Abstract
The aim of this study was to identify class 1 integrons from extended-spectrum and metallo-β-lactamase-negative, multidrug-resistant Pseudomonas aeruginosa clinical isolates from Hungary and to characterize the isolates by phenotypic and molecular methods. Fourteen selected P. aeruginosa isolates resistant to ceftazidime, gentamicin, and ciprofloxacin were subjected to serotyping, random amplification of polymorphic DNA (RAPD), integron content analysis, and a phenotypic test to detect high-level production of AmpC. Four representative isolates were further analyzed by multilocus sequence typing. Two P. aeruginosa multidrug-resistant clonal lineages were identified with a countrywide distribution. The first lineage is characterized by serotype O4, RAPD genotype A, sequence type ST175, and the presence of a class 1 integron harbouring aadB and aadA13 gene cassettes in its variable region. The second lineage is characterized by serotype O6, RAPD genotype B, sequence type ST395, and a class 1 integron carrying a single aadB cassette. The corresponding isolates were recovered from altogether 11 towns in Hungary. ST175 and ST395 are the presently calculated founders of two distinct P. aeruginosa clonal complexes that appear to have a wide geographical distribution also outside Hungary. The multidrug-resistant phenotype associated with these two clonal lineages might have contributed to an increase in their frequency and to their subsequent diversification. Both P. aeruginosa lineages displayed ≥8-fold synergy with boronic acid/ceftazidime combinations, suggesting an AmpC-mediated resistance to ceftazidime. Our observations underscore the role of class 1 integrons in the spread of aminoglycoside resistance by clonal dissemination among P. aeruginosa clinical isolates in Hungary.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pseudomonas aeruginosa is one of the most important clinical pathogens in the nosocomial setting and a common causative agent of bacteremia, pneumonia, and urinary tract infections. Multidrug-resistance among nosocomial isolates of P. aeruginosa is a matter of major concern [4]. Several different factors can contribute to multidrug-resistant (MDR) phenotype in P. aeruginosa, including the overproduction of the chromosomal AmpC cephalosporinase, upregulated efflux systems, and the acquisition of various resistance determinants [8, 14]. The occurrence of acquired metallo-β-lactamase (MBL) and extended-spectrum β-lactamase (ESBL) determinants has been examined among P. aeruginosa clinical isolates in Hungary in earlier works [11–13, 20]. According to these observations, MBL producers occur at a rate below 1% and ESBL producers occur only sporadically among P. aeruginosa clinical isolates in Hungary. The aim of this study was to detect and characterize class 1 integrons from MBL and ESBL-negative, MDR P. aeruginosa clinical isolates from Hungary and to analyze these isolates by phenotypic and molecular techniques.
Materials and Methods
Bacterial Strains
The MDR P. aeruginosa clinical isolates were provided by Hungarian microbiological laboratories between March 2005 and January 2007. Isolates producing MBLs or ESBLs were excluded from this study. Fourteen isolates were selected for characterization displaying resistance to ceftazidime (minimal inhibitory concentration (MIC) ≥32 μg/mL), ciprofloxacin (MIC ≥4 μg/mL), and gentamicin (MIC ≥256 μg/mL) and a balanced geographical distribution throughout Hungary (Table 1). P. aeruginosa strains PA1975 and PA2040 [21] were used as positive controls in phenotypic detection of AmpC. The ATCC 27853 P. aeruginosa strain was used as the quality control strain.
Antibacterial Susceptibility Tests and Detection of AmpC-Mediated Resistance
Antimicrobial susceptibility tests were performed as recommended by the Clinical and Laboratory Standards Institute (CLSI). MICs were determined by agar dilution and interpreted according to the current CLSI breakpoints [2]. The agar dilution method was performed using phenylboronic acid as the AmpC inhibitor at the concentration of 200 mg/L alone and in combination with ceftazidime to detect high-level AmpC-mediated resistance, as recommended [1, 9]. With this method a ≥8-fold synergy (MIC potentiation ratio) with the boronic acid/ceftazidime combination was reported for AmpC-producing Pseudomonas spp. [9].
