Abstract
The mutant strain PN-120 of Cellulomonas flavigena produces a ß-glucosidase that is 10-fold more active than the corresponding enzyme isolated from the parental strain. These enzymes were partially purified through Q Sepharose and Bio-Gel filtration. A single protein band was detected on polyacrylamide–gel electrophoresis/zymogram using 4-methylumbelliferyl-β-D-glucoside. On sodium dodecyl sulfate–PAGE, the enzyme displayed three protein bands, suggesting that in C. flavigena the enzyme is oligomeric with a molecular mass of 210 kDa. On purification, the specific activity of ß-glucosidase isolated from PN-120 was increased 16-fold and showed three times more affinity for cellobiose than the enzyme of the parental strain; nevertheless, the optimum pH and temperature were similar for both enzymes. The kinetic parameters suggested that the increase in the activity of the enzyme, from the mutant strain, was caused by a mutation that affects the catalytic site of the enzyme. The partial amino-acid sequence of the isolated enzyme confirmed that it is a β-glucosidase because of its homology with other β-glucosidases produced by cellulolytic bacteria and fungi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The use of cellulosic residues available every year as a source of glucose to produce ethanol and other fuel products is creating major interest because they represent an alternative source of energy [1, 2]. The degradation of the cellulose involves the action of an enzymatic complex consisting of at least three types of enzymes where the β-glucosidase (EC3.2.1.21) hydrolyzes the last step, i.e., turning cellobiose into glucose [3]. Most of the β-glucosidases are inhibited by glucose accumulation during the reaction, thus limiting the cellulose degradation rate [4]. Two alternatives to solve this problem can be used: (1) the addition of an external β-glucosidase to commercial cellulase preparations [5, 6] or (2) finding a suitable β-glucosidase produced by micro-organisms through strain selection with improved catalytic properties.
C. flavigena is an excellent source of cellulose degrading enzymes because it is able to grow using different types of agriculture wastes as carbon source [7]. A mutant strain (PN-120) was isolated that had 10-fold more β-glucosidase activity than the wild-type strain when both strains were grown on sugar cane bagasse [8]. To characterize the enzyme and to gain insight into the reason for the high activity in the mutant strain, we performed partial purification and characterization of the β-glucosidase produced from the wild-type strain and its mutant, PN-120, when both strains were grown on sugar cane bagasse as carbon source.
Materials and Methods
Micro-organisms and culture conditions
C. flavigena wild-type (CDBB531) strain and its mutant PN-120 stain [8] were used in all experiments as source of β-glucosidase. Cells were grown in Erlenmeyer flasks containing saline medium supplemented with biotin (10 μg·l−1), thiamine (1 mg·l−1), and 1% w/v alkali-pretreated sugar cane bagasse in an orbital shaker at 100 rpm (New Brunswick Science, Edison, NJ) at 37°C for 48 hours.
Production and purification of β-glucosidase
The strains of C. flavigena, wild-type strain, and mutant PN-120 strain were grown in 2-l Erlenmeyer flasks, and the procedures were carried out in parallel as described later. The residual substrate was removed by vacuum filtration through a GD 120 glass fiber filter disc (MFS, Dublin, CA), and the biomass was recovered by centrifugation and washed three times with 0.02 M Tris-HCl pH 6.2 at 5000 x g for 30 minutes at 4°C. The pellet was resuspended in 40 ml of the same buffer supplied with 10 mM ethylenediaminetetraacetic acid (EDTA); 20 mg lysozyme (Research Organics) were added to the bacterial suspension and subsequently incubated at 37°C at 40 rpm for 30 minutes. The final viscous solution was centrifuged at 5000 x g for 30 minutes at 4°C, and the clear supernatant (called “crude extract”) was used for the next purification steps. An aliquot, 3 ml, of the crude extract was applied onto a Q Sepharose anion exchange column (1.5 × 3 cm) (Amersham Bioscience) pre-equilibrated with 0.02 M Tris-HCl pH 6.2. The β-glucosidase was eluted using a nonlinear gradient of KCl ranging from 0.2 to 1 M in the same elution buffer, and p-NPGase active fractions (see later) were collected and pooled. The sample was desalted and concentrated by ultrafiltration using a PM30 membrane (Amicon). The ultrafiltrate was chromatographed through a Bio Gel P60 column (1.5 × 4 cm) (Bio-Rad) pre-equilibrated with 0.02 M Tris-HCl pH 6.2. The active fractions were pooled and applied to a Bio Gel P100 column (1.5 × 4 cm) (Bio-Rad). p-NPGase fractions were pooled, concentrated, and applied to a native PAGE. The active band protein over 4-methylumbelliferyl-β-D-glucoside (MUG) (see below) was electroeluted (Bio-Rad) and used as purified enzyme for β-glucosidase characterization. All purification steps were monitored by SDS-PAGE and performed according to Laemmli [9]. Protein bands were visualized with Coomassie brilliant blue G250 or by silver staining.
