Abstract
Acute myeloid leukemia (AML) is the most common hematopoietic malignancy with abnormal lipid metabolism. However, currently available information on the involvement of the alterations in lipid metabolism in AML development is limited. In this study, we demonstrate that FABP5 expression facilitates AML cell viability, protects AML cells from apoptosis, and maintains triglyceride production. Our bioinformatics analysis revealed that FABP5 expression was upregulated and correlated with unfavorable overall survival of AML patients. FABP5 expression may be used to distinguish normal and AML with high accuracy. FABP5-based risk score was an independent risk factor for AML patients. AML patients with highly expressed FABP5 predicted resistance to drugs. In vitro study showed that FABP5 expression was remarkably elevated in primary AML blasts and an AML cell line. Silencing FABP5 expression attenuated AML cell viability, reduced triglyceride production and lipid droplet accumulation, and induced apoptosis. We utilized AutoDock online tool to identify lycorine as an FABP5 inhibitor by binding FABP5 at amino acid residues Ile54, Thr56, Thr63, and Arg109. Lycorine treatment downregulated the expression levels of FABP5 and its target PPARγ, impaired AML cell viability, triggered apoptosis, and reduced triglyceride production in AML cells. These results demonstrate that FABP5 is critical for AML cell survival and highlight a novel metabolic vulnerability for AML.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute myeloid leukemia (AML) is a heterogeneously hematopoietic malignancy with poor survival [1]. In spite of recent advances in therapy, there is a lack of effective means to prevent or control AML, leading to the fact that the relapse rate is more than 60% in AML patients with high risk [2, 3]. Thus, it is urgent to identify novel and promising targets for AML therapy.
AML cells highly rely on fatty acid oxidation (FAO) as their main source of energy [4]. FAO requires the support of de novo FA synthesis (FAS) in human leukemia [4]. Some genes involved in the metabolism of long-chain polyunsaturated fatty acids (PUFAs), such as linoleic acid and arachidonic acid, are significantly elevated in myeloid leukemia [5]. Additionally, the levels of long-chain PUFAs such as linoleic acid, eicosatrienoic acid, and arachidonic acid were significantly upregulated in plasma in AML patients [6]. These PUFAs are not only the major structural components of cell membranes but also the inter-metabolites for de novo FA synthesis and precursors of eicosanoids that promote cancer development [7]. AML cells with higher levels of fatty acids have been reported to be resistant to venetoclax with azacitidine [8]. Reducing fatty acid synthase (FASN) expression which catalyzes de novo FA synthesis sensitizes AML cells to differentiation therapy [9]. These studies highlight that de novo FA synthesis (FAS) is critical to AML cells. However, ω3 and ω6 polyunsaturated FAs (PUFAs) such as linoleic acid and α-linolenic acid cannot be synthesized de novo in human cells [10]. They are from the diet. Therefore, identifying the factors that mediate the uptake and transportation of PUFAs in AML is critical to understand AML lipid metabolism.
CD36 and FA binding proteins (FABPs) are major players in the uptake and transportation of FAs [11]. FABP5, one member of the FABP family, has been reported to have a high affinity with oleic acid and stearic acid, and silencing FABP5 reduced the uptake of oleic acid and stearic acid [12, 13]. In addition, FABP5 uptakes FAs from the extracellular microenvironment and traffics them to cellular compartments such as the endoplasmic reticulum, mitochondria, nucleus, and peroxisomes for de novo FAS [5]. Moreover, FABP5 was also identified as one of the upregulated lipid-associated essential genes in AML using CRISPR screen [14], and FABP5 mediates lipid metabolism in some solid cancers such as prostate cancer, cervical cancer, and liver cancer [15,16,17]. These studies indicate that FABP5 mediates cancer development in some solid cancers as an oncogene, but its role in AML is still unclear.
In this study, we performed bioinformatics analysis and found that FABP5 was remarkably elevated and positively associated with worse survival of AML patients in the TCGA-LAML dataset. Therefore, in this study, we investigated FABP5’s roles in AML cells. We found that FABP5 expression was significantly upregulated in AML primary blasts and AML cell lines. Inhibiting FABP5 expression by RNAi reduces cellular triglyceride and lipid droplet accumulation, decreases cell viability, and induces AML apoptosis. Importantly, we identified lycorine as an FABP5 inhibitor. Our results suggest that FABP5 may present a potential prognostic biomarker for AML patients and provide a novel target for AML therapy.
Materials and methods
Public AMLdata collection
ggplot2 in Xiantaoxueshu (https://www.xiantao.love/) was performed on cleaned RNA sequencing data from TCGA-LAML and Genotype-Tissue Expression (GTEx) to compare FABP5 expression between AML (n = 173) and normal samples (n = 70). Packages dplyr, survival, and survminer of R language (version 3.6.3) were conducted on TCGA-LAML, Gene Expression Omnibus (GEO) datasets (GSE71014 (n = 104), GSE12417 (n = 79), and GSE37642 (n = 124)) for overall survival (OS) analysis. Samples were divided by FABP5-High (> its median expression) and FABP5-Low (≤ its median expression). OS curves were visualized by ggsurvplot.
Receiver operating characteristic (ROC) curve analysis was performed on TCGA-LAML or TCGA-LAML & GTEx datasets using Xiantaoxueshu (https://www.xiantao.love/).
Univariate and multivariate regression analyses
A total of 140 AML patients with complete clinical data in the TCGA-LAML dataset (https://www.xiantao.love/) were selected for univariate and multivariate regression analyses in order to examine whether FABP5 expression was an independent prognostic risk factor for AML. P < 0.05 is considered as an independent prognostic factor.
