Abstract
The phyllosphere is one of the largest habitats for terrestrial microorganisms. To gain a better insight into the factors underlying the composition of bacterial communities inhabiting leaf surfaces we performed culture-dependent and independent (Denaturing Gradient Gel Electrophoresis) analyses on the bacteria associated with the leaves of three plant species: Amygdalus communis, Citrus paradisi, and Nicotiana glauca. We found that the culturable classes Bacilli and Actinobacteria were the predominant classes on the phyllosphere of all three plant species. In contrast to this consistency on the bacterial class level, we found a significant variation on the bacterial species-level based on the culturable methods. Although some variation was detected among individual plants within one plant species, the inter-specific variability exceeded the intra-specific variability. C. paradisi leaf surface had the highest predicted total species richness (Chao 2 and ICE) and the highest species diversity (βw) among the three plant species. Our findings demonstrate that environmental conditions, mainly the plant species within a site, govern the bacterial community composition on leaf surfaces.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The traditional hypothesis regarding bacterial community composition (BCC) is that “everything is everywhere, but the environment selects” [1]. The rationale behind this hypothesis is that microorganisms are extremely abundant, proliferate rapidly, and disperse easily. As recently reviewed by Lindström and Langenheder [2], this approach has been increasingly challenged. Here, we examine the role of environmental factors in determining the BCC of the phyllosphere of three plant species in northern Israel.
The phyllosphere, which is the microbial habitat found on the aerial surface of plants, is known to be colonized by diverse assemblages of microorganisms, including filamentous fungi, yeasts, bacteria, and bacteriophages [3–7]. The phyllosphere in general, and plant leaf surfaces specifically, represent a hostile environment for residing bacteria due to stressful conditions such as temperature shifts and UV light [5, 9, 10]. Despite these environmental constraints, microbes have been found to flourish on leaf surfaces. Although some of these bacteria were shown to be highly important for plants through both their antagonistic and mutualistic effects on plant health and growth [5, 10, 11], little is known about the diversity, biogeography, and composition of the bacterial communities (BCC) in the phyllosphere [7].
Recent studies on the role of environmental factors in shaping phyllosphere BCC revealed mixed results. Junker et al. [8] showed that the BCCs on Saponaria officinalis and Lotus corniculatus leaves were relatively similar and thus not host-dependent. Redord et al. [7] found that inter-specific variability of BCC exceeded intra-specific variability and thus BCC depended on specific tree species within a site. Interestingly, they reported that BCC on the same plant species (Pinus ponderosa needles) did not become more phylogenetically distinct with increasing geographic distance, even across thousands of kilometers. Furthermore, the similarity of phyllosphere bacterial communities that was found among tropical tree species showed a significant tendency to follow host-plant phylogeny, with more similar communities on more closely related hosts [12]. On the other hand, Knief et al. [13] demonstrated that site-specific factors had a stronger impact on the Methylobacterium community composition on plant leaves than did plant species-specific factors (see also [14–16].
The aim of the present study was to expand our knowledge on the importance of local ecological factors by comparing bacterial community composition (BCC) and diversity on the leave surface of three plant species in northern Israel. Specifically, we addressed the following objectives: (a) to quantify and compare the inventory and diversity of bacterial classes and species on the leaf surface of the different plants; and (b) to explore whether BCC is host-specific.
Materials and Methods
Plant Sampling
Three plant species [Nicotiana glauca (Tree Tobacco), Amygdalus communis (Almond), and Citrus paradisi (Grapefruit)] were sampled from March to June 2009, all plants were located within a 400 km2 area in northern Israel. Five individual plants were sampled for each species with different distances between them. The minimal and maximal distances between two individuals of A. communis were 0.5 and 9.5 km, respectively, of N. glauca 1 and 10 km, respectively, and of C. paradisi 2 and 8 km, respectively.
Leaf Phyllosphere Sampling
Three gr. leaves (equals to 2–20 leaves per individual plant) were picked from each individual sampled plant using ethanol washed gloves, and transferred into a sterile 50 ml Falcon tube. Fifteen ml sterile saline water supplemented with 0.1 % Glycerol and 0.15 % Tween 80 were added and the tubes sonicated for 4 min at 22 °C in an ultrasonic cleaning bath (40 kHz; Bransonic 32, MRC, Israel) to dislodge bacteria from the leaves. The resulting suspension was used for culturing and DNA extraction.
