Abstract
A total of eight strains of bacteria were isolated from the root nodule of Vicia faba on the selective media of Rhizobium. Two of these strains produced phenotypically distinct mucoid colonies (one slow growing and the other fast growing) and were examined using a polyphasic approach for taxonomic identification. The two strains (MTCC 7405 and MTCC 7406) turned out to be new strains of biovar 1 Agrobacterium rather than Rhizobium, as they showed growth on alkaline medium as well as on 2% NaCl and neither catabolized lactose as the carbon source nor oxidized Tween-80. The distinctness between the two strains was marked with respect to their growth on dextrose and the production of lysine dihydrolase, ornithine decarboxylase and DNA G + C content. 16S rDNA sequencing and their comparison with the 16S rDNA sequences of previously described agrobacteria as well as rhizobia strains confirmed the novelty of the two strains. Both of the strains clustered with strains of Agrobacterium tumefaciens in the 16S rDNA-based phylogenetic tree. The phenotypic and biochemical properties of the two strains differed from those of the recognized biovar of A. tumefaciens. It is proposed that the strains MTCC 7405 and MTCC 7406 be classified as novel biovar of the species A. tumefaciens (Type strains MTCC 7405 = DQ383275 and MTCC 7406 = DQ383276).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Agrobacterium, a member of family Rhizobiaceae has been included in the α-2 subclass of Proteobacteria mainly on the basis of ribosomal characteristics [16, 27]. Agrobacteria are soil microorganisms, some of which induce crown gall tumors primarily at the crown or on roots [4, 5]. The ability of strains to produce crown gall depends on the presence of a fragment called T-DNA present on the Ti plasmid, which is stably integrated in the nuclear genome during infection in wounded plant cells [26]. The expression of T-DNA leads to synthesis of plant hormones and some unusual compounds called opines, which play a significant role in the epidemiology of crown gall [7, 26]. Although tumor-inducing agrobacteria are common, some non-tumorogenic strains have also been reported from aerial tumors and root nodules [19, 28].
There has been a great deal of dispute over the classification and nomenclature of Agrobacterium and Rhizobium because of a number of characteristics they share in common [8, 29, 30]. Two major lineages were distinguished: One included Agrobacterium rhizogenes along with most of the Rhizobium sp. and the other included A. tumefaciens [29]. A polyphasic approach, which includes both phenotypic as well as phylogenetic parameters such as 16S rDNA sequence data, has, nevertheless, been useful in the delimitation of Agrobacterium from that of the genus Rhizobium [8].
Previous findings suggest that the genus Agrobacterium is polyphyletic [12]. Keane et al. [15], based on some specific phenotypic and biochemical characteristics, suggested that the genus Agrobacterium be subdivided into two biovar, 1 and 2. An additional biovar 3 was subsequently described from the isolates from grapevine [21]. Whereas biovar 1 contains several strains of A. radiobacter and A. tumefaciens, biovar 2 contains many strains of A. rhizogenes and biovar 3 corresponds to A. vitis [11, 21]. Different strains of A. tumefaciens have been further classified in biovar 1, 2, and 3 on the basis of characteristic physiological and biochemical differences [13]. This implies that A. tumefaciens alone shows a great deal of diversity within species and isolation of novel strains of this bacterium cannot be ruled out.