Primary AML samples, AML cell lines, and reagents
AML patients’ specimens were conducted in accordance with the Declaration of Helsinki. Normal peripheral blood was obtained from anonymous age-matched healthy donors. Bone marrow (BM) aspirate specimens and peripheral blood of AML were obtained from Guizhou Provincial People’s Hospital (institutional review board [IRB] no. 2016(58)). Non-AML BM samples were collected from patients with suspected leukemia who were subsequently shown to have no leukemia involvement by a series of examinations. All specimens were handled according to legal and ethical standards. Informed consent was obtained from all patients for being included in the study.
Murine C1498 and human MV4-11 AML cell lines were purchased from ATCC. C1498 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 100 μg/mL streptomycin, and 100 μg /mL penicillin. MV4-11 cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% FBS, 100 μg/mL streptomycin, and 100 μg /mL penicillin. Cells were incubated in a humidified atmosphere of 5% CO2 at 37 °C. Lycorine was purchased from MedChemExpress LLC.
Western blotting
ACK lysis buffer (Solarbio Life Sciences, China) was used to lyse red blood cells in peripheral blood and BM aspirates. The resulting cells were washed with pre-chilled 1 × phosphate buffer saline (PBS), then lysed with RIPA buffer supplemented with protease and phosphatase inhibitor cocktails (Roche, Indianapolis, IN). Protein samples were fractionated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking, PVDF membranes were incubated with indicated primary antibodies against FABP5, PPARγ, GAPDH, β-Actin, and Tubulin, respectively. Antibodies were bought from Cell Signaling. After being washed, the membranes were incubated with horseradish peroxidase-labeled anti-rabbit or anti-mouse antibody for 1 h at room temperature. Protein bands were detected by an enhanced chemiluminescence detection system (Millipore), and the protein band signals were visualized using a chemiluminescence imaging system (Genegenome XRQ). Image J was applied to quantify the signal intensity of protein bands.
Development andvalidationof the risk score
First, 1540 differentially expressed genes (DEGs) co-expressed with FABP5 were obtained by Venn diagram to intersect 2000 genes co-expressed with FABP5 (spearman correlation of |R|> 0.431, P < 0.05) with 6240 DEGs (|logFC|≥ 2, adjusted P < 0.05). TCGA-LAML samples were randomly divided into a training set and an internal set at a ratio of 7:3. Univariate Cox survival was run on 1540 DEGs expressed with FABP5 to get 142 survival-related genes. In the training set, the least absolute shrinkage and selection operator (LASSO) analysis was applied to 142 survival-related genes to identify the optimal genes with the best discriminative capability to get prognostic signature genes. The risk score was developed based on those optimal candidate genes and LASSO coefficients as follows:
where n is the total number of the risk score; β \(i\) is the coefficient of the \(i\)−th gene; \(Ei\) indicates the z-score transformed relative expression value of \(i\)-th gene. In the validation dataset, “β \(i\)” used was the same as that in the TCGA-LAML training set. AML patients were classified into high- and low-risk score groups by the median of the risk score. ROC R package was used to plot ROC curves.
Overall, 125 AML patients with mutation information and survival information in TCGA-LAML were downloaded from the web (https://portal.gdc.cancer.gov/) and used to develop a nomogram using the R language nomogram function, rms, and survival packages. Calibration curves were generated using the R language calibration function and rms package.
Drug sensitivity
pRRophetic package was used to predict clinical drug responses using gene expression data and in vitro drug sensitivity in cell lines provided by Cancer Genome Project [18]. Wilcoxon test was applied to calculate P values.
Transfection
Cells were cultured in a medium without antibiotics for 8 h and transfected with 50 nM siRNA against FABP5 or negative control (siNC) using EntransterTM-R4000 Transfection Reagent (Engreen Biosystem) for 8 h, then supplemented with 10% FBS for 40 h. The sequences were as follows: human siFABP5 5'-UCAUUCUCUUAAUUAACUCCU-3', mouse siFABP55'-GAUCCCUUAUGACUGAUAUAU-3', siNC 5'-ACGUGACACGUUCGGAGAATT-3'. Silencing efficacy was verified by Western blotting.
Cell count kit (CCK8) assay
Cell viability was measured by optical density (OD) values at 450 nm using a CCK8 assay kit (Shanghai Yeasen Biotechnology Co., Ltd., Shanghai, China) following the instructions.
Apoptosis
Apoptosis was determined using Annexin V/PI Kit (Shanghai Yeasen Biotechnology Co., Ltd., Shanghai, China). Briefly, 1 × 106 cells were collected, washed with 1 × PBS, suspended in 200 μl binding buffer plus 5 μl Annexin V-FITC, incubated at room temperature for 15 min in the dark, and then added 2 μl propidium iodide (PI) and 200 μl binding buffer. Cellular apoptotic rates were evaluated by flow cytometry (BD Biosciences, USA) and analyzed using Flowjo 10.
BODIPY staining
Lipid droplet accumulation was detected using BODIPY 493/503 staining (Thermo Fisher Scientific, Molecular Probes™) following the protocol [5]. BODIPY 493/503 staining fluorescent dye emits bright green fluorescence. Cells were transfected with siNC (50 nM), siFABP5 (50 nM), pCMV (empty vector, EV) (1 μg), and pCMV-FABP5 (overexpressing FABP5, OE-FABP5) (1 μg) for 48 h, respectively. Cells were collected, washed with PBS to remove media/serum, stained with 2 μM BODIPY 493/503 staining solution in PBS in the dark for 15 min at 37 °C, washed with PBS to remove the staining solution, and resuspended in PBS supplemented with 2% FBS to obtain a single cell suspension for flow cytometry.