Culturing and Enumeration of Bacteria from Leaves
Leaf samples (after sonication) were serially diluted and 0.1 ml aliquots spread on R2A agar (Himedia) and R2A agar supplemented with 20 % sucrose. The plates were incubated under aerobic conditions at 30 °C for 48 h. Individual colonies representing isolates with different morphologies and appearances were picked from the different dilutions agar plates and sub-cultured again at least five times before identification. Bacterial isolates were stored in LB with 30 % glycerol (−80 °C).
Identification of Isolates Using the 16S rRNA Gene
Universal bacterial primers 11F (5′-CAC GGA TCC AGA CTT TGA T(C/T)(A/C) TGG CTC AG-3′) and 1512R (5′-GTG AAG CTT ACG G(C/T)T AGC TTG TTA CGA CTT-3′), based on Escherichia coli positions, were used to amplify internal fragments of 16S rRNA genes [17]. Bacterial isolates were suspended in PCR-grade water (Sigma, USA) until the first visible sign of turbidity. Eight microliter of this suspension were transferred to a sterile, thin-walled PCR tube. One microliter of each primer (20 pmol/μl) and 10 μl of the PCR master mixture (ReddyMix, ABgene, UK) were added to the tube to make up a final reaction volume of 20 μl. Cell breakage and initial DNA denaturation were performed at 94 °C for 4 min, followed by 33 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 50 s, and elongation at 72 °C for 2 min and then a final elongation step at 72 °C for 10 min. To confirm amplicon production (~1,500 bp), the mixture was analyzed by electrophoresis on 1.5 % agarose gel, followed by staining with ethidium bromide and visualization under ultraviolet light. The amplified PCR products were sequenced in the MCLAB laboratories (California, USA).
Sequences Analysis
For the identification of closest relatives, newly determined sequences were compared to those available in the EZtaxon software, version 2.1 (http://www.eztaxon.org; [18]).
DNA Extraction
Phyllosphere suspensions after sonication were centrifuged at 20,000 g for 30 min, and the pellet resuspended in 200 μl saline (0.85 % NaCl). Total DNA was extracted using a DNA isolation kit (DNeasy Blood and Tissue, Qiagene, Germany) according to the manufacturer’s description.
Denaturing Gradient Gel Electrophoresis (DGGE) Analysis
The extracted DNA, in a final concentration of 1 ng/μl, was amplified using ReddyMix (AB gene, UK). A 193-bp fragment, of the 16S rRNA gene corresponding to the V3 region, was amplified using 341F GC-clamp (5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC GCC TAC GGG AGG CAG CAG-3′) and 534R (5′ ATT ACC GCG GCT GCT GG-3′) primers [19]. The thermal cycling conditions were 95 °C for 4 min; 40 cycles at 94 °C for 30 s, at 55 °C for 50 s, at 72 °C for 120 s, and finally at 72 °C for 10 min. PCR was performed on the MyCycler Personal Thermal Cycler (Bio-Rad, Hercules, CA, USA).
For DGGE analysis, the Dcode universal mutation detection system (Bio-Rad, Hercules, CA, USA) was used to separate the V3 region’s PCR products. Forty microliters of each product were electrophoresed on 8 % (wt/vol) polyacrylamide gels containing a denaturating gradient of 40–60 % urea and formamide [a 100 % denaturant corresponds to 7 M urea and 40 % (vol/vol) formamide] in 1× TAE running buffer (2 M Tris base, 1 M glacial acetic acid, 50 mM EDTA). Migration was performed at 90 V for 16 h, and the running buffer temperature was kept constant at 60 °C. Gels were stained with GelStar Nucleic Acid Stain (0.1 μl/ml) (Cat. No. 50535 Cambrex Bio Science, Rockland, USA), and photographed by UV GelDoc (Bio-Rad, Hercules, CA, USA). DGGE bands were excised from the gel and eluted by incubation in 30 μl sterile distilled water at 4 °C overnight. Next, the primers 341F (without the GC-clamp) and 534R were used for amplification. The PCR products (193-bp) were sequenced by MCLAB laboratories (California). Sequence analysis was performed with the EZtaxon software, version 2.1 (http://www.eztaxon.org/; [18]), and the NCBI database (http://blast.ncbi.nlm.nih.gov).