During routine screening for studying bacterial diversity in the fertile cropland located south of the river Ganges in the state of Bihar, India, for testing the potential of selected strains as a biofertilizer/biomineralizer, we isolated two strains of A. tumefaciens from root nodules of Vicia faba when plants had just started flowering. Because these strains differed mainly with respect to morphological (colony configuration and cell size) and biochemical characteristics, they were subjected to a polyphasic taxonomic study including partial 16S rRNA (rrs) gene sequence analysis. A phylogenetic tree based on the results thus obtained conclusively suggests that the two isolates are novel strains/biovar of A. tumefaciens.Footnote 1
Materials and Methods
Isolation and Culturing of Strains
The root nodule of V. faba was used as the starting material. It was surface sterilized using 0.1% mercuric chloride for 5 min, followed by washing with sterile distilled water three times. The nodule was crushed in sterile normal saline [prepared by dissolving 0.87% (w/v) NaCl in distilled water, followed by autoclaving at 121°C for 15 min]. 0.1 mL of serially diluted suspension (10−5) was plated over yeast extract mannitol agar (YEMA) containing (per liter) 0.5 g K2HPO4, 0.2 g MgSO4·7H2O, 0.1 g NaCl, 10 g mannitol, 1 g yeast extract, and 1.5% (w/v) agar, a nutrient medium commonly used for selective isolation of rhizobia [25]. Colonies were randomly selected after 36 h of incubation at 30 ± 2°C based on differences in margin; they were then purified and maintained on YEMA slants at 4°C. Growth was studied spectrophotometrically [optical density at 600 nm (OD600)] using broth culture at an interval of 2 h up to 24 h. All subsequent growth conditions were the same as above.
Preliminary Characterization of Strains
The pattern of growth of both the strains was studied on Hofer’s alkaline medium (pH 11.0) described for differentiating Rhizobium from Agrobacterium [1]. Gram staining was performed as described elsewhere [10]. Cell morphology was studied under an Olympus CX41 research microscope at ×1000, with cells grown for 48 h at 30°C. Growth of strains was studied at a temperature range between 4°C and 65°C. Tolerance of the strains to NaCl was determined on YEMA supplemented with different concentrations of the salt between 2% and 10% (w/v). The pH requirements of the strains were tested in the range from 4.0 to 12.0.
Biochemical Tests
MTCC 7405 and MTCC 7406 were further examined for biochemical characteristics such as hydrolysis of casein, gelatin, starch, urea, and Tween-20. Growth and acid production were studied in basal synthetic media containing (per liter) 2 g (NH4)2SO4, 0.24 g K2HPO4, 0.24 g MgSO4 7H2O, 0.1 g KCl, 0.1 g yeast extract, and 8–10 drops of 0.2% aqueous solution of bromocresol purple (pH 7.2). Tests were conducted using disks of different carbon sources in duplicate (Table 1) as described in the standard protocol for taxonomic characterization of bacteria [16, 17].
Test for Resistance to Antibiotics
The antibiotic resistance profile was studied in the presence of a fixed concentration (15 μg) of six different antibiotics (erythromycin, ofloxacin, norfloxacin, tetracycline, gatifloxacin, and clarithromycin) at 30 ± 2°C on YEMA by the disk diffusion method [3].
PCR Amplification and Sequencing of 16S rDNA
Genomic DNA from strains MTCC 7405 and MTCC 7406 was isolated using a genomic DNA isolation kit (Qiagen) and was quantified spectrophotometrically (Lambda 35, Perkin-Elmer). Polymerase chain reaction (PCR) amplification of 16S rDNA was performed with universal primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′) of the Escherichia coli 16S rDNA numbering system as described by Brosius et al. [6]. The reaction mixture of 25 μL contained 70 ng of chromosomal DNA, 1 U of Deep Vent DNA polymerase, 1X Thermopol reaction buffer, 200 μM of each deoxynucleoside triphosphate (New England Biolabs), and 20 pmol of each primer (BioBasic Inc). PCR cycling parameters included an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min and extension at 75°C for 2 min, and a final extension for 10 min at 75°C. An ~1.5-kb amplicon was separated by gel electrophoresis, eluted by a Qiaquick gel extraction kit (Qiagen), and sequenced using 27F and 1492R primers. The 16S rDNA sequence was determined following the dideoxy chain-termination method using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit as directed in the manufacturer’s protocol. Sequence reactions were electrophoresed and analyzed by an ABI 310 Genetic Analyzer (Applied Biosystems, USA). The complete sequences were submitted to GenBank.
Phylogenetic Analysis
The 16S rDNA sequences of closely related validly published taxa were retrieved from the GenBank database using BLASTN [2] and aligned using the CLUSTAL X program [23]. For the neighbor-joining analysis [22], the distances between the sequences were calculated based on the Juke and Cantor method [14] and the phylogenetic tree was constructed using the TREECON program [24]. Bootstrap analysis of 1000 replications was performed to assess the confidence limits of the branching [9].