Triglyceride measurement
Triglyceride production was measured using the Adipogenesis Detection Assay kit (colorimetric/Fluorometric, ab102513). Briefly, cells were collected after cells were transfected with siFABP5 or siNC or treated with 2 μM lycorine for 48 h, washed with PBS, added with 100 μl lipid extraction solution, sealed, incubated at 95 °C for 30 min, cooled at room temperature, and shaken to mix solution for 1 min to release triglycerides. The reaction was set up by following the instruction provided by the manufacturer. OD values were read at 570 nm. Triglyceride production was calculated following the instructions.
Molecular docking
Lycorine structure (pubChem CID:72,378) was downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and saved it as sdf format, then converted its sdf format to mol2 format using OpenBable software. The crystal structure of the FABP5 protein (PDB: 5HZ5) was downloaded from the PDB database. Lycorine and FABP5 structures were processed by AutoDock (version 1.5.6) [19]. PyMOL was used to visualize lycorine-FABP5 binding.
Statistical analyses
R language (v3.6.3) was used for the statistical analysis of public datasets. The data are represented as means ± SD. P values of the difference in gene expression between normal tissues and AML were calculated using Wilcoxon signed rank test. P values of Kaplan–Meier survival analysis were calculated using the log-rank test. The differences between the two groups were compared using Student’s t test. The differences among more than two groups were compared using one-way ANOVA plus Bonferroni post-test. P < 0.05 was considered statistically significant. P values are indicated in the figures with asterisks: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns: not significant. Data were analyzed using GraphPad Prism software (GraphPad Prism, version 5.01, GraphPad Software.Inc., San Diego, CA, USA).
Results
FABP5 expression was upregulated and predicted poor overall survival of AML patients
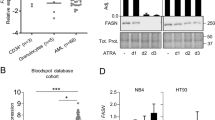
Since FABP5 has been shown to be one of the upregulated lipid-associated essential genes in AML using CRISPR screens [14], we investigated the expression pattern of FABP family members in TCGA-LAML samples. FABPs 2/3/5/7 had alterations > 2.5% (Supplementary Fig. 1A). The heatmap revealed that FABP4 and FABP5 were upregulated in TCGA-LAML (Fig. 1A). FABPs 1/6/7 were downregulated, while FABPs 2/5 were upregulated in TCGA-LAML patient samples relative to normal samples (Fig. 1B). FABP2 expression was not associated with OS (Supplementary Fig. 1B), while FABP5 was associated with unfavorable OS (Fig. 1C). We verified that FABP5 expression was upregulated in primary AML blasts and AML cell line (Fig. 1D, Supplementary Table 1). To assess the power of FABP5 expression to predict the OS of AML patients, ROC curves were constructed. AUCs were 0.526 at 1 year, 0.636 at 3 years, and 0.658 at 5 years, respectively (Fig. 1E). AUC in distinguishing normal and AML was 0.896 (Fig. 1F). These results indicate that high FABP5 expression may predict AML patient OS and diagnose AML.
FABP5 expression is upregulated and correlates with poor survival in AML patients. (A) The expression pattern of FABP family members between normal and AML samples in the TCGA-LAML dataset. (B) mRNA expression of FABP family members between normal and AML samples in TCGA-LAML dataset. P value was calculated using the Wilcoxon test. (C) Association of FABP5 expression with OS of AML patients. (D) FABP5 expression in MV4-11 cells, white blood cells from healthy donors, primary AML blasts, and BM. Non-AML BM samples represent the samples from the patients with suspected leukemia who were subsequently shown to have no leukemia involvement. Western blot was used to detect FABP5 expression. β-actin or GAPDH served as loading controls. (E) AUCs of FABP5 expression in 1-year, 3-year, and 5-year survival of AML patients (n = 173). (F) ROC curve of FABP5 as a marker to distinguish normal (n = 70) and AML (n = 173)
Correlation analysis was applied to assess the association between FABP5 protein expression and clinicopathological factors in AML. When BM blasts ≤ 20%, the percentage of AML patient number between low FABP5 vs. high FABP5 was 24.5% vs. 15.2% (P = 0.026; Supplementary Table 2), indicating that the less BM AML burden, the more AML patients with low FABP5 expression. In contrast, when BM blasts > 20%, the percentage of AML patient numbers between low FABP5 vs. high FABP5 was 25.2% vs. 35.1% (P = 0.026; Supplementary Table 2), indicating that the more BM AML burden, the more AML patients with high FABP5 expression. Between low-FABP5 vs. high-FABP5, BM blasts were 33.72% vs. 44%, and PB blasts were 61.76% vs. 69.83%, indicating that AML patients with high FABP5 had higher BM blasts and PB blasts. These results indicate that AML burden increase in BM is corresponding to high FABP5 expression. Moreover, multivariate regression analysis revealed that age, gender, FLT3 mutation, IDH1 R172 mutation, NPM1 mutation, and FABP5 were unfavorable prognostic factors for AML (Supplementary Table 3). Taken together, FABP5 expression predicts the AML blast amount and worse OS for AML patients.
Establishment ofFABP5-related risk score in AML
To look for candidate genes for establishing FABP5-related risk score in AML, a Venn diagram was run on FABP5 co-expressed genes (Supplementary Table 4) and DEGs (Supplementary Table 5) to identify DEGs co-expressed with FABP5 (Supplementary Table 6), which were analyzed by univariate Cox survival to generate 142 survival-related DEGs co-expressed with FABP5(Supplementary Table 7), further analyzed using LASSO to obtain ASCC1, AIFM1, MPI, ETFB, ACOT7, CAMK1D, NACA, and ATXN7 for constructing a risk score (Supplementary Fig. 2A). A highly dense state of death was distributed in high-risk score area, indicating that risk score was reliable in predicting survival status of AML patients (Fig. 2A, left panel). The high-risk group correlated with an unfavorable OS of AML patients, and risk score AUCs for 1 year, 3 years, and 5 years were 0.838, 0.811, and 0.818, respectively (Fig. 2A, right panel). Similar patterns were validated in an internal set (Fig. 2B) and an external set (Fig. 2C). These results demonstrated that the risk score in predicting prognosis was reliable.