Nucleotide Sequence Accession Numbers
All the sequences from this study were deposited in the GenBank database under accession numbers HQ284832-HQ284868; HQ284907-HQ284947, and HQ284971-HQ284990.
Comparison of BCC on Leaves of Different Plant Species (Culture Dependent)
We calculated the proportional similarity index (PSI, ranges from 0 to 1) to estimate the similarity between the frequency distributions of bacterial classes on the leaf surfaces of the three different plant species. It was calculated by:
where pi and qi represent the proportion of bacteria class i [20]. We used Sørensen’s similarity index (QS) to estimate the similarity of bacterial species composition between the leaf surfaces of the three plant species. It was calculated based on the pooled inventory of bacterial species on all plants of the same species by:
where A and B are the number of species in samples A and B, respectively, and C is the number of species shared by the two samples; QS ranges from 0 to 1.
Rarefaction analysis provides estimates of the expected number of species (richness) with increasing sample size. Observed sample species richness was estimated from the sample-based rarefaction curves using the Mau Tau (Sobs) moment based interpolation method. Sampling is considered to be adequate if the rarefaction curve approaches an asymptote. We used rarefaction analysis also to estimate the expected total bacterial species richness on the leaf surface of each of the three plant species using Chao2 and ICE (Incidence-based Coverage Estimator of species richness [21, 22]) based on 100 randomizations without replacement. These two indices are recommended as the best estimate for incidence-based richness when Chao’s estimated CV for incidence distribution is >0.5 as was found in the case of our three plant species. Observed and estimated (rarefaction) species accumulation curves were calculated and plotted using the software EstimateS 7.52 [23].
We calculated beta diversity (based on presence/absence data) to measure the difference in bacterial species composition on each plant species between each individual plant using Whittaker’s βw:
where S is the total number of species and α is the average species richness of the samples. As beta diversity increases, individual localities differ more markedly from one another and display a smaller proportion of the species occurring in the region [24]. As has long been recognized, βw thus captures a fundamental facet of the spatial pattern of biodiversity [25].
BCC of the leaf surface of the three plant species based on the results of the cultural-dependent method was examined by canonical correspondence analysis (CCA) using the CANOCO program [26]. CCA is a multivariate ordination technique which constructs those linear combinations of environmental variables along which the distributions of the species are maximally separated. The eigenvalues produced by CCA measure this separation. Plant species were used as the environmental factor. The ordination diagram generated by CCA visualizes not only a pattern of community variation but also the main features of the genera distribution along the environmental variables. The length of the arrow in the diagram is a measure for the contribution of the variable to the total explained variance, the angle in between two arrows is a measure for the correlation between the two variables (small angle means high correlation), and the projection of a species point on an arrow is a measure for the relative value of that point; in other words, for the position of that point on the gradient described by the arrow. The significance of the ordinations and of each variable’s contribution to the total variance was investigated with 999 permutations using the Monte Carlo test.
Uncultured (DGGE) based BCC: Cluster analysis was used to compare the relationships among the DGGE profiles of two plant species (A. communis and C. paradisi). DGGE bands were manually scored as present (+) or absent (−) to generate a data set that could be analyzed on the software program. The average-linkage-between-groups UPGMA (unweighted pair-group method using arithmetic averages) was calculated using SPSS version 19. The UPGMA defines the distance between two clusters as the average of the distances between all pairs of cases in which one member of the pair is from each of the clusters. Two samples in which only a very low number of bands could be observed were not analyzed in the cluster analysis.
Comparison of BCC on Leaves Within One Plant Species
We used QS also to estimate the similarity of the bacterial species composition on the leaf surfaces of the five individuals collected from each plant species, based on the results of the culture-dependent methods. QS was calculated for each pair of plants in each plant species.
