env9Δ cells in poor carbon sources that are fermentable (galactose) and non-fermentable (ethanol). In both scenarios, WT cells form many LDs. Under galactose loading, env9Δ cells continue to form fewer LDs compared to WT cells, and this phenotype is further accentuated under ethanol loading (Fig. 3c, d). Thus, poor carbon source triggers LD proliferative morphology, and ENV9 may have a specific role in this cellular response.
As deletion of ENV9 did not significantly affect LD morphology, we assessed LD morphology upon ENV9 overexpression from its own regulatory sequence in a 2 μ multi copy plasmid. Our results showed that moderate ENV9 overexpression did not significantly affect lipid droplet morphology under normal growth conditions (Fig. 3e, f). We also assessed LD morphology under high level of ENV9 overexpression from a constitutive PGK promoter (Env9-HA). Our microscopic analysis showed that WT cells have LD numbers ranging from 3 to 7, while cells that highly overexpress ENV9 form fewer and larger LDs (Fig. 3g, h). High-level overexpression of ENV9 in env9Δ background also leads to the same phenotype (Supplementary Figs. 20, 21). Taken together, these results indicate that ENV9 is involved in LD size and number. Its involvement may include regulation of LD fusion or expansion and carbon stress-induced LD proliferation events.
Env9 contains an oxidoreductase domain and is conserved in all domains of life
For further investigation into Env9’s possible functions, we used the reciprocal BLASTp method to search for potentially orthologous sequences in other organisms. Thirty-four such sequences were found among both prokaryotes and eukaryotes. The top-scoring BLASTp results from representative species were used to construct a multiple sequence alignment (Fig. 4A) and Bayesian phylogenetic tree (Fig. 5). Protein annotation and multiple sequence alignment revealed that Env9 possesses an oxidoreductase domain containing NADPH-binding residues and active site residues that are conserved in SDRs (Fig. 4A). A glycine-rich NADPH-binding motif (T22GGNTGIG29) and key active site residues (Y202AMSK206) place Env9 in a large, classical family of SDRs (Kavanagh et al. 2008).
Env9 contains conserved key oxidoreductase domains. A An annotated multiple sequence alignment of Env9 amino acid sequence and representative orthologs and homologs shows well-aligned features. Eight regions either homologous or orthologous to Env9 were aligned using MAFFT alignment and visualized with geneious. The similarity of the aligned residues is indicated by a black-to-white graded scale (black corresponding to the most similarity and white to the least). Portions of the alignment containing homology-based predictions of active site and/or NADPH-binding residues are magnified below the complete multiple sequence alignment. Active site residues are indicated on Env9 by forward-slash bars underneath them, and NADPH-binding residues have black, downward-pointing triangles beneath them. B Three-dimensional representations of S. cerevisiae Env9 and H. sapiens RDH12 highlighting important structures and residues. a, b Show space-filling models of Env9, but b is rotated 180o about a longitudinal axis. c, d show ribbon models of Env9 in the same orientations as a, b, respectively. e Depicts the ribbon structure for RDH12, the human ortholog of Env9. The I-TASSER online server was used to generate a three-dimensional model in silico from the amino acid sequence for Env9. The predicted membrane-associated domain is colored green. Active site and NADPH-binding residues specific to SDRs were assigned based on multiple sequence alignment similarity or identity. Active site residues are shaded red, and NADPH-associated residues are colored blue. Two residues that are predicted to be part of both the active site and NADPH-binding region are shaded purple. N- and C-termini are labeled on the ribbon representations if visible on the model