Risk score, the distributions of survival status, and OS. Left panel: risk score distribution, survival status, and heatmaps of 8 genes (ASCC1, AIFM1, MPI, ETFB, ACOT7, CAMK1D, NACA, and ATXN7). Upper right panel: upper- Kaplan–Meier OS curves of patients in high- and low-risk groups. Lower right panel: time-dependent ROC curves verified the prognostic performance of risk score. (A) TCGA-LAML-training set; (B) TCGA-LAML-internal set; (C) GSE37642
The association of the risk score with clinicopathological characters
To examine whether risk score was an independent clinical factor in AML, univariate and multivariate COX regression analyses were conducted. Both age and risk score were independent factors influencing the prognosis of AML patients. The hazard ratio (HR) of the risk score was 8.03 with 95% CI (3.97 ~ 16.24, P = 6.53E-09), indicating that the risk score is a superior predicator for the prognosis of AML patients (Supplementary Table 8). To explore the clinical significance of risk score, we developed a nomogram showing a high-risk score on the side of low OS (Fig. 3A), indicating that a high-risk score predicts low OS. The calibration curves showed that the nomogram-predicted OS was highly consistent with the actual 1-year OS (Fig. 3B), highlighting the risk reliability.
Prediction of drug resistance based on FABP5 expression
Inhibiting FABP5 synergized taxane-mediated the cytotoxic and tumor-suppressive effects in prostate cancer cells [20], suggesting that FABP5 is involved in drug sensitivity. Therefore, we examined whether FABP5 expression was implicated in drug sensitivity in AML. AML patients with high FABP5 expression had higher IC50s of some targeted drugs (dasatinib, erlotinib, imatinib, lapatinib) and chemo-drugs (AZD0530, AZD6482, AKT inhibitor VIII, etc.) than AML patients with low FABP5 expression (Fig. 4, Supplementary Fig. 3), indicating that high FABP5 expression was correlated with in drug resistance in AML.
InhibitingFABP5 expressionreducedlipidaccumulation, impaired AML cell viability, and induced apoptosis
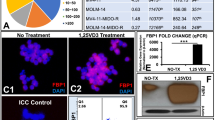
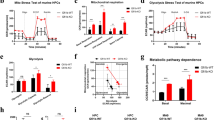
To investigate the role of FABP5 expression in AML cells, FABP5 expression was silenced (Fig. 5A). Since FABP5 has been shown to promote de novo FA synthesis and the uptake and transport of FAs [21, 22], and cellular lipids store as triglycerides (TAGs) in lipid droplets, therefore, we examined the effect of FABP5 expression on TAG production in AML cells. Inhibiting FABP5 expression remarkably reduced TAG production in AML cells (Fig. 5B). Next, we examined the effect of FABP5 expression on lipid droplet accumulation. As illustrated in Fig. 5C, inhibiting FABP5 expression remarkably reduced lipid droplet accumulation, while overexpressing FABP5 increased lipid droplet accumulation. Since TAG is one of the major energy sources to support cell viability, cell survival, and anti-apoptosis, we examined the effect of FABP5 expression on AML cell viability and apoptosis. As expected, silencing FABP5 expression reduced cell viability (Fig. 5D) and induced apoptosis (Fig. 5E). These results indicated that FABP5 expression facilitates lipid accumulation in AML cells and promotes AML cell viability and anti-apoptosis.
The effect of FABP5 expression on triglyceride production, lipid droplet accumulation, AML cell viability, and apoptosis. (A) Inhibiting FABP5 expression by RNAi. Cells were transfected with siRNA against FABP5 (siFABP5) or negative control (siNC) for 48 h. Western blot was applied to detect FABP5 expression. GAPDH served as an internal loading control. (B) The effect of silencing FABP5 expression on triglyceride production. At 48 h after transfection, triglyceride production was measured using the Adipogenesis Detection Assay kit. P value was calculated using Student’s t test. **P < 0.01, ***P < 0.001. (C) The effect of FABP5 expression on cellular lipid accumulation. Cells were transfected with siNC, siFABP5, empty vector(EV), and OE-FABP5, respectively. At 48 h after transfection, cells were stained for BODIPY 493/503 fluorescent dye for detecting lipid accumulation. P values were calculated using one-way ANOVA. *P < 0.05. (D) The effect of FABP5 expression on AML cell viability. Cells were transfected with siFABP5 or siNC. Cell viability was determined by CCK8 assay. P values were calculated by one-way ANOVA. (E) The effect of FABP5 expression on apoptosis. At 48 h after transfection, cells were collected and stained with Annexin V/PI. Apoptosis was determined by flow cytometry. P values were calculated using Student’s t test. (F) The effect of silencing FABP5 on PPARγ expression. Western blot was applied to detect PPARγ expression. Tubulin served as a loading control. (G) PPARγ mRNA expression levels between normal and AML samples in TCGA-LAML dataset. P value was calculated using the Wilcoxon test. (H) ROC curve of PPARγ in TCGA-LAML
Since FABP5 facilitates the uptake of FAs from the outside of cells, thus increasing FAs in cells, cellular FAs can stimulate peroxisome proliferator-activated receptor-γ (PPARγ) [23], which mediates lipid metabolism [24] and FA transport [25], we inferred that FABP5 might affect PPARγ expression. Silencing FABP5 expression significantly downregulated PPARγ expression (Fig. 5F), indicating that FABP5 may mediate lipid metabolism through PPARγ in AML cells. PPARγ expression was upregulated in AML (Fig. 5G), and its AUC was 0.92 in distinguishing normal and AML (Fig. 5H), indicating that PPARγ is clinically relevant to AML. Taken together, these results indicate that lipid-related energy is critical for AML cell viability and anti-apoptosis and targeting FABP5 disrupts lipid metabolism is promising for AML therapy.