Results and Discussion
Culturable Inter-specific Variability of Phyllospheric Flora: The Bacteria Class Level
The most dominant classes/phyla of bacteria that were previously reported to colonize leaves of other plant species were Alpha- and Gammaproteobacteria, followed by representatives of Betaproteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria [7, 10, 12, 51]. Using culture-independent methods, Yang et al. [27] reported that Pseudomonadaceae and Microbacteriaceae were the most abundant families on leaves. In the present study, species belonging to all of the above-mentioned classes/phyla were cultured from all different plant species phyllospheres, but interestingly, representatives of the Bacilli class dominated (47–81 %) the culturable leaf phyllosphere bacterial communities of all three plant species (Fig. 1). Members of the Actinobacteria class were the second most abundant (Fig. 1), while members of the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Sphingobacteria classes were cultured from some but not all of the plants’ phyllosphere (Fig. 1). Accordingly, the PSI (similarity index of bacterial class level) between A. communis and N. glauca was 0.56, between A. communis and C. paradisi was 0.64 and between C. paradisi and N. glauca was 0.89, demonstrating the high similarity on the bacterial class level.
Relative abundance of culturable bacterial species from leaf phyllosphere at class phylogenetic level. Bacterial species were identified from three plant species (n = 5 plants per species): Amygdalus communis, Citrus paradisi and Nicotiana glauca. The y axis is the percentage of the identified species that were affiliated to a specific class from the total of the different species that were identified from leaf phyllosphere of a certain plant species
Culturable Inter-specific Variability of Phyllospheric Flora: The Bacteria Species Level
Ninety-four isolates were cultured and identified from A. communis (37), C. paradisi (35), and N. glauca (22) leaf phyllosphere (Table 1). The most abundant culturable species belonged to the Burkholderia (in A. communis) and the Bacillus genera (in C. paradisi and N. glauca), similarly to previously described taxa from leaves of various plant species worldwide [3, 4, 8, 27–30]. Noticeably, 14 % of the isolates were novel species (Table 1). Citrus paradisi isolates had representatives from 12 genera, A. communis from 9 genera, and N. glauca from 4 genera. The number of species found in C. paradisi (32) was much higher than that found in A. communis (14 species) or N. glauca (11 species). A similar trend was found when the number of obtained species was set into relation with the total number of isolates analyzed with 91 % (32/35), 50 % (11/22), and 38 % (14/37) for each plant species, respectively. Observed sample-based rarefaction analyses (Mau Tau, Sobs) demonstrated that although the curves of N. glauca and A. communis were less steep than that of C. paradisi, the sampling of five individual plants within each of the three plant species was not suffice to reach asymptote (Fig. 2) and bacterial species accumulation curves were nearly a straight line. The shape of the rarefaction curves suggests that the rate of new species accumulation is higher for C. paradisi (approximately seven new species for every sampled individual plant that were 22 % of the total analyzed isolates) than for A. communis and N. glauca (~2.5 and 2 new species for every sampled individual plant, respectively that were 7 and 9 % of the total analyzed isolates, respectively).
The ICE and Chao 2 estimators of the total species richness seem stabilized to some extent (Fig. 3). Whereas there was a large difference in the predicted total species richness in the leaf bacterial community of C. paradisi according to ICE and Chao 2 (~387 and 177 species, respectively), in the other two species the difference was much smaller (N. glauca: 21 and 14 species and A. communis: 45 and 37 species, respectively). Based on ICE and Chao 2 estimators, the actual species richness (11 species) detected for N. glauca was 52–79 % of the expected species richness (11/21–11/14, respectively), for A. communis (14 species) was 31–38 % (14/37–14/45, respectively) and for C. paradisi (32 species) was only 8–18 % (32/387–32/177, respectively). Thus, among the three studied plant species, C. paradisi leave surface had the steeper bacteria species accumulation curve, the highest observed and predicted total species richness (Chao 2 and ICE) and the highest species diversity. The specific environmental conditions of C. paradisi that allow for such a rich and diverse BCC are unknown and call for further studies.