A Bayesian phylogenetic tree of Env9 and 34 of its homologs suggests its conservation in Archaea, Bacteria, and Eukaryota. The amino acid sequences of Env9 and 34 of its homologs were aligned by MAFFT multiple sequence alignment software, and this alignment was submitted to the MEGA6 program to determine an appropriate evolutionary model. The Bayesian phylogenetic tree, using H. volcanii HVO_2590 as the outgroup, was then generated from the alignment (fixed rate matrix: WAG; rate variation: invariant/gamma; gamma categories: 4; number of heated chains: 2; heated chain temperature: 0.25; chain length: 1,000,000; ASDSF: 0.05526). Sequences are shaded according to their membership to a domain or kingdom. The scale bar represents 0.4 amino acid changes. Thick branches indicate more than 90% posterior probability
The Bayesian tree supports Env9 as a homolog of human retinol dehydrogenase (RDH12) (Fig. 5). This short-chain dehydrogenase is highly expressed in retina and is involved in the visual cycle in photoreceptor cells. In vitro, RDH12 reportedly acts on retinoid substrates with a high affinity toward NADPH and is implicated in detoxification of unsaturated aldehydes produced from lipid peroxidation during oxidative stress (Belyaeva et al. 2005; Marchette et al. 2010). RDH12 mutations are associated with different types of retinal dystrophies (Gong et al. 2015; Huang et al. 2016; Zolnikova et al. 2016) including Leber congenital amaurosis (LCA), a severe childhood-onset autosomal recessive retinal dystrophy characterized by early visual loss (Janecke et al. 2004; Thompson et al. 2005; Mackay et al. 2011).
Three-dimensional in silico modeling of Env9 revealed that it contains a Rossman-fold-like scaffold conducive to cofactor binding (Fig. 4B). This fold is present in many other SDRs (Kavanagh et al. 2008) including RDH12. The model also shows extensive structural similarities between RDH12 and Env9 particularly in the active and NADPH-binding sites (Fig. 4B—c, e). While the active site residues and NADPH-binding residues appear to be somewhat buried in the enzyme, the membrane association domain is located on a highly accessible loop that extends away from the bulk of the structure (Fig. 4B—c, d). However, this stretch of 25 amino acids (241–265) is not well conserved outside of yeast and is mostly absent in RDH12. The fact that this domain is not well conserved but important for Env9 membrane association (Fig. 2b–g) suggests that the Env9 membrane association mechanism may be unique in yeast.
Env9 shows oxidoreductase activity in vitro
Based on our bioinformatics analysis of Env9 with its orthologs, we hypothesized that Env9 is an oxidoreductase and assayed for its oxidoreductase activity using an in vitro approach. P13 or LD-enriched fractions isolated from env9Δ or ENV9-HA overexpressing cells (ENV9-HA) were used as the enzyme sources and tested on various substrates including 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA), the substrate for the rate-limiting enzyme (HMG-CoA reductase) in sterol biosynthesis, and 4-hydroxynonenal (4-HNE), one of the established in vitro substrates of RDH12 (Belyaeva et al. 2005; Marchette et al. 2010). Control reactions containing pure HMG-CoA reductase showed large decreases in NADPH absorbance, confirming its activity on its specific substrate HMG-CoA in these experiments (Fig. 6a). P13 membrane fractions from ENV9 overexpressing cells (ENV9-HA) showed significantly higher activity on HMG-CoA than env9Δ cells (Fig. 6b). We believe that the residual activity on HMG-CoA in env9Δ membranes is due to the presence of other known oxidoreductases in the P13 fraction. Previously identified LD oxidoreductases include Ayr1(Athenstaedt and Daum 2000), Hfd1 (Nakahara et al. 2012; Currie et al. 2014), and Tsc10 (Beeler et al. 1998; Currie et al. 2014). Interestingly, this residual activity significantly decreased when 4-HNE was used as a substrate (Fig. 6c), indicating that Env9 oxidoreductase activity on this aldehyde substrate, which is also a substrate of RDH12, may be minimally redundant in yeast. Due to this higher specificity, we also used LD-enriched fraction as enzyme source and tested Env9 oxidoreductase activity on this aldehyde substrate. LD-enriched fraction from ENV9-HA overexpressing cells showed more activity on 4-HNE compared to P13 fraction (Fig. 6d), which may be due to high concentration of Env9-HA in LD-enriched fraction.