Characterisation of Class 1 Integrons and Serotyping
Polymerase chain reaction (PCR) amplification and sequencing of integrons were performed as described earlier [22]. The anti-P. aeruginosa in vitro agglutinating sera (Bio-Rad, Marnes-la-Coquette, France) were used for serotyping. The nucleotide sequences of variable regions of class 1 integrons A and B were deposited in GenBank under accession Nos. EU863269 and EU863270.
Random Amplification of Polymorphic DNA (RAPD)
Random Amplification of Polymorphic DNA (RAPD) typing was performed as described previously [15] and analyzed by Fingerprinting II Informatix™ software (Bio-Rad) using a cutoff value of 80% similarity by the Dice coefficient to identify RAPD genotypes [6, 13]. The statistical significance of the clusters was tested by cophenetic correlation (CC) analysis [17], performed in Fingerprinting II Informatix.
Multilocus Sequence Typing (MLST)
Multilocus Sequence Typing (MLST) was performed according to the protocol published by Curran et al. [3]. Nucleotide sequences were searched against the MLST database (www.pubmlst.org/paeruginosa) for assignment of sequence types (STs). The eBURST software was used for phylogenetic analysis as described [5]. Clonal complexes were defined as a group of isolates with either identical STs or STs that varied at one or two loci (single- or double-locus variants) [3].
Mating-Out Assays
Mating-out assays were carried out on MH agar plates using isolates 05-140 and 05-340 as donors and the RifR Pseudomonas putida strain UWC1 as recipient [7]. Transconjugants were selected on plates containing 16 μg/mL gentamicin and 300 μg/mL rifampicin. The initial donor/recipient ratio was 0.2. Mating plates were incubated at 37°C for 14 h.
Results
Detection and Sequencing of Class 1 Integrons
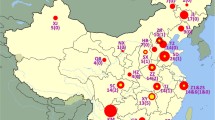
Various features for the analyzed 14 MDR P. aeruginosa isolates are shown in Table 1. PCR experiments identified class 1 integrons in all isolates. In seven isolates, a variable region of about 1.5 kb was detected, whereas in the remaining seven isolates, a variable region of about 0.7 kb was detected (Table 1). The full variable regions of these integrons were sequenced for all isolates.
The 1.5-kb integron harbored two cassettes: an aadB gene encoding an aminoglycoside 2′-O-adenylyltransferase that inactivates gentamicin and tobramycin, followed by an aadA13 cassette encoding and aminoglycoside-3′-adenylyltransferase that confers resistance to streptomycin and spectinomycin (Integron A, Table 1) [18, 19]. The 0.7-kb integron carried a single aadB gene cassette (Integron B). One isolate, 05-310, harbored a second class 1 integron of about 1.65 kb. Partial sequencing demonstrated that a bla OXA-4 gene [16] is located next to its 3’conserved sequence (3’CS).
Serotyping and RAPD Analysis
All isolates harboring integron A could be assigned to serotype O4 and those carrying integron B could be assigned to serotype O6. The RAPD experiment established two clusters within the isolates in good correlation with serotyping (RAPD genotypes A and B, respectively; Table 1). Within the RAPD, genotypes isolates not sharing an identical pattern were assigned to RAPD subtypes displaying ≥80% identity by the Dice coefficient (Table 1). The CC coefficient value of the dendrogram was 97%.
MLST Analysis
Two isolates from each RAPD genotype were selected for MLST typing, representing two different subtypes and towns of origin of RAPD genotypes A and B, respectively. Isolates 05-314 and 06-154, showing the lowest level of identity (that is 80%) by the Dice coefficient to other isolates within RAPD genotypes A and B, respectively, were included in the MLST analysis. Isolates 05-140 and 05-314 displayed sequence type ST175, whereas isolates 05-340 and 06-154 could be assigned to ST395. Both STs were already present in the P. aeruginosa MLST database (http://pubmlst.org/paeruginosa/).