Estimation of Genomic G + C Content
The G + C content of genomic DNA was determined spectrophotometrically (Lambda 35, Perkin-Elmer) using the thermal denaturation method [18].
Culture Collection and Gene Bank ID for the Strains
The two strains of A. tumefaciens after identification were deposited in MTCC and Gene Bank, IMTECH, Chandigarh, India with the ID MTCC 7405 and MTCC 7406. The Accession Numbers of the complete 16S rRNA gene sequence available at NCBI, Maryland are DQ383275 for MTCC7405 and DQ383276 for MTCC 7406.
Results and Discussion
Isolation and Growth Characteristics of Strains
Two (MTCC 7405 and 7406) out of eight isolates obtained from surface-sterilized root nodules on selective media appeared visibly distinct with respect to the margin of the colonies and size of the cells (MTCC 7405: margin irregular, size 1.0–2.0 μm; MTCC 7406: margin entire, size 1.5–2.5 μm). On YEMA plates, MTCC 7405 produced slow-growing, small colonies, whereas MTCC 7406 showed very fast growth and large colonies. When growing in liquid medium, the two strains produced a large amount of slime, making the cultures very viscous. The growth curve profile of the two strains also differed significantly.
Interestingly both of the isolates produced mucoid colonies like most of the strains of Rhizobium. However, growth of these isolates on Hofer’s alkaline medium (pH 11.0) and 2% NaCl as well as the inability to catabolize lactose, unlike most of the strains of Rhizobium and biovar 1 Agrobacterium [28, 29], led to a suspicion that the two isolates under investigation could be different strains of Agrobacterium. The identity of each of the suspected Agrobacterium isolates was determined by the appropriate physiological and biochemical tests [12, 19, 20]. Both of the strains were Gram-negative, none spore-forming motile rods producing circular, creamy, raised, opaque, and smooth mucoid colonies. None of them grew under anaerobic conditions. The growth of both the strains occurred well between 15°C and 42°C and from pH 5.0 to 12.0. Analysis of the carbon sources oxidized by the strains revealed that both could oxidize a wide range of carbon compounds similar to those reported previously for other strains of Agrobacterium [4]. However, they were differentiated from known agrobacteria by their inability to oxidize Tween-80 and by being nonlactose fermenters (NLFs) (Table 1). In addition, a marked difference was also observed in the ability to oxidize dextrose by MTCC 7405 (no growth) and MTCC 7406 (normal growth) as the sole source of carbon. These findings clearly indicated that the two strains were distinct from hitherto described biovar of A. tumefaciens.
Test for Antibiotic Resistance
The antibiotic-resistance profile in the presence of a fixed amount (15 μg) of six different antibiotics revealed that strain MTCC 7405 was more resistance to all antibiotics tested compared to MTCC 7406. Although the two strains were significantly resistant to norfloxacin, a marked difference was observed in terms of zones of inhibition in the presence of erythromycin and ofloxacin.
16S rRNA Gene Sequencing and Phylogenetic Tree
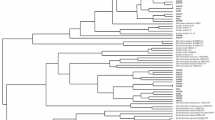
The distinct nature of MTCC 7405 and MTCC 7406 was further confirmed by 16S rDNA sequencing and comparing it to the 16S rRNA gene sequences of previously described biovar and strains of A. tumefaciens and Rhizobium. It is evident from the phylogenetic tree (Fig. 1) that strains MTCC 7405 (1440 nt; DQ383275) and MTCC 7406 (1432 nt; DQ383276) were distinct but closely resembled the A. tumefaciens ATCC 23308T (AB247617) with a 16S rDNA sequence homology of 99.0% and 98.5%, respectively. However, neither of the two strains had resemblances with either the strains of other biovar of Agrobacterium or those of Rhizobium used in comparison.
Phylogenetic neighbor-joining tree based on 16S rDNA sequences showing relationship between A.tumefaciens MTCC 7405 and A.tumefaciens MTCC 7406 and other type strains of related taxa. Sphingomonas paucimobilis DSM 1098T was used as the outgroup. Numbers at nodes indicate levels of bootstrap support ≥ 50% based on a neighbor-joining analysis of 1000 resampled datasets. GenBank accession numbers are given in parentheses. Bar = 10 nucleotides substitution per 100 nucleotides.