Env9 is an oxidoreductase and requires conserved oxidoreductase domains for its enzymatic activity in vitro. Reductase activity is expressed as the rate of decrease of NADPH absorbance and expressed in percentage of the initial value. a Control reactions contained purified HMG-CoA reductase (2.5–3.5 μg), 400 μM NADPH, and 0.3 μg/μl HMG-CoA incubated for 30 min at 30 °C. P13 fractions (containing 27–30 μg protein) with or without Env9-HA were incubated with 400 μM NADPH and 0.3 μg/μl HMG-CoA (b) or 0.3 μg/μl 4-hydroxynonenal (c) for 30 min at 30 °C and rate of decrease in absorbance at 340 nm was recorded every 15 min for 30 min. d LD-enriched fractions (containing 12 μg protein) with or without Env9-HA were incubated with 400 μM NADPH and 0.3 μg/μl 4-HNE for 120 min at 30 °C. Rate of decrease in absorbance at 340 nm was recorded at 0, 30, 60, and 120 min. P13 fractions (containing 27–30 μg protein) from ENV9-overexpressing cells (ENV9-HA) or indicated mutants were assayed for reductase activities using 0.3 μg/μl HMG-CoA (e) or 0.3 μg/μl 4-hydroxynonenal (f) as described in b, c. g LD-enriched fractions (containing 12 μg protein) from ENV9-overexpressing cells (ENV9-HA) or indicated mutants were assayed for reductase activities using 0.3 μg/μl 4-HNE as described in d. Data shown are average of at least two independent experiments
Env9 oxidoreductase domain mutants are enzymatically inactive in vitro
SDRs contain highly conserved glycine residues (GXXXGXG) in the cofactor-binding domain and a conserved asparagine residue in the active site. Mutations in these conserved residues have resulted in diminished reductase activity in another member of SDRs (Takahashi et al. 2009). Sequence alignment of Env9 with its homologs suggests that the glycine-rich motif (GGNTGIG) and asparagine residue (N146) may be crucial for Env9 reductase activity as well (Fig. 4A). To confirm this hypothesis, we generated two separate functional domain mutants using PCR-based site-directed mutagenesis as described previously (Manandhar et al. 2013). In one mutant, two glycine codons (G23–24) in the predicted cofactor-binding domain were mutated into alanine codons (G23–24A), and in the other, the asparagine codon (N146) in the predicted active site was mutated into a leucine codon (N146L).
Oxidoreductase activity of these Env9 mutants was determined using the method and substrates described for wild-type Env9. P13 fractions containing overexpressed HA-tagged Env9N146L and Env9G23–24A showed only residual activity on HMG-CoA (Fig. 6e) that was comparable to those from env9Δ cells. Furthermore, oxidoreductase activity of both Env9N146L-HA and Env9G23–24A-HA decreased significantly, relative to WT Env9-HA, on 4-HNE substrate (Fig. 6f). LD-enriched fraction containing mutants Env9-HA also showed statistically significant decline in oxidoreductase activity on 4-HNE substrate (Fig. 6g). Taken together, these suggest that the conserved asparagine residue of the active domain and glycine residues in the NADPH-binding domain are required for Env9 catalytic activity and further confirm Env9 function as a typical SDR.
Env9 oxidoreductase defective mutants are also defective in directing LD morphology in vivo
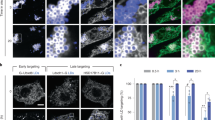
Formation of fewer and larger LDs upon high level of ENV9 overexpression indicates Env9 involvement in LD morphology in cells (Fig. 3g, h). To further investigate whether Env9 oxidoreductase activity is required for its cellular role in LD morphology, LD size and number of Env9 oxidoreductase defective mutants was assessed using a confocal microscopic approach as before. Overexpression of WT Env9 (ENV9-HA) led to formation of fewer and larger LDs as consistently observed; however, this phenotype was not observed upon overexpression of Env9G23–24A-HA nor Env9N146L-HA (Fig. 7a, b). This suggests that Env9 oxidoreductase activity is essential for Env9’s function in LD morphology in vivo. As env9Δ cells are defective in glycerol-induced LD proliferation, we attempted to investigate LD morphology of ENV9 oxidoreductase defective mutants in the presence of glycerol as sole carbon source. However, these plasmid-carrying WT cells and mutants were not viable in glycerol-containing SM media, and we were unable to study their LD morphology.