Lycorine was identified as an FABP5 inhibitor
Although inhibiting FABP5 expression by a genetic approach such as RNAi is feasible in bench experiments, systemic administration of RNAi against genes of interest for cancer therapy faces many challenges such as serum degradation. Currently, there are two commercially available, but very expensive FABP5 inhibitors: FABP5-IN-1 and SBFI26. FABP5-IN-1 was shown to have an antinociceptive effect [26]. SBFI26 suppressed malignant characteristics of PC3-M cells through inhibiting FABP5/PPARγ signaling [14]. Therefore, we searched for potential lipid-lowering compounds to target FABP5. We found that lycorine m1ay bind to FABP5 using AutoDock [27]. The chemical structure of lycorine was shown in Fig. 6A. Lycorine bound to FABP5 at amino acid residues Ile54, Thr56, Thr63, and Arg109 in β-barrel consisting of ten anti-parallel β-strands capped by two α-helices (Fig. 6B). The binding affinity between lycorine-FABP5 vs. SBFI26-FABP5 was − 9.13 vs. − 8.66 kcal/mol (Supplementary Table 9), indicating that lycorine has a higher affinity with FABP5 than SBFI26.
Lycorine-targeted FABP5. (A) Lycorine chemical structure. (B) Lycorine-FABP5 binding. Ala-Alaine, Thr-Threonine, Arg-arginine. (C, D) The effect of lycorine on the expression levels of FABP5 and PPARγ. Cells were treated with 2 μM lycorine for the indicated time. Western blot was applied to detect protein expression. P values were calculated by one-way ANOVA. (E) The effect of lycorine on cell viability. Cell viability was determined by CCK8 assay. P values were calculated by two-way ANOVA. (F) The effect of lycorine on triglyceride production. Cells were treated with 2 μM lycorine for 48 h. Triglyceride production was measured using the Adipogenesis Detection Assay kit. P value was calculated using Student’s t test. (G) The effect of lycorine on apoptosis. Cells were collected and stained with Annexin V/PI. Apoptosis was determined by flow cytometry. P values were calculated using two-way ANOVA
To verify whether lycorine may inhibit FABP5, we examined the effect of lycorine on FBAP5 expression. Lycorine treatment downregulated FABP5 and its target PPARγ expression in a time-dependent manner (Fig. 6C,D). Like siFABP5, lycorine decreased cell viability (Fig. 6E), reduced triglyceride production (Fig. 6F), and induced apoptosis in dose- and time-dependent manners (Fig. 6G). These results indicated that lycorine exerted its action via targeting FABP5/PPARγ pathway.
Discussion
AML cells with elevated PUFAs acquire energy in an FAO-dependent manner. FAO is supported by de novo FAS, which relies on FA uptake and transport. However, what factors in AML cells mediate the uptake and transportation of FAs remains unclear. Here, our study demonstrates that FABP5 expression plays important roles in AML cell triglyceride production and lipid accumulation, viability, and anti-apoptosis. Inhibiting FABP5 by RNAi inhibited AML cell viability, induced apoptosis, and reduced triglyceride production. Importantly, we identified lycorine as an FABP5 inhibitor.
FABP5 expression clinically correlates with AML prognosis and predicts drug resistance in AML
Our results revealed that FABP5 expression was remarkably elevated in TCGA-LAML samples, an AML cell line, and primary AML blasts in peripheral blood and bone marrow. Elevated FABP5 mRNA expression was correlated with poor overall survival of TCGA-LAML patients, supporting that highly expressed FABP5 expression is associated with worse overall survival in several cancers such as prostate cancer [28], cervical cancer [16], and clear cell renal cell carcinoma [5]. Additionally, FABP5 expression may distinguish normal and AML samples with high AUC values. Moreover, FABP5 alone and FABP5-based risk score independently predicted the OS of AML patients with relatively high accuracy. Our results indicate that FABP5 expression was clinically correlated with AML.
Our study showed that FABP5 expression was implicated in drug resistance in AML. The samples with highly expressed FABP5 had higher IC50 values in multi-drugs than the samples with lowly expressed FABP5 in TCGA-LAML dataset, implying that FABP5 expression in AML cells is involved in drug resistance, consistent with the study that inhibiting FABP5 augmented taxane-mediated the cytotoxic and tumor-suppressive effects in prostate cancer cells [15], and pancreatic ductal adenocarcinoma cells with highly expressed FABP5 were significantly resistant to all-trans retinoic acid (ATRA) relative to PCDA cells without FABP5 expression [29], curcumin-sensitized retinoic acid-resistant MDA-MB-468 cells to ATRA by downregulating FABP5 expression [30]. Therefore, it is plausible that FABP5 expression in AML might contribute to drug resistance.