Enzyme assays
β-Glucosidase activity was determined using p-nitrophenyl-β-D-glucopyranoside (p-NPG) (Sigma-Aldrich) as substrate [10]. When cellobiose was used as substrate, the released glucose was measured by the glucose–oxidase method [11]. One IU activity was defined as the amount (μmol) of p-NP or glucose liberated per minute under standard assay conditions. All determinations were made in three independent experiments in triplicate. The maximum difference among the three values was < 5% of the mean.
Zymogram analysis
For localization of β-glucosidase activity in gel, samples were run in PAGE. After washing with distilled water, the gels were overlaid with 3 mM MUG (Sigma-Aldrich) in 0.02 M Tris-HCl pH 6.2 and incubated at 40°C for 30 minutes or until fluorescent bands appeared (360 nm).
Temperature and pH optima
Optimum pH was estimated for the β-glucosidase at pH values between 5 and 8 using citrate phosphate buffer (pH range 5 to 6) and Tris-HCl (pH range 6 to 8). p-NPG and cellobiose were used as substrates for these assays. The effect of temperature on the activity of the β-glucosidase was determined in the range of 20°C to 70°C in 0.02 M Tris-HCl pH 6.2. The substrate solution was previously equilibrated at each temperature.
Enzyme kinetic analysis
The linear region for the β-glucosidase activity was calculated at different enzyme concentrations and different times of incubation. The effect of substrate concentration was estimated using cellobiose and p-NPG (1 to 20 mM and 1 to 10 μM, respectively), and the samples were incubated as described previously. The K m and V max values were calculated from the slopes and intercepts of regression lines of Lineweaver-Burk plots. The assays were performed by triplicate.
Protein sequencing
The only band detected under native PAGE and with β-glucosidase activity was electroeluted and separated by SDS-PAGE. The prominent protein band was isolated and sequenced at the W. M. Keck Biomedical Mass Spectrometry Laboratory, University of Virginia, according to Kinter and Sherman [12]. The peptides were matched in the databases to find homology with other β-glucosidases.
Results
Enzyme purification
The mutant PN-120 was isolated after two rounds of mutagenesis and selected for high specific growth rate using cane sugar bagasse as sole carbon source [8]. The initial characterization of PN-120 demonstrated that it produces more cellulose-degrading enzymes than its parental wild-type strain. The activity of β-glucosidase present in the mutant is 10-fold higher than in the wild-type strain, and we were interested in knowing the molecular basis of such a difference. The β-glucosidase from C. flavigena, wild-type, and PN-120 strains was isolated from cell extracts and purified by column chromatography using Q Sepharose followed by gel filtration using P60 and P100 columns. After the final step, the β-glucosidase from the wild-type and PN-120 strains was purified 28.8- and 15.9-fold for a final recovery of 6.4% and 5.1%, respectively. After all purification steps, the PN-120 β-glucosidase showed approximately 6-fold more specific activity than that of the wild-type strain (Table 1).
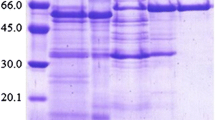
The fractions with ß-glucosidase activity, eluted from the P100 column, were run in a nondenaturing PAGE. We detected only one band with activity toward MUG (Fig. 1A). This band was cut off from the gel, electroeluted, and loaded into an SDS-PAGE. Three bands of 90, 70, and 50 kDa came out for both Wt and PN-120 enzymes, suggesting that the β-glucosidase from C. flavigena is an oligomeric protein with a relative molecular weight of approximately 210 kDa (Fig. 1B).