Env9 reductase activity is required for its cellular function in LD dynamics. a Overexpression of Env9 oxidoreductase functional domain mutants did not promote LD enlargement/fusion. env9Δ cells carrying control plasmid, ENV9 overexpressing plasmid (ENV9-HA), or mutated ENV9 overexpressing plasmids (ENV9G23-24A-HA or ENV9N146L-HA), were grown to stationary phase in SM-Ura media, stained with Nile Red, and viewed by confocal microscopy for LD numbers and morphology. b Quantification of data presented in a. Data shown are representative of three independent experiments. *P < 0.0001 compared to env9Δ cells under the same conditions
Discussion
Here, we report that Env9 is a novel LD oxidoreductase involved in LD morphology. Its association with LDs is via a C-terminal hydrophobic stretch that is also essential for its stability and may involve GET-dependent mechanism. We also present phylogenetic support that Env9 is an ortholog of RDH12, a member of SDR superfamily. Lastly, we show that both in vitro oxidoreductase activity and in vivo role in LD morphology of Env9 are dependent on its conserved oxidoreductase domains.
LD localization mechanism of Env9
Our results establish that Env9 localization to LDs is dependent on its single C-terminal hydrophobic domain. Our results also establish that the hydrophobic stretch, and presumably LD membrane association, is essential for Env9 stability. The localization mechanism of LD membrane-associated proteins remains poorly defined. Most recently, at least two types of membrane association have been proposed. One proposes initial association at the ER via hairpin hydrophobic domains, while the other proposes direct association with LDs from the cytoplasm through (1) amphipathic helix and hydrophobic domains, (2) lipid anchors, or (3) binding to LD proteins (Kory et al. 2016). Our results in GET deletion mutants suggest a tail insertion GET-dependent mechanism but don’t distinguish between initial ER and direct LD association. More extensive investigations on Env9 membrane association are currently in progress. Human RDH12 does not contain a putative membrane association domain, and immunostaining analysis of fixed cells has shown its localization to the ER (Keller and Adamski 2007). This is interesting as the most accepted model of LD biogenesis is biogenesis directly from the ER.
A conserved cellular role for Env9
RDH12 can catalyze the reduction of medium-chain aldehyde 4-HNE to non-toxic alcohol in vitro (Belyaeva et al. 2005). 4-HNE is a product of lipid peroxidation and can induce apoptosis; RDH12 is shown to decrease toxic 4-HNE-protein adduct formation and cell death in mouse retina in an NADPH-dependent reaction, which supports the role of RDH12 in 4-HNE detoxification (Marchette et al. 2010). We show specific oxidoreductase activity of Env9 on 4-HNE in vitro. In yeast, 4-HNE treatment results in longer lag phase, cell cycle arrest as indicated by retention in unbudded state, and reduction in incorporation of thymidine, uridine, and amino acids into DNA, RNA, and protein, respectively (Wonisch et al. 1998). Env9 may physiologically protect against cytotoxicity effects of 4-HNE including cell cycle arrest. Furthermore, Env9 putative NADPH-binding domain and active site are essential to its oxidoreductase activity on 4-HNE in vitro. These results are consistent with those reported in another retinol dehydrogenase, RDH10, in which point mutations of key residues in cofactor-binding and catalytic domains diminish its activity (Takahashi et al. 2009). RDH10 is also shown to relocalize to LDs and display highest activity there during LD formation (Jiang and Napoli 2013). Our results implicate Env9 in LD morphology and support a role for it in LD biogenesis. Specifically, our results suggest that Env9 positively regulates LD fusion/expansion under normal growth conditions and LD proliferation under carbon stress. In normal growth media, deletion of ENV9 did not significantly affect LD morphology. There may be LD residents that can compensate for the loss of ENV9 in maintaining wild-type LD dynamics under normal conditions. Under carbon stress conditions, however, Env9 role may be less redundant. Future lipidomic analyses to quantitatively measure phospholipid levels in ENV9 deletion mutant, ENV9 overexpressing strain, and Env9 oxidoreductase defective mutants would shed further light on Env9 role in LD biogenesis.