In contrast to our findings of the similarity of the three plant species BCC on the bacterial class level, similarity on the bacterial species level was relatively low. The QS between A. communis and N. glauca was 0.16, between A. communis and C. paradisi was 0.17, and between C. paradisi and N. glauca was 0.37. The Beta Diversity index was much higher for C. paradisi (βw = 3.39) than for N. glauca and A. communis (βw = 2.67 and 2.26, respectively).
The results from the canonical correspondence analysis using the CANOCO software also demonstrated a significant difference between leaf phyllosphere bacterial communities from the three different plant species (Fig. 4). The distribution of the bacterial species along the ordinates was not random according to the Monte Carlo test (F = 1.31, P = 0.006) (Fig. 4) and thus can be explained by originating from different plant species. The horizontal and the vertical axes explained 62 and 35 % of the variation, respectively.
Ordination diagram (calculated with CANOCO software) showing the variation and the relationships between bacterial genus isolates from the leaf phyllosphere of the three plant species. The environmental variables (plant species) are displayed as arrows radiating from the center of the diagram. The length of the arrows represents the contribution of each plant species to the variation of the sample. The angle in between two arrows is a measure for the correlation between the two variables (small angle means high correlation), and the projection of a taxa point on an arrow is a measure for the relative value of that point; in other words, for the position of that point on the gradient described by the arrow. The triangles represent different bacterial species. The identity of the species is as follows: group 1: Hymenobacter xinjiangensis, Kocuria turfanensis; group 2: Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus pasteuri; group 3: Staphylococcus warneri, Bacillus simplex, Bacillus endophyticus, Bacillus niacini; group 4: Bacillus megaterium; group 5: Bacillus stratosphericus; group 6: Kocuria kristinae, Rhodococcus erythropolis, Micrococcus yunnanensis, Methylobacterium fujisawaense, Exiguobacterium mexicanum, Burkholderia caledonica, Burkholderia cenocepacia, Burkholderia fungorum, Paenibacillus validus; group 7: Burkholderia oxyphila; group 8: Bacillus methanolicus, Lysinibacillus macroides; group 9: Bacillus drentensis, Bacillus thioparans, Bacillus subtilis subsp. inaquosorum, Bacillus firmus, Bacillus infantis, Bacillus foraminis, Bacillus licheniformis Bacillus nanhaiensis, Brevibacterium casei, Curtobacterium flaccumfaciens, Paenibacillus urinalis, Paenibacillus xylanilyticus, Paenibacillus amylolyticus, Paenibacillus peoriae, Sporosarcina aquimarina, Acinetobacter johnsonii, Aerococcus urinaeequi, Aerococcus urinaeequi, Arthrobacter arilaitensis, Arthrobacter globiformis, Arthrobacter phenanthrenivorans
Thus, although the phyllospheric BCC at the class level was relatively similar in all three studied plants (see above), the species level was fundamentally different in the three plant species and thus clearly demonstrated host-specificity. However, a bulk of studies that have examined the distribution of species of bacteria across natural and cultivated plant species, found that BCC on leaves are not host-specific. For example, the BCCs on Saponaria officinalis and Lotus corniculatus leaves were relatively similar [8]. Low diversity of phyllospheric nitrogen fixing bacterial species was detected among Tillandsia species inhabiting different environments where, for example, Bacillus megatherium was detected in five Tillansia species, each inhabit a unique habitat [31]. Relatively similar BCCs were reported for ice nucleation-active bacteria [32] and pink-pigmented facultative methylotrophs [33]. In general, such studies have found the same species of bacteria present on a broad range of plant species and hence indicate a lack of host-specificity. The conclusion has been drawn that these groups are therefore well adapted to the unique properties of the leaf surfaces [34].
On the other hand, several other studies support our finding of clear host-specificity. For example, Opelt et al. [35] found a high degree of host-specificity for associated bacteria and two species of Sphagnum from six temperate and boreal bogs in Europe independent of the geographical region. Other studies also showed that different plant species select for distinct BCC (e.g., [12, 27, 36, 37]).