PAGE of the protein eluted from the P100 column. (A) Protein separated under native conditions from wild-type (Lanes 1 and 2) and PN-120 (Lanes 3 and 4) strains of C. flavigena stained with Coomassie (Lanes 2 and 4) and zymogram of β-glucosidase using MUG as substrate (Lanes 1 and 3). (B) Protein separated under denaturizing conditions from wild-type and PN-120 strains stained with Coomassie. MW = molecular weight standards.
Biochemical properties
The optimum pH for hydrolysis of p-NPG was 6 for both enzymes, showing no significant difference in any buffer used. When cellobiose was used as substrate, the optimum pH changed to values close to 6.5 (Table 2). In the same mode, both enzymes had an optimum temperature for catalysis at 37°C for p-NPG and 40°C for cellobiose (Table 2).
The amount of protein recovered was approximately 40% higher for the wild-type than the PN-120 mutant strain (Table 1), but the specific activity was approximately 80% higher for the PN-120 than for the wild-type strain. We evaluated the biochemical properties of the two enzymes and found linear behavior of the β-glucosidase activity in the range of 1 to 10 mM for p-NPG and 1 to 80 mM for cellobiose using 1 μg protein and a time range between 10 and 40 minutes. The K m values determined were 7.10 and 5.25 mM using p-NPG and 1 μg of the enzymes isolated from the wild-type and PN-120 strains, respectively; when the substrate for the enzyme was cellobiose, the K m values were 114 and 37 mM. From these analyses, the V max values were 90.1 and 250 mmol·min−1·mg−1 of protein for the wild-type and PN-120 strains, respectively, using p-NPG as substrate. When cellobiose was used as substrate, the values were 0.04 and 0.06 mmol·min−1·mg−1 of protein for the wild-type and PN-120 strains, respectively (Table 2).
Protein sequence
The band of 90 kDa, distinguished in SDS-PAGE from the β-glucosidase isolated from the wild-type strain, was excised from the gel and partially sequenced. We recovered 12 peptides, and one of them, called “peptide 10,” showed high homology with other β-glucosidases produced by some cellulolytic bacteria (Fig. 2).
Alignment of peptide 10 obtained after the sequencing the ß-glucosidase isolated from C. flavigena. Accession number of corresponding enzymes are Tm NP227841, Ct CAA42814, Sc NP624778, Sa AAC12650, Sv AAC68679, Sr NP851452, Tf AAZ55642, Cg BAA03152, Bt NP812226, Yp NP668455, Xc NP636465, Pp NP743562, Ec NP754551, and Pa NP250417.
Discussion
C. flavigena produces intracellular β-glucosidases as part of the cellulase enzyme system when grown on sugar cane bagasse [13]. The contribution of β-glucosidase to cellulose hydrolysis is significant because cellobiose is a feedback inhibitor of both endoglucanases and exoglucanases, and it must be removed to allow efficient and complete saccharification of cellulose [3, 4]. The role of β-glucosidase in the saccharification process requires high enzyme activity for large-scale production. The mutant strain, PN-120, as stated previously, was obtained after two rounds of mutagenesis, and we anticipate that it incorporates numerous mutations associated with the production and activity of the enzymes required for the use of cellulose as carbon source. Previous results from our laboratory suggested that some of these enzymes could be partially deregulated because the amount of enzymes produced under nonstimulated conditions is higher in the mutant [13]
Native and mutant β-glucosidases were partially purified from the wild-type and PN-120 strains of C. flavigena. The capacity of the enzyme to use p-NPG and cellobiose as substrates validates its identity as β-glucosidase. The fact that some of the peptides displayed identities against β-glucosidases of other organisms supports the nature of the enzyme isolated. One of the sequenced peptides showed strong homology with bacterial β-glucosidases from family 3 of the glycosylhydrolases [14]. As viewed on nondenaturized PAGE, the β-glucosidases appear as a single band, but on SDS-PAGE, three bands were observed for both normal and mutant enzymes. The molecular weight (210 kDa) of the β-glucosidase, as purified in this study, is higher than the majority of the fungal and bacterial β-glucosidases reported either monomers or dimers [1]. Nevertheless, it is related to the MW of the β-glucosidases tetramers isolated from C. biazotea [15] and Pyrococcus horikoshii [16].