Recently, numerous studies have focused on mutations in RDH12, as they have been genetically linked to early-onset retinal dystrophy and LCA. Yet, the cellular role of RDH12 remains undefined. Our report establishes the yeast ortholog of RDH12 as an LD membrane resident that displays oxidoreductase activity in vitro and is involved in LD morphology in vivo. Among identified proteins involved in LD morphology, Env9 is the first oxidoreductase. Taken together, our results provide insight into significance of SDRs in maintaining homeostasis in endomembrane dynamics.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. doi:10.1016/S0022-2836(05)80360-2
Athenstaedt K, Daum G (2000) 1-Acyldihydroxyacetone-phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame YIL124w is a major component of lipid particles. J Biol Chem 275:235–240. doi:10.1074/jbc.275.1.235
Barbosa AD, Savage DB, Siniossoglou S (2015) Lipid droplet–organelle interactions: emerging roles in lipid metabolism. Curr Opin Cell Biol 35:91–97. doi:10.1016/j.ceb.2015.04.017
Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T (1998) The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Δ mutant. J Biol Chem 273:30688–30694
Belyaeva OV, Korkina OV, Stetsenko AV, Kim T, Nelson PS, Kedishvili NY (2005) Biochemical properties of purified human retinol dehydrogenase 12 (RDH12): catalytic efficiency toward retinoids and C9 aldehydes and effects of cellular retinol-binding protein type I (CRBPI) and cellular retinaldehyde-binding protein (CRALBP) on the oxidation and reduction of retinoids. Biochemistry 44:7035–7047. doi:10.1021/bi050226k
Belyaeva OV, Johnson MP, Kedishvili NY (2008) Kinetic analysis of human enzyme RDH10 defines the characteristics of a physiologically relevant retinol dehydrogenase. J Biol Chem 283:20299–20308. doi:10.1074/jbc.M800019200
Binns D et al (2006) An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol 173:719–731. doi:10.1083/jcb.200511125
Cartwright BR, Binns DD, Hilton CL, Han S, Gao Q, Goodman JM (2015) Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol Biol Cell 26:726–739. doi:10.1091/mbc.E14-08-1303
Chen W, Zhou H, Saha P, Li L, Chan L (2014) Molecular mechanisms underlying fasting modulated liver insulin sensitivity and metabolism in male lipodystrophic Bscl2/seipin-deficient mice. Endocrinology 155:4215–4225. doi:10.1210/en.2014-1292
Choudhary V, Ojha N, Golden A, Prinz WA (2015) A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J Cell Biol 211:261–271. doi:10.1083/jcb.201505067
Conibear E, Stevens TH (2000) Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell 11:305–323
Connerth M, Czabany T, Wagner A, Zellnig G, Leitner E, Steyrer E, Daum G (2010) Oleate inhibits steryl ester synthesis and causes liposensitivity in yeast. J Biol Chem 285:26832–26841. doi:10.1074/jbc.M110.122085
Currie E et al (2014) High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J Lipid Res 55:1465–1477. doi:10.1194/jlr.M050229
Dugail I (2014) Lysosome/lipid droplet interplay in metabolic diseases. Biochimie 96:102–105. doi:10.1016/j.biochi.2013.07.008
Farjo KM, Moiseyev G, Nikolaeva O, Sandell LL, Trainor PA, Ma JX (2011) RDH10 is the primary enzyme responsible for the first step of embryonic Vitamin A metabolism and retinoic acid synthesis. Dev Biol 357:347–355. doi:10.1016/j.ydbio.2011.07.011
Fei W et al (2008) Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol 180:473–482. doi:10.1083/jcb.200711136
Fujimoto Y et al (2006) Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of Acyl-CoA synthetase. Biol Pharm Bull 29:2174–2180