FABP5 expression promotes AML TAG production, lipid droplet accumulation, cell viability, and anti-apoptosis
Obesity is a risk factor for myeloid leukemia [31]. Understanding the molecular mechanism leading to elevated FAs and subsequent obesity in AML patients is crucial for guiding nutritional support for AML patients. Upregulated FABP5 may uptake more PUFAs into AML cells for de novo FAS, supporting elevated FAs in AML cells. We showed that silencing FABP5 expression decreased TAG production and lipid droplet accumulation in AML cells, indicating that FABP5 expression promotes lipid accumulation in AML cells. Given that FABP5 promotes diet-induced obesity [32] and the uptake of oleic acid and stearic acid for de novo FAS [12, 13], a diet with less oleic acid and stearic acid should be beneficial for AML patients to prevent obesity.
Our results were consistent with the study showing that stable silencing FABP5 expression decreased lipid droplets in gastric cancer cells [33]. In contrast, transient silencing FABP5 expression increased TAG production in ARPE-19 cells (a human retinal pigment epithelial cell line) [34] and increased lipid droplets in prostate cancer cells [5]. These studies indicate that FABP5 mediates lipid metabolism in a context-dependent manner. In addition, silencing FABP5 expression led to lipid deprivation, thus inhibiting AML cell viability and inducing apoptosis. Our results support that inhibiting FABP5 blunted the viability of liver cancer cells [17] and induced apoptosis in clear cell renal cell carcinoma cells [35], T regulatory cells [36], and gastric cancer cells [37]. Collectively, our results indicate that FABP5 is a potential target for AML therapy.
Lycorine is an FABP5 inhibitor
Our results demonstrated lycorine as an FABP5 inhibitor. Lycorine is bound to the FABP5 β-barrel, the binding pocket for FABP5 inhibitors [38]. The binding affinity of lycorine to FABP5 was very close to that of 5M7 (an FABP5 inhibitor) [38], − 9.13 versus − 9.46. Lycorine bound to FABP5 residues at Ile54, Thr56, Thr63, and Arg109. Lycorine has two OH groups carrying negative charges, which may generate strong electrostatic interactions with a positive charge of Arg109, thus maintaining lycorine-FABP5 structural stability. We identified SBFI-26 binding at Ala78 and Arg109 in FABP5, and SBFI-26 was reported to bind at Arg109, Arg129, and Tyr 131 in FABP5 [39]. The discrepancy of binding sites between SBFI-26 and FABP5 might stem from different methods, in which Arg109, Arg129, and Tyr131 were identified based on the SBFI-26-FABP5 complex formed with the addition of SBFI-26 before FABP5 protein was crystallized [39]. In spite of the differences, Arg109 is a commonly shared binding site for FABP5 inhibitors, thus supporting that lycorine is an FABP5 inhibitor. As expected, lycorine exerts its function via inhibiting FABP5/PPARγ signaling in AML cells. In agreement with our study, lycorine induced human leukemia cell apoptosis [15] and exerted a lowering lipid effect [15]. Taken together, our studies indicate that lycorine exerts a lipid-lowering effect in AML cells via targeting the FABP5 pathway.
Limitations and conclusion
There are several limitations to this study. First, FA metabolism mediates myeloid differentiation in leukemogenesis. We did not examine whether FABP5 expression would mediate AML cell differentiation. Second, we did not determine the association of serum FABP5 levels with AML patient prognosis due to limited available AML samples. Third, the experiments we performed were in vitro study. In spite of these limitations, we demonstrate that FABP5 expression is clinically relevant to AML and a promising target for AML therapy. In addition, our study provides insight to guide nutritional support for AML patients. Importantly, we identify lycorine as an FABP5 inhibitor. FABP5 is oncogenic in several cancer types, and lycorine is cheap and affordable. Therefore, targeting FABP5 signaling might be a promising therapeutic target for AML and feasible.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Marando L, Huntly BJP (2020) Molecular landscape of acute myeloid leukemia: prognostic and therapeutic implications. Curr Oncol Rep 22(6):61. https://doi.org/10.1007/s11912-020-00918-7
Molica M, Breccia M, Foa R, Jabbour E, Kadia TM (2019) Maintenance therapy in AML: the past, the present and the future. Am J Hematol 94(11):1254–1265. https://doi.org/10.1002/ajh.25620
Pepe S, Scalzulli E, Colafigli G, Di Prima A, Diverio D, Mancini M, Latagliata R, Martelli M, Foà R, Breccia M (2020) Predictive factors for response and survival in elderly acute myeloid leukemia patients treated with hypomethylating agents: a real-life experience. Ann Hematol 99(10):2405–2416. https://doi.org/10.1007/s00277-020-04217-w
Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, Andreeff M (2010) Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Investig 120(1):142–156. https://doi.org/10.1172/jci38942