Detection of AmpC-Mediated Resistance
None of the isolates were inhibited in their growth by 200 mg/L boronic acid alone. All isolates showed a ≥8-fold MIC potentiation ratio (MPR) with ceftazidime in the presence of boronic acid (Table 2). In 71% (10/14) of the tested isolates, susceptibility to ceftazidime (MIC ≤ 4 μg/mL) could be restored by boronic acid. The control AmpC overexpressing isolates PA2040 and PA1975 showed MPRs higher than the proposed cutoff value of 8 [9]. Isolate 05-310 produces an integron-borne OXA-4 β-lactamase, which has no ceftazidime hydrolyzing activity [16]. Thus, the presence of OXA-4 did not have an impact on the outcome of the AmpC synergy test performed.
Mating-Out Assays
No transconjugant colonies were obtained with representative donor isolates 05-140 and 05-340 in our mating-out assays under the experimental conditions applied. These results suggest that clonal spread rather than horizontal gene transfer might be the fundamental mechanism behind the countrywide dissemination of integrons A and B in Hungary among MDR P. aeruginosa isolates.
Discussion
Isolates concurrently resistant to ceftazidime, ciprofloxacin, and gentamicin constituted about 4% of all P. aeruginosa clinical isolates in 2006 in Hungary according to the Hungarian Bacteriological Surveillance Database (http://www.oek.hu ). We selected 14 ESBL and MBL-negative isolates displaying this phenotype for analysis. Two large clusters were identified among these isolates based on their serotype (O4 or O6), RAPD genotype (A or B), and integron content (A or B), respectively (Table 1).
Our data demonstrated a strong association between the MDR phenotype and the presence of class 1 integrons. A low level of diversity was found among the integrons identified in the analyzed isolates. aadB was previously shown to be the most prevalent aminoglycoside-resistance mechanism among gentamicin-resistant P. aeruginosa isolates [18], and in our study it was present in all isolates. The aminoglycoside 2′-O-adenylyltransferase encoded by aadB inactivates gentamicin and tobramycin but not netilmicin and amikacin [18]. This was in accordance with the observation that 12 of the 14 tested isolates remained susceptible to amikacin (data not shown). We detected no integron-borne ceftazidime- and ciprofloxacin-resistance determinants in this study. Further experiments revealed that all isolates displayed a ≥8-fold MIC potentiation ratio with boronic acid/ceftazidime combinations (Table 2), suggesting an AmpC-mediated resistance to ceftazidime.
Both MLST-typed representative isolates of the serotype O4 cluster could be assigned to ST175. According to eBURST analysis of the currently available sequence types in the MLST database, ST175 is the founder sequence type of an earlier not described P. aeruginosa clonal complex (Fig. 1a). STs belonging to this clonal complex were also identified in the United Kingdom and Canada (Fig. 1a; http://pubmlst.org/paeruginosa/). Representative isolates of the serotype O6 cluster could be assigned to ST395 by MLST. ST395 is the founder sequence type of another P. aeruginosa clonal complex that contains isolates from the United Kingdom and the United States as well (Fig. 1b; http://pubmlst.org/paeruginosa/) [10]. Thus, these two P. aeruginosa clonal complexes appear to have a wide geographical distribution also outside Hungary. The MDR phenotype associated with isolates belonging to ST175 and ST395 might have contributed to an increase in their frequency and to their subsequent diversification [5, 13].
eBURST diagram of sequence types belonging to two distinct P. aeruginosa clonal complexes with founder sequence types ST175 (a) and ST395 (b) generated by software available at http://eburst.mlst.net/ [5]. Numbers indicate STs, with ST175 and ST395 in the centers as the presently calculated founder STs, respectively. Numbers around the founders, connected to them by black lines, represent single-locus variant STs (SLVs) of the founders. These SLVs share six identical alleles with the founders out of the seven housekeeping genes analyzed [5]
In conclusion, our work identified two MDR P. aeruginosa clonal lineages with a countrywide distribution in Hungary and uncovered an important role of the AmpC and aadB determinants in the emergence of MDR clinical isolates. The corresponding isolates were recovered from altogether 11 towns in Hungary. We also provide supporting evidence for the role of integrons in the spread of aminoglycoside resistance by clonal dissemination, presumably through the transfer of colonized patients between different epidemiological settings. Further studies are warranted to investigate the different factors that are involved in and influence this process.