Gancedo C, Gancedo JM, Sols A (1968) Glycerol metabolism in yeasts. Pathways of utilization and production. Eur J Biochem 5:165–172
Gao Q, Goodman JM (2015) The lipid droplet-a well-connected organelle Front Cell. Dev Biol 3:49. doi:10.3389/fcell.2015.00049
Gong J et al (2011) Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol 195:953–963. doi:10.1083/jcb.201104142
Gong B et al (2015) Exome sequencing identified a recessive RDH12 mutation in a family with severe early-onset retinitis pigmentosa. J Ophthalmol 2015:942740. doi:10.1155/2015/942740
Grahn TH et al (2013) FSP27 and PLIN1 interaction promotes the formation of large lipid droplets in human adipocytes. Biochem Biophys Res Commun 432:296–301. doi:10.1016/j.bbrc.2013.01.113
Greenspan P, Mayer EP, Fowler SD (1985) Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100:965–973. doi:10.1083/jcb.100.3.965
Henikoff S, Henikoff JG (1993) Performance evaluation of amino acid substitution matrices. Proteins 17:49–61. doi:10.1002/prot.340170108
http://zhanglab.ccmb.med.umich.edu/I-TASSER/. Accessed 26 Sept 2013 (last modified: July 10, 2014)
Huang L et al (2016) Molecular genetics of cone-rod dystrophy in Chinese patients: new data from 61 probands and mutation overview of 163 probands. Exp Eye Res 146:252–258. doi:10.1016/j.exer.2016.03.015
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425:686–691. doi:10.1038/nature02026
Jambunathan S, Yin J, Khan W, Tamori Y, Puri V (2011) FSP27 promotes lipid droplet clustering and then fusion to regulate triglyceride accumulation. PLoS One 6:1–12. doi:10.1371/journal.pone.0028614
Janecke AR et al (2004) Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat Genet 36:850–854. doi:10.1038/ng1394
Jiang W, Napoli JL (2013) The retinol dehydrogenase Rdh10 localizes to lipid droplets during acyl ester biosynthesis. J Biol Chem 288:589–597. doi:10.1074/jbc.M112.402883
Johnson N, Powis K, High S (2013) Post-translational translocation into the endoplasmic reticulum. Biochim Biophys Acta Mol Cell Res 1833:2403–2409. doi:10.1016/j.bbamcr.2012.12.008
Jonikas MC et al (2009) Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323:1693–1697. doi:10.1126/science.1167983
Kallberg Y, Oppermann U, Jornvall H, Persson B (2002) Short-chain dehydrogenase/reductase (SDR) relationships: a large family with eight clusters common to human, animal, and plant genomes. Protein Sci 11:636–641. doi:10.1110/ps.26902
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Katoh K, Kuma K, Miyata T, Toh H (2005) Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform 16:22–33
Kavanagh KL, Jörnvall H, Persson B, Oppermann U (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 65:3895–3906. doi:10.1007/s00018-008-8588-y
Kearse M et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi:10.1093/bioinformatics/bts199
Keller B, Adamski J (2007) RDH12, a retinol dehydrogenase causing Leber’s congenital amaurosis, is also involved in steroid metabolism. J Steroid Biochem Mol Biol 104:190–194. doi:10.1016/j.jsbmb.2007.03.015
Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K (2012) Flexibility of a eukaryotic lipidome-insights from yeast lipidomics. PLoS One 7:e35063. doi:10.1371/journal.pone.0035063
Klug L, Daum G (2014) Yeast lipid metabolism at a glance. FEMS Yeast Res 14:369–388. doi:10.1111/1567-1364.12141
Kory N, Farese RV, Walther TC (2016) Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol. doi:10.1016/j.tcb.2016.02.007
Kweon Y, Rothe A, Conibear E, Stevens TH (2003) Ykt6p is a multifunctional yeast R-SNARE that is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell 14:1868–1881. doi:10.1091/mbc.E02-10-0687