Our above findings indicate that leaves of each plant species represent distinct microbial environments. Different plant species could offer diverse micro-environmental conditions that may govern BCC such as the availability of nutrients, amino acids, water, and the occurrence of secondary metabolites [38]. Hence, leaves of different plant species could be expected to harbor distinct bacterial communities which were adapted to the specific unique environmental conditions characterizing the specific-plant species. The three plants in the present study are well-known for their alkaloid content. N. gluaca is rich in nicotine and anabasine [39], C. paradisi is rich in caffeine [40] and A. communis is rich in amygdalin [41]. Alkaloids have been found in leaf epicuticular wax [42] and may stimulate both positive and negative effects on bacteria, depending on their concentration and on specific-environmental conditions (e.g., [37, 43]). Several bacteria may utilize these alkaloids as a carbon and/or nitrogen source while other bacteria are deterred by the same compound. For example, Pseudomonas sp. Nic22 degrades nicotine [44], but nicotine also acts as an antibacterial agent [45]; several Pseudomonas species utilize caffeine [46], but caffeine can also act as antibacterial [46, 47]; and some Actinomyces species are able to utilize amygdalin [48], but this cyanide-containing compound has antibacterial qualities as well [45]. Further study is necessary to reveal the role of leaf alkaloids on the bacterial composition.
Plant Species Intraspecific Variability in Phyllospheric BCC
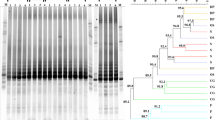
Microorganisms residing on leaves of plants of a single species could be exposed to highly variable environmental factors. Thus, the surface of leaves may represent distinct habitat characteristics for bacterial growth and development not only in the inter-specific level but in the intra-specific level as well. Intra-specific variation of host-plant microclimate conditions include temperature, relative humidity, UV radiation, wind speed, moisture, leaf age, and location to name a few (e.g., [34, 38]). Many of those could affect community composition of the phyllospheric bacteria, thus each plant within a plant species may display different BCC (e.g., [49]). Indeed, the similarity of bacterial species on the leaves of the five individual plants within each plant species was relatively low. The average (±SD) QS among all pairs of N. glauca was 0.10 ± 0.18 (n = 10). The BCC of one individual plant was totally different from the other four plants, thus it had QS = 0. The average (±SD) QS among all pairs of C. paradisi was 0.10 ± 0.08 (n = 10) and of A. communis was 0.27 ± 0.19 (n = 10). However, comparison of the DGGE patterns from leaf phyllosphere bacterial communities revealed two distinct clusters of the two studied plants (C. paradisi and A. communis) (Fig. 5). Buchnera sp. and Candidatus Arsenophonus arthropodicus were identified from A. communis leaf surfaces, while an Enterobacteriaceae symbiont was identified from C. paradisi leaf surfaces. Thus, a high plant-by-plant variation of leaf-associated bacterial communities was found in the three plant populations we studied, yet the inter-specific variation was higher than the intra-specific variation (Fig. 5).
Dendrogram based on cluster analyses (using unweighted pair group method with arithmetic mean) shows the similarities of BCC on the leaves of two plant species and between individual plants within each plant species. The dendrogram is based on the DGGE patterns of bacterial communities in two plant species: Amygdalus communis (AC) and Citrus paradisi (CP). The scale indicates the degree of distance (distance metric) between BCCs. Buchnera sp. (1) and Candidatus Arsenophonus arthropodicus (2) were identified from A. communis leaf surfaces, while an Enterobacteriaceae symbiont (3) was identified from C. paradisi leaf surfaces. Two samples were omitted from the DGGE analysis since, for unknown reasons, no bands were apparent
To conclude, this study demonstrated that plant phyllosphere is colonized by complex and highly divers bacterial communities and that there is a pronounced variability in BCCs on different plant species. Some variability occurred between plants within the same plant population. Other possible variation sources of BCC among plant could be dependent upon time of the year, growth conditions etc. Thus, further studies are needed to explore other factors that contribute to the variability of leaf BCC among plant species and to explore the mechanisms that the underlie phyllosphere bacterial species presence and absence in space and time [50].