Genomic GC Content
The DNA G + C contents of strains MTCC 7405 and MTCC 7406 were estimated to be 62.6 and 63.0 mol% (mean of three replications), respectively—values within the range (57–63 mol%) known in A. tumefaciens [17] but different from their closest phylogenetic neighbor A. tumefaciens ATCC 23308T (61.0 mol%) [21]. Based on the data obtained from phylogenetic analysis and the genomic GC ratio, both of the strains represented novel biovar of A. tumefaciens.
Comparison of Characteristics of Different Biovar of Agrobacterium
Almost all of the strains of A. tumefaciens described so far were isolated from either crown gall or root nodules of dicotyledonous angiosperms or from soil [28–30]. We report the isolation of two new strains of A. tumefaciens from the root nodule of the cultivated leguminous plant of V. faba. Table 2 summarizes the characteristic differences of these strains and their comparison with the existing biovar of A. tumefaciens [13]. Growth on alkaline medium (pH 11.0) and 2% NaCl differentiated these strains from the known strains of Rhizobium. The ability to grow at 35°C (up to 42°C), the failure to oxidize lactose as the sole carbon source, and being catalase negative suggested the novelty of the two strains within the species of A. tumefaciens. The inability to utilize dextrose as the carbon source by MTCC 7405 and the ability of MTCC 7406 to use this monosaccharide as the energy source apart from some biochemical parameters (lysine dihydrolase and ornithine decarboxylase) as well as genomic G + C content differentiated the two strains. Moreover, both of the strains showed resistance to norfloxacin.
The presence of nontumorogenic agrobacteria in crown gall or root nodules has been reported earlier [19, 28]. The isolated strains have, however, been shown to be incapable of nodulating on their original or alternate host [28]. Such strains are thought to be opportunistic bacteria from soil invading the nutrient-rich tumor environment. Our finding that A. tumefaciens might inhabit root nodules of legumes itself points toward an important question pertaining to the role of this soil bacterium. Do some novel biovar play some critical role in nodulation and/or nitrogen fixation in association with Rhizobium? Our assumption is that, if not all, at least some nonpathogenic biovar of A. tumefaciens might be involved in the process of root nodule or tumor formation, the induction of which requires some component, yet unknown, produced by this bacterium as a symbiont.
The type of strains have been identified as two novel biovar of A. tumefaciens: MTCC 7405 (DQ383275) and MTCC 7406 (DQ383276).
Notes
The GenBank accession numbers for the 16S rRNA gene sequences of the two biovar deposited in NCBI, Bethesda, MD, USA is DQ383275 for MTCC7405 and DQ383276 for MTCC 7406, which canbe found online at http://www.ncbi.nlm.nih.gov. The sequence is also available in EMBL in Europe and the DNA Data Bank of Japan.