Lee SA, Belyaeva OV, Popov IK, Kedishvili NY (2007) Overproduction of bioactive retinoic acid in cells expressing disease-associated mutants of retinol dehydrogenase 12. J Biol Chem 282:35621–35628. doi:10.1074/jbc.M706372200
Lee SA, Belyaeva OV, Kedishvili NY (2008) Effect of lipid peroxidation products on the activity of human retinol dehydrogenase 12 (RDH12) and retinoid metabolism. Biochim Biophys Acta 1782:421–425. doi:10.1016/j.bbadis.2008.03.004
Lin JL, Wheeldon I (2014) Dual N- and C-terminal helices are required for endoplasmic reticulum and lipid droplet association of alcohol acetyltransferases in Saccharomyces cerevisiae. PLoS One 9:e104141. doi:10.1371/journal.pone.0104141
Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Ory DS, Schaffer JE (2003) Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA 100:3077–3082. doi:10.1073/pnas.0630588100
Mackay DS et al (2011) RDH12 retinopathy: novel mutations and phenotypic description. Mol Vis 17:2706–2716
Mallorqui-Fernandez N et al (2008) A new autocatalytic activation mechanism for cysteine proteases revealed by Prevotella intermedia interpain A. J Biol Chem 283:2871–2882. doi:10.1074/jbc.M708481200
Manandhar SP, Gharakhanian E (2014) ENV7 and YCK3, which encode vacuolar membrane protein kinases, genetically interact to impact cell fitness and vacuole morphology. FEMS Yeast Res 14:472–480. doi:10.1111/1567-1364.12128
Manandhar SP, Ricarte F, Cocca SM, Gharakhanian E (2013) Saccharomyces cerevisiae Env7 is a novel serine/threonine kinase 16-related protein kinase and negatively regulates organelle fusion at the lysosomal vacuole. Mol Cell Biol 33:526–542. doi:10.1128/MCB.01303-12
Manandhar SP, Calle EN, Gharakhanian E (2014) Distinct palmitoylation events at the amino-terminal conserved cysteines of Env7 direct its stability, localization, and vacuolar fusion regulation in S. cerevisiae. J Biol Chem 289:11431–11442. doi:10.1074/jbc.M113.524082
Marchette LD, Thompson DA, Kravtsova M, Ngansop TN, Mandal MNA, Kasus-Jacobi A (2010) Retinol dehydrogenase 12 detoxifies 4-hydroxynonenal in photoreceptor cells. Free Radical Biol Med 48:16–25. doi:10.1016/j.freeradbiomed.2009.08.005
Nakahara K et al (2012) The Sjögren–Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol Cell 46:461–471. doi:10.1016/j.molcel.2012.04.033
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, Oxford
Oldenburg KR, Vo KT, Michaelis S, Paddon C (1997) Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 25:451–452
Ricarte F et al (2011) A genome-wide immunodetection screen in S. cerevisiae uncovers novel genes involved in lysosomal vacuole function and morphology. PLoS One 6:e23696. doi:10.1371/journal.pone.0023696
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi:10.1038/nprot.2010.5
Saarikangas J, Barral Y (2016) Protein aggregation as a mechanism of adaptive cellular responses. Curr Genet 62:711–724. doi:10.1007/s00294-016-0596-0
Sandell LL et al (2007) RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev 21:1113–1124. doi:10.1101/gad.1533407
Schrader M, Godinho LF, Costello JL, Islinger M (2015) The different facets of organelle interplay-an overview of organelle interactions. Front Cell Dev Biol 3:56. doi:10.3389/fcell.2015.00056
Schuldiner M et al (2008) The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134:634–645. doi:10.1016/j.cell.2008.06.025
Schüller H-J (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43:139–160. doi:10.1007/s00294-003-0381-8
Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27