References
Baas-Becking LGM (1934) Geobiologie of inleiding tot de milieukunde. WP Van Stockum & Zoon, The Hague
Lindström ES, Langenheder S (2011) Local and regional factors influencing bacterial community assembly. Environ Microbiol Rep 4:1–9
Halpern M, Raats D, Lev-Yadun S (2007) Plant biological warfare: infecting pathogenic bacteria into herbivores by thorns. Environ Microbiol 9:584–592
Halpern M, Raats D, Lev-Yadun S (2007) The potential antiherbivory role of microorganisms on plant thorns. Plant Signal Behav 2:503–504
Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69:1875–1883
Lindow SE, Leveau JH (2002) Phyllosphere microbiology. Curr Opin Biotechnol 13:238–243
Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N (2010) The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12:2885–2893
Junker RR, Loewel C, Gross R et al (2011) Composition of epiphytic bacterial communities differs on petals and leaves. Plant Biol 13:918–924
Whipps JM, Hand P, Pink D, Bending GD (2008) Phyllosphere microbiology with special reference to diversity and plant genotype. J Appl Microbiol 105:1744–1755
Delmotte N, Knief C, Chaffron S et al (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci USA 106:16428–16433
Fürnkranz M, Wanek W, Richter A et al (2008) Nitrogen fixation by phyllosphere bacteria associated with higher plants and their colonizing epiphytes of a tropical lowland rainforest of Costa Rica. ISME J 2:561–570
Kim M, Singh D, Lai-Hoe A et al (2012) Distinctive phyllosphere bacterial communities in tropical trees. Microb Ecol 63:674–681
Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA (2010) Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J 4:719–728
Finkel OM, Burch AY, Lindow SE, Post AF, Belkin S (2011) Phyllosphere microbial communities of a salt-excreting desert tree: geographical location determines population structure. Appl Environ Microbiol 7:7647–7655
Rasche F, Marco-Noales E, Velvis H et al (2006) Structural characteristics and plant-beneficial effects of bacteria colonizing the shoots of field grown conventional and genetically modified T4-lysozyme producing potatoes. Plant Soil 289:123–140
van Overbeek L, van Elsas JD (2008) Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol Ecol 64:283–296
Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans AD (1997) Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 143:2983–2989
Chun J, Lee JH, Jung Y et al (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Rosef O, Kapperud G, Lauwers S, Gondrosen B (1985) Serotyping of Campylobacter-jejuni, Campylobacter coli and Campylobacter-lardis from domestic and wild animals. Appl Environ Microbiol 49:1507–1510
Chao A, Shen TJ (2010) Program SPADE (Species Prediction and Diversity Estimation). Program and User’s Guide. http://chao.stat.nthu.edu.tw
Magurran AE (2004) Measuring biological diversity. Blackwell Publishing, Oxford
Colwell RK (2009) EstimateS: statistical estimation of species richness and shared species from samples, version 7.52. User’s Guide and application published online. http://viceroy.eeb.uconn.edu/estimates
Koleff P, Gaston KJ, Lennon JJ (2003) Measuring beta diversity for presence–absence data. J Anim Ecol 72:367–382
Whittaker RH (1960) Vegetation of the Siskiyou Mountains, Oregon and California. Ecol Monogr 30:279–338
ter Braak CJF (1986) Canonical correspondence analysis: a new eigenvector method for multivariate direct gradient analysis. Ecology 67:1167–1179
Yang CH, Crowley DE, Borneman J, Keen NT (2001) Microbial phyllosphere populations are more complex than previously realized. Proc Natl Acad Sci USA 98:3889–3894
Halpern M, Waissler A, Dror A, Lev-Yadun S (2011) Biological warfare of the spiny plant: introducing pathogenic microorganisms into Herbivore’s tissues. Adv Appl Microbiol 74:97–116