Senga S, Kobayashi N, Kawaguchi K, Ando A (1863) Fujii H (2018) Fatty acid-binding protein 5 (FABP5) promotes lipolysis of lipid droplets, de novo fatty acid (FA) synthesis and activation of nuclear factor-kappa B (NF-κB) signaling in cancer cells. Biochim Biophys Acta 9:1057–1067. https://doi.org/10.1016/j.bbalip.2018.06.010
Pabst T, Kortz L, Fiedler GM, Ceglarek U, Idle JR, Beyoğlu D (2017) The plasma lipidome in acute myeloid leukemia at diagnosis in relation to clinical disease features. BBA clinical 7:105–114. https://doi.org/10.1016/j.bbacli.2017.03.002
Wang D, Dubois RN (2010) Eicosanoids and cancer. Nature reviews. Cancer 10(3):181–193. https://doi.org/10.1038/nrc2809
Stevens BM, Jones CL, Pollyea DA, Culp-Hill R, D’Alessandro A, Winters A, Krug A, Abbott D, Goosman M, Pei S, Ye H, Gillen AE, Becker MW, Savona MR, Smith C, Jordan CT (2020) Fatty acid metabolism underlies venetoclax resistance in acute myeloid leukemia stem cells. Nature cancer 1(12):1176–1187. https://doi.org/10.1038/s43018-020-00126-z
Humbert M, Seiler K, Mosimann S, Rentsch V, Sharma K, Pandey AV, McKenna SL, Tschan MP (2021) Reducing FASN expression sensitizes acute myeloid leukemia cells to differentiation therapy. Cell Death Differ 28(8):2465–2481. https://doi.org/10.1038/s41418-021-00768-1
Chen YQ, Edwards IJ, Kridel SJ, Thornburg T, Berquin IM (2007) Dietary fat-gene interactions in cancer. Cancer Metastasis Rev 26(3–4):535–551. https://doi.org/10.1007/s10555-007-9075-x
Hajri T, Abumrad NA (2002) Fatty acid transport across membranes: relevance to nutrition and metabolic pathology. Annu Rev Nutr 22:383–415. https://doi.org/10.1146/annurev.nutr.22.020402.130846
Lee GS, Pan Y, Scanlon MJ, Porter CJH, Nicolazzo JA (2018) Fatty acid-binding protein 5 mediates the uptake of fatty acids, but not drugs, into human brain endothelial cells. J Pharm Sci 107(4):1185–1193. https://doi.org/10.1016/j.xphs.2017.11.024
Armstrong EH, Goswami D, Griffin PR, Noy N, Ortlund EA (2014) Structural basis for ligand regulation of the fatty acid-binding protein 5, peroxisome proliferator-activated receptor β/δ (FABP5-PPARβ/δ) signaling pathway. J Biol Chem 289(21):14941–14954. https://doi.org/10.1074/jbc.M113.514646
Adamson J, Morgan EA, Beesley C, Mei Y, Foster CS, Fujii H, Rudland PS, Smith PH, Ke Y (2003) High-level expression of cutaneous fatty acid-binding protein in prostatic carcinomas and its effect on tumorigenicity. Oncogene 22(18):2739–2749. https://doi.org/10.1038/sj.onc.1206341
Liu XS, Jiang J, Jiao XY, Wu YE, Lin JH, Cai YM (2009) Lycorine induces apoptosis and down-regulation of Mcl-1 in human leukemia cells. Cancer Lett 274(1):16–24. https://doi.org/10.1016/j.canlet.2008.08.029
Zhang C, Liao Y, Liu P, Du Q, Liang Y, Ooi S, Qin S, He S, Yao S, Wang W (2020) FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. Theranostics 10(15):6561–6580. https://doi.org/10.7150/thno.44868
Seo J, Jeong DW, Park JW, Lee KW, Fukuda J, Chun YS (2020) Fatty-acid-induced FABP5/HIF-1 reprograms lipid metabolism and enhances the proliferation of liver cancer cells. Commun Biol 3(1):638. https://doi.org/10.1038/s42003-020-01367-5
Geeleher P, Cox NJ, Huang RS (2014) Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol 15(3):R47. https://doi.org/10.1186/gb-2014-15-3-r47
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Carbonetti G, Converso C, Clement T, Wang C, Trotman LC, Ojima I, Kaczocha M (2020) Docetaxel/cabazitaxel and fatty acid binding protein 5 inhibitors produce synergistic inhibition of prostate cancer growth. Prostate 80(1):88–98. https://doi.org/10.1002/pros.23921
Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O’Malley JT, Gehad A, Teague JE, Divito SJ, Fuhlbrigge R, Puigserver P, Krueger JG, Hotamisligil GS, Clark RA, Kupper TS (2017) Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543(7644):252–256. https://doi.org/10.1038/nature21379
Pan Y, Scanlon MJ, Owada Y, Yamamoto Y, Porter CJ, Nicolazzo JA (2015) Fatty acid-binding protein 5 facilitates the blood-brain barrier transport of docosahexaenoic acid. Mol Pharm 12(12):4375–4385. https://doi.org/10.1021/acs.molpharmaceut.5b00580
Bao Z, Malki MI, Forootan SS, Adamson J, Forootan FS, Chen D, Foster CS, Rudland PS, Ke Y (2013) A novel cutaneous fatty acid-binding protein-related signaling pathway leading to malignant progression in prostate cancer cells. Genes Cancer 4(7–8):297–314. https://doi.org/10.1177/1947601913499155
Evans RM, Barish GD, Wang YX (2004) PPARs and the complex journey to obesity. Nat Med 10(4):355–361. https://doi.org/10.1038/nm1025
Khan D, Ara T, Ravi V, Rajagopal R, Tandon H, Parvathy J, Gonzalez EA, Asirvatham-Jeyaraj N, Krishna S, Mishra S, Raghu S, Bhati AS, Tamta AK, Dasgupta S, Kolthur-Seetharam U, Etchegaray JP, Mostoslavsky R, Rao PSM, Srinivasan N, Sundaresan NR (2021) SIRT6 transcriptionally regulates fatty acid transport by suppressing PPARγ. Cell reports 35(9):109190. https://doi.org/10.1016/j.celrep.2021.109190