References
Allen EK, Allen ON (1958) Biological aspects of symbiotic nitrogen fixation. In: Rubland W (ed) Hand buch der Pflanzenphysiologie, Vol. B. Springer-Verlag, Berlin pp 48–118
Altschul SF, Madden TL, Schaffer AA, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Atlas RM, Brown AE, Dobra KW, et al. (1973) Experimental microbiology: fundamentals and applications. Macmillan, New York
Bouzar H, Jones JB, Hodge NC (1993) Differential characterization of Agrobacterium species using carbon source utilization pattern and fatty acid profiles. Phytopathology 83:733–739
Bouzar H, Moore LW (1987) Isolation of different Agrobacterium biovar from a natural oak savanna and tallgrass prairie. Appl Environ Microbiol 53:717–721
Brosius J, Palmer ML, Kennedy PJ, et al. (1978) Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA 75:4801–4805
Dessaux Y, Petit A, Tempé J (1992) Opines in Agrobacterium biology. In Verma DPS (ed) Molecular signals in plant-microbe communication, CRC Press, Boca Raton, FL pp 109–136
Farrand SK, van Berkum P, Oger P (2003) Agrobacterium is a definable genus of the family Rhizobiaceae. Int J Syst Evol Microbiol 53:1681–1687
Felsenstein J (1985) Confidence limits on phylogenetics: an approach using the bootstrap. Evolution 39:783–791
Gerhardt P, Murray RGE, Wood WA, et al. (1994) Methods for general and molecular bacteriology. American Society for Microbiology, Washington DC
Holmes B (1988) The taxonomy of Agrobacterium. Acta Hortic (The Hague) 225:47–52
Holmes B, Roberts P (1981) The classification, identification and nomenclature of agrobacteria. Incorporating revised descriptions for each of Agrobacterium tumefaciens (Smith & Townsend) Conn 1942, Agrobacterium rhizogenes (Ricker et al.) Conn 1942, and Agrobacterium rubi (Hildebrand) Starr & Weiss 1943. J Appl Bacteriol 50:443–467
Holt JG, Krieg NR, Sneath PHA, et al. (1993) Bergey’s manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore
Juke TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism, Vol. 3, Academic Press, New York, pp 21–132
Keane PJ, Kerr A, New PB (1970) Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci 23:585–595
Kersters K, De Ley J, Sneath PHA, et al. (1973) Numerical taxonomic analysis of Agrobacterium. J Gen Micrbiol 78:227–239
Kersters K, De Ley J (1984) Genus III. Agrobacterium Conn 1942. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology, Vol. 1. Williams & Wilkins, Baltimore, pp 244–254
Mandel M, Marmur J (1968) Use of ultraviolet absorbance-temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol 12B:195–206
Moore LW, Bouzar H, Burr T (1988) Agrobacterium. In: Schaad NW (ed) Laboratory Guide for identification of plant pathogenic bacteria, 2nd ed. APS Press, St Paul, MN, pp 16–36
Mougel C, Cournoyer B, Nesme X (2001) Novel tellurite-ammended media and specific chromosomal and Ti plasmid probes for direct analysis of soil populations of Agrobacterium biovars 1 and 2. Appl Env Microbiol 67:65–74
Ophel K, Kerr A (1990) Agrobacterium vitis sp. nov. for strains of Agrobacterium biovars 3 from grapevines. Int J Syst Evol Microbiol 40:236–241
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 44:406–425
Thompson JD, Gibson TJ, Plewniak F, et al. (1997) The clustalx windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Van de Peer Y, De Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10:569–570
Vincent GM (1970) A manual for the practical study of the root nodule bacteria. IBH Handbook No. 15. Blackwell Scientific, Oxford
Winans SC (1992) Two-way chemical signaling in Agrobacterium–plant interactions. Microbiol Rev 56:12–31
Woese CR, Stackebrandt E, Weisburg WG, et al. (1984) The phylogeny of purple bacteria: the alpha subdivision. Syst Appl Microbiol 5:315–326
Wong LL, Wong ET, Liu J, et al. (2006) Endophytic occupation of root nodules and root of Melilotus dentatus by Agrobacterium tumefaciens. Microbial Ecol 52:436–443
Young JM, Bull CT, De Boer SH, et al. (2001) Classification, nomenclature and plant pathogenic bacteria: a clarification. Phytopathology 91:617–620
Young JM, Kuykendall LD, Martinez-Romero E, et al. (2003) Classification and nomenclature of Agrobacterium and Rhizobium: a reply to Farrand et al. (2003). Int J Syst Evol Microbiol 53:1689–1695
Acknowledgments
This work was supported by Research Grant No. BT/PR-4191/PID/06/182/2003 (Microbial Biodiversity Consortium Network of Bihar) from the Department of Biotechnology, Government of India. We thank Ms. Vartika Joshi of the Institute of Microbial Technology, Chandigarh for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tiwary, B.N., Prasad, B., Ghosh, A. et al. Characterization of Two Novel Biovar of Agrobacterium tumefaciens Isolated from Root Nodules of Vicia faba . Curr Microbiol 55, 328–333 (2007). https://doi.org/10.1007/s00284-007-0182-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-007-0182-2