Sprague GF, Cronan JE (1977) Isolation and characterization of Saccharomyces cerevisiae mutants defective in glycerol catabolism. J Bacteriol 129:1335–1342
Szymanski KM et al (2007) The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA 104:20890–20895. doi:10.1073/pnas.0704154104
Takahashi Y, Moiseyev G, Farjo K, Ma JX (2009) Characterization of key residues and membrane association domains in retinol dehydrogenase 10. Biochem J 419:113–122. doi:10.1042/BJ20080812 (111 p following 122)
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tan JSY, Seow CJP, Goh VJ, Silver DL (2014) Recent advances in understanding proteins involved in lipid droplet formation, growth and fusion. J Genet Genom 41:251–259. doi:10.1016/j.jgg.2014.03.003
Thiam AR, Foret L (2016) The physics of lipid droplet nucleation, growth and budding. Biochim Biophys Acta 1861:715–722. doi:10.1016/j.bbalip.2016.04.018
Thompson DA et al (2005) Retinal degeneration associated with RDH12 mutations results from decreased 11-cis retinal synthesis due to disruption of the visual cycle. Hum Mol Genet 14:3865–3875. doi:10.1093/hmg/ddi411
Walther TC, Farese RV (2012) Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81:687–714. doi:10.1146/annurev-biochem-061009-102430
Wang CW (2015) Lipid droplet dynamics in budding yeast. Cell Mol Life Sci 72:2677–2695. doi:10.1007/s00018-015-1903-5
Wang F, Whynot A, Tung M, Denic V (2011) The mechanism of tail-anchored protein insertion into the ER membrane. Mol Cell 43:738–750. doi:10.1016/j.molcel.2011.07.020
Wang CW, Miao YH, Chang YS (2014) Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J Cell Sci 127:1214–1228. doi:10.1242/jcs.137737
Wilfling F et al (2013) Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24:384–399. doi:10.1016/j.devcel.2013.01.013
Wilfling F, Haas JT, Walther TC, Farese RV (2014a) Lipid droplet biogenesis. Curr Opin Cell Biol 29:39–45. doi:10.1016/j.ceb.2014.03.008
Wilfling F et al (2014b) Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife 3:e01607. doi:10.7554/eLife.01607
Winzeler EA et al (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906
Wonisch W et al (1998) Treatment of the budding yeast Saccharomyces cerevisiae with the lipid peroxidation product 4-HNE provokes a temporary cell cycle arrest in G1 phase. Free Radic Biol Med 25:682–687
Yadav KK, Rajasekharan R (2016) The transcription factor GCN4 regulates PHM8 and alters triacylglycerol metabolism in Saccharomyces cerevisiae. Curr Genet 62:841–851. doi:10.1007/s00294-016-0590-6
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinform 9:40. doi:10.1186/1471-2105-9-40
Zolnikova IV et al (2016) Stargardt disease-associated mutation spectrum of a Russian Federation cohort. Eur J Med Genet. doi:10.1016/j.ejmg.2016.12.002
Acknowledgements
We thank Dr. Greg Payne (University of California Los Angeles) for the gift of single-gene deletion library and Dr. Joel Goodman (University of Texas Southwestern Medical Center) for the gift of Erg6-dsRed plasmid. This work was supported by NIH AREA #R15 GM85794-02 to EG and by NSF-MRI grant DBI0722757 for confocal microscopy. VC was supported by National Institute of Minority Health and Health Disparities Grant #5R25MD006851-03 and by NIH-RISE Award #R25GM071638.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Siddiqah, I.M., Manandhar, S.P., Cocca, S.M. et al. Yeast ENV9 encodes a conserved lipid droplet (LD) short-chain dehydrogenase involved in LD morphology. Curr Genet 63, 1053–1072 (2017). https://doi.org/10.1007/s00294-017-0702-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-017-0702-y