Yan S, Elmes MW, Tong S, Hu K, Awwa M, Teng GYH, Jing Y, Freitag M, Gan Q, Clement T, Wei L, Sweeney JM, Joseph OM, Che J, Carbonetti GS, Wang L, Bogdan DM, Falcone J, Smietalo N, Zhou Y, Ralph B, Hsu HC, Li H, Rizzo RC, Deutsch DG, Kaczocha M, Ojima I (2018) SAR studies on truxillic acid mono esters as a new class of antinociceptive agents targeting fatty acid binding proteins. Eur J Med Chem 154:233–252. https://doi.org/10.1016/j.ejmech.2018.04.050
Seeliger D, de Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24(5):417–422. https://doi.org/10.1007/s10822-010-9352-6
Carbonetti G, Wilpshaar T, Kroonen J, Studholme K, Converso C, d’Oelsnitz S, Kaczocha M (2019) FABP5 coordinates lipid signaling that promotes prostate cancer metastasis. Sci Rep 9(1):18944. https://doi.org/10.1038/s41598-019-55418-x
Gupta S, Pramanik D, Mukherjee R, Campbell NR, Elumalai S, de Wilde RF, Hong SM, Goggins MG, De Jesus-Acosta A, Laheru D, Maitra A (2012) Molecular determinants of retinoic acid sensitivity in pancreatic cancer. Clin Cancer Res 18(1):280–289. https://doi.org/10.1158/1078-0432.Ccr-11-2165
Thulasiraman P, McAndrews DJ, Mohiudddin IQ (2014) Curcumin restores sensitivity to retinoic acid in triple negative breast cancer cells. BMC Cancer 14:724. https://doi.org/10.1186/1471-2407-14-724
Mazzarella L, Botteri E, Matthews A, Gatti E, Di Salvatore D, Bagnardi V, Breccia M, Montesinos P, Bernal T, Gil C, Ley TJ, Sanz M, Bhaskaran K, Coco FL, Pelicci PG (2020) Obesity is a risk factor for acute promyelocytic leukemia: evidence from population and cross-sectional studies and correlation with FLT3 mutations and polyunsaturated fatty acid metabolism. Haematologica 105(6):1559–1566. https://doi.org/10.3324/haematol.2019.223925
Shibue K, Yamane S, Harada N, Hamasaki A, Suzuki K, Joo E, Iwasaki K, Nasteska D, Harada T, Hayashi Y, Adachi Y, Owada Y, Takayanagi R, Inagaki N (2015) Fatty acid-binding protein 5 regulates diet-induced obesity via GIP secretion from enteroendocrine K cells in response to fat ingestion. Am J Physiol Endocrinol Metab 308(7):E583-591. https://doi.org/10.1152/ajpendo.00543.2014
Dong SR, Ju XL, Yang WZ (2019) STAT5A reprograms fatty acid metabolism and promotes tumorigenesis of gastric cancer cells. Eur Rev Med Pharmacol Sci 23(19):8360–8370. https://doi.org/10.26355/eurrev_201910_19147
Wu T, Tian J, Cutler RG, Telljohann RS, Bernlohr DA, Mattson MP, Handa JT (2010) Knockdown of FABP5 mRNA decreases cellular cholesterol levels and results in decreased apoB100 secretion and triglyceride accumulation in ARPE-19 cells. Lab Investig 90(6):906–914
Lv Q, Wang G, Zhang Y, Han X, Li H, Le W, Zhang M, Ma C, Wang P, Ding Q (2019) FABP5 regulates the proliferation of clear cell renal cell carcinoma cells via the PI3K/AKT signaling pathway. Int J Oncol 54(4):1221–1232. https://doi.org/10.3892/ijo.2019.4721
Field CS, Baixauli F, Kyle RL, Puleston DJ, Cameron AM, Sanin DE, Hippen KL, Loschi M, Thangavelu G, Corrado M, Edwards-Hicks J, Grzes KM, Pearce EJ, Blazar BR, Pearce EL (2020) Mitochondrial integrity regulated by lipid metabolism is a cell-intrinsic checkpoint for treg suppressive function. Cell Metab 31(2):422-437.e425. https://doi.org/10.1016/j.cmet.2019.11.021
Wang W, Liu Z, Chen X, Lu Y, Wang B, Li F, Lu S, Zhou X (2021) Downregulation of FABP5 suppresses the proliferation and induces the apoptosis of gastric cancer cells through the hippo signaling pathway. DNA Cell Biol 40(8):1076–1086. https://doi.org/10.1089/dna.2021.0370
Yan F, Liu X, Zhang S, Su J, Zhang Q, Chen J (2018) Molecular dynamics exploration of selectivity of dual inhibitors 5M7, 65X, and 65Z toward fatty acid binding proteins 4 and 5. International journal of molecular sciences 19 (9) https://doi.org/10.3390/ijms19092496
Hsu HC, Tong S, Zhou Y, Elmes MW, Yan S, Kaczocha M, Deutsch DG, Rizzo RC, Ojima I, Li H (2017) The antinociceptive agent SBFI-26 binds to anandamide transporters FABP5 and FABP7 at two different sites. Biochemistry 56(27):3454–3462. https://doi.org/10.1021/acs.biochem.7b00194
Funding
This work was supported by the National Natural Science Foundation of China (31960137, 32260171) and the Guizhou Provincial Natural Science Foundation (Qiankehejichu [2016]1416).
Author information
Authors and Affiliations
Contributions
Q.F.Z. and H.B. conceived and designed the study. B.X.G diagnosed AML patients and provided samples. X.M.L., W.L.F., Y.H.P, J.J.D., T.Z, D.G.F., and W.H. obtained and assembled data. X.L.Q., C.X.W., Y.H., and W.F.Y. analyzed and interpreted the data. Y.H. and C.J.W. did the statistical analysis. The paper was written by Q.F.Z. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Consent to participate
Informed consent was obtained from all patients for being included in the study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liang, X., Fu, W., Peng, Y. et al. Lycorine induces apoptosis of acute myeloid leukemia cells and inhibits triglyceride production via binding and targeting FABP5. Ann Hematol 102, 1073–1086 (2023). https://doi.org/10.1007/s00277-023-05169-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-023-05169-7