At the end of purification, the specific activity of PN-120 β-glucosidase (14.1 IU·mg−1) was approximately 6-fold higher than the β-glucosidase from the wild-type strain, and it showed higher affinity for p-NPG and cellobiose. Both enzymes were more active in hydrolyzing p-NPG than cellobiose, and according to these results, the β-glucosidase from C. flavigena has broad substrate specificity capable of hydrolyzing aryl-alkyl-β-D-glycosides as well as disaccharides [1, 14]. The kinetic parameters (V max and K m ) suggested that the PN-120 strain has a mutation that influences the catalytic properties of the enzyme. This characteristic supports the fact that the mutant produces a more efficient β-glucosidase than the wild-type strain when grown on sugar cane bagasse [8]. Currently we are working on the cloning and the characterization of the gene that codifies for the 90-kDa protein from the wild-type and PN-120 strains. From the partial amino-acid sequence of the protein, we designed oligonucleotides to amplify a DNA fragment to be used as a probe for library screening.
Literature Cited
Bathia Y, Mishra S, Bisaria VS (2002) Microbial β-glucosidases: Cloning, properties, and applications. Crit Rev Biotechnol 22:375–407
Pack SP, Park K, Yoo YJ (2002) Enhancement of β-glucosidase stability and cellobiose-usage using surface-engineered recombinant Saccharomyces cerevisiae in ethanol production. Biotechnol Lett 24:1919–1925
Lynd LR, Weimer PJ, van Zyl WH, et al. (2002) Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol Mol Biol Rev 3:506–577
Woodward J, Wiseman A (1982) Fungal and other β-glucosidases: Their properties and applications. Enzyme Microbiol Technol 4:73–79
Golias H, Dumsday GJ, Stanley GA, et al. (2000) Characteristics of cellulase preparations affecting the simultaneous saccharification and fermentation of cellulose to ethanol. Biotechnol Lett 22:617–621
Rajoka MI, Durrani IS, Khalid AM (2004) Kinetics of improved production and thermostability of an intracellular β-glucosidase from a mutant-derivative of Cellulomonas biazotea. Biotechnol Lett 26:281–285
De la Torre M, Casas C (1984) Isolation and characterization of a symbiotic cellulolytic mixed bacteria culture. Appl Microbiol Biotechnol 19:430–439
Ponce-Noyola T, de la Torre M (1995) Isolation of a high-specific-growth-rate mutant of Cellulomonas flavigena on sugar cane bagasse. Appl Microbiol Biotechnol 42:709–712
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Kim HK, Pack MY (1989) Cloning and expression of Cellulomonas fimi β-glucosidase genes in Escherichia coli. Enzyme Microbiol Technol 11:313–316
Ghose TK (1987) Measurement of cellulase activities. Pure Appl Chem 59:257–268
Kinter M, Sherman NE (2000) Protein sequencing and identification using tandem mass spectrometry. New York, NY: Wiley Interscience
Ponce-Noyola T, de la Torre M (2001) Regulation of cellulases and xylanases from a depressed mutant of Cellulomonas flavigena growing on sugar-cane bagasse. Bioresource Technol 78:285–291
Henrissat B, Bairoch A (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem J 316:695–696
Siddiqui KS, Rashid MH, Ghauri TM, et al. (1997) Purification and characterization of an intracellular β-glucosidase from Cellulomonas biazotea. World J Microbiol Biotechnol 13:245–247
Matsui I, Sakai Y, Matsui E, et al. (2000) Novel substrate specificity of a membrane-bound β-glycosidase from the hyperthermophilic archeon Pyrococcus horikoshii. FEBS Lett 467:195–200
Acknowledgments
This work was supported by CONACyT-México(45678-Z). A. Barrera-Islas received a scholarship from CONACyT- México. The investigators thank O. Pérez-Avalos and M. Mercado-Morales for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Barrera-Islas, G., Ramos-Valdivia, A., Salgado, L. et al. Characterization of a β-Glucosidase Produced by a High-Specific Growth-Rate Mutant of Cellulomonas flavigena . Curr Microbiol 54, 266–270 (2007). https://doi.org/10.1007/s00284-006-0105-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-006-0105-7