References
Beesley T, Gascoyne N, Knott-Hunziker V et al (1983) The inhibition of class C beta-lactamases by boronic acids. Biochem J 209:229–233
Clinical and Laboratory Standards Institute (CLSI) (2008) Standards for antimicrobial susceptibility testing. Eighteenth informational supplement M100-BS18. CLSI, Wayne, PA
Curran B, Jonas D, Grundmann H et al (2004) Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42:5644–5649
Driscoll JA, Brody SL, Kollef MH (2007) The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351–368
Feil EJ, Li BC, Aanensen DM et al (2004) eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530
Giske CG, Libisch B, Colinon C et al (2006) Establishing clonal relationships between VIM-1-like metallo-β-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J Clin Microbiol 44:4309–4315
Gotz A, Smalla K (1997) Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl Environ Microbiol 63:1980–1986
Hocquet D, Berthelot P, Roussel-Delvallez M et al (2007) Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob Agents Chemother 51:3531–3536
Hope R, Warner M, Hill R et al. (2008) Phenotypic AmpC detection: which inhibitor is best? Clin Microbiol Infect 14(S7):P876
Johnson JK, Arduino SM, Stine OC (2007) Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J Clin Microbiol 45:3707–3712
Libisch B, Lepsanovic Z, Krucso B et al (2007) Characterization of PER-1 extended-spectrum ß-lactamase producing Pseudomonas aeruginosa clinical isolates from Hungary and Serbia. Clin Microbiol Infect 13(s1):S162
Libisch B, Muzslay M, Gacs M et al (2006) Molecular epidemiology of VIM-4 metallo-β-lactamase-producing Pseudomonas sp. isolates in Hungary. Antimicrob Agents Chemother 50:4220–4223
Libisch B, Watine J, Balogh B et al (2008) Molecular typing indicates an important role for two international clonal complexes in dissemination of VIM-producing Pseudomonas aeruginosa clinical isolates in Hungary. Res Microbiol 159:162–168
Livermore DM (2002) Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis 34:634–640
Mahenthiralingam E, Campbell ME, Foster J et al (1996) Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol 34:1129–1135
Marumo K, Takeda A, Nakamura Y et al (1999) Detection of OXA-4 β-lactamase in Pseudomonas aeruginosa isolates by genetic methods. J Antimicrob Chemother. 43:187–193
McEllistrem MC, Kolano JA, Pass MA et al (2004) Correlating epidemiologic trends with the genotypes causing meningococcal disease, Maryland. Emerg Infect Dis 10:451–456
Poole K (2005) Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:479–487
Revilla C, Garcillán-Barcia MP, Fernández-López R et al (2008) Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob Agents Chemother 52:1472–1480
Szabó D, Szentandrássy J, Juhász Z et al (2008) Imported PER-1 producing Pseudomonas aeruginosa, PER-1 producing Acinetobacter baumanii and VIM-2-producing Pseudomonas aeruginosa strains in Hungary. Ann Clin Microbiol Antimicrob 7:12
Tam VH, Schilling AN, LaRocco MT et al (2007) Prevalence of AmpC over-expression in bloodstream isolates of Pseudomonas aeruginosa. Clin Microbiol Infect 13:413–418
White PA, McIver CJ, Rawlinson WD (2001) Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother 45:2658–2561
Acknowledgments
We would like to acknowledge financial support from the EU through the DRESP2 FP6 grant. This publication made use of the Pseudomonas aeruginosa MLST website (http://pubmlst.org/paeruginosa/) sited at the University of Oxford. The development of this site has been funded by the Wellcome Trust. We also thank Dr Vincent Tam for strains PA1975 and PA2040.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Libisch, B., Balogh, B. & Füzi, M. Identification of Two Multidrug-Resistant Pseudomonas aeruginosa Clonal Lineages with a Countrywide Distribution in Hungary. Curr Microbiol 58, 111–116 (2009). https://doi.org/10.1007/s00284-008-9285-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-008-9285-7