Abstract
The genetic predisposition to familial hematological malignancies has been previously reported highlighting inherited gene mutations. Several genes have been reported but genetic basis remains not well defined. In this study, we extended our investigation to a potential candidate GATA2 gene which was analyzed by direct sequencing in 119 cases including familial aggregations with a variety of hematological malignancies and sporadic acute leukemia belonging to Tunisian and French populations. We reported a deleterious p.Arg396Gln GATA2 mutation in one patient diagnosed with both sporadic acute myeloid leukemia (AML) and breast cancer. We also reported several GATA2 variations in familial cases. The absence of deleterious mutations in this large cohort of familial aggregations of hematological malignancies may strengthen the hypothesis that GATA2 mutations are an important predisposing factor, although as a secondary genetic event, required for the development of overt malignant disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Several studies have reported that genetic predisposition to familial hematological malignancies (FHM) may lead to an increased risk of this pathology. Owing to the molecular progress in the last decades, many genes have been identified in association with familial aggregations of hematological malignancies. Due to the heterogeneity of the identified genes, the predisposition to FHM remains difficult to identify. In fact, the incomplete penetrance of the involved genes leads to overlapping and variable phenotypes depending on the studied familial aggregation. The lack of large families with multiple cases of hematological malignancies and unavailability of complete familial history enhance the complexity of the inherited factor identification. The important advances in molecular tests have led to the discovery of several genes recurrently mutated in familial hematological malignancies such as GATA2 [1