Krimm U, Abanda-Nkpwatt D, Schwab W, Schreiber L (2005) Epiphytic microorganisms on strawberry plants (Fragaria ananassa cv. Elsanta): identification of bacterial isolates and analysis of their interaction with leaf surfaces. FEMS Microbiol Ecol 53:483–492
Thompson IP, Bailey MJ, Fenlon JS et al (1993) Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar-beet (Beta vulgaris). Plant Soil 150:177–191
Brighigna L, Montaini P, Favilli F, Trejo AC (1992) Role of the nitrogen-fixing bacterial microflora in the epiphytism of Tillandsia (Bromeliaceae). Am J Bot 79:723–727
Lindow SE, Arny DC, Upper CD (1978) Distribution of ice nucleation-active bacteria on plants in nature. Appl Environ Microbiol 36:831–838
Corpe WA, Rheem S (1989) Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol Ecol 62:243–250
Hirano SS, Upper CD (2000) Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64:624–653
Opelt K, Berg C, Schonmann S, Eberl L, Berg G (2007) High specificity but contrasting biodiversity of Sphagnum-associated bacterial and plant communities in bog ecosystems independent of the geographical region. ISME J 1:502–516
Lambais MR, Crowley DE, Cury JC, Bull RC, Rodrigues RR (2006) Bacterial diversity in tree canopies of the Atlantic forest. Science 312:1917
Ruppel S, Krumbein A, Schreiner M (2008) Composition of the phyllospheric microbial populations on vegetable plants with different glucosinolate and carotenoid compositions. Microb Ecol 56:364–372
Hallman J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW (1997) Bacterial endophytes in agricultural crops. Can J Microbiol 43:895–914
Saitoh F, Noma M, Kawashima N (1985) The alkaloid contents of sixty Nicotiana species. Phytochemistry 24:477–480
Kretschmar JA, Baumann TW (1999) Caffeine in Citrus flowers. Phytochemistry 52:19–23
London-Shafir I, Shafir S, Eisikowitch D (2003) Amygdalin in almond nectar and pollen-facts and possible roles. Plant Syst Evol 238:87–95
Athayde ML, Coelho GC, Schenkel EP (2000) Caffeine and theobromine in epicuticular wax of Ilex paraguariensis A. St.-Hil. Phytochemistry 55:853–857
Bednarerk P, Osbourn A (2009) Plant-microbe interactions: chemical diversity in plant defense. Science 324:746–748
Chen C, Li X, Yang J, Gong X, Li B, Zhang K (2008) Isolation of nicotine-degrading bacterium Pseudomonas sp. Nic22, and its potential application in tobacco processing. Int Biodeter Biodegr 62:226–231
Zakaria ZA, Fatimah CA, Mat Jais AM et al (2006) The in vitro antibacterial activity of Muntingia calabura extracts. Int J Pharm 2:439–442
Mazzafera P (2002) Degradation of caffeine by microorganisms and potential use of decaffeinated coffee husk and pulp in animal feeding. Sci Agric 59:815–821
Ibrahim SA, Salameh MM, Phetsomphou S, Yang H, Seo CW (2006) Application of caffeine, 1,3,7- trimethylxanthine, to control Escherichia coli O157:H7. Food Chem 99:645–650
Wang X, Liu Z, Ma H, Liu Z (2008) Screening of amygdalin-degrading microorganisms and relevant antagonistic microorganisms from peach rhizosphere soil. Acta Hortic 764:145–150
Mengoni A, Pini F, Huang LN, Shu WS, Bazzicalupo M (2009) Plant-by-plant variations of bacterial communities associated with leaves of the nickel hyperaccumulator Alyssum bertolonii Desv. Microb Ecol 58:660–667
Meyer KM, Leveau JHJ (2012) Microbial of the phyllosphere: a playground for testing ecological concepts. Oecologia 168:621–629
Lv D, Ma A, Bai Z, Zhuang X, Zhuang G (2012) Response of leaf-associated bacterial communities to primary acyl-homoserine lactone in the tobacco phyllosphere. Res Microbiol 163:24–119
Acknowledgments
This study was supported by a grant from the Israel Science Foundation (ISF, Grant Nos. 189/08 and 1094/12).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Izhaki, I., Fridman, S., Gerchman, Y. et al. Variability of Bacterial Community Composition on Leaves Between and Within Plant Species. Curr Microbiol 66, 227–235 (2013). https://doi.org/10.1007/s00284-012-0261-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-012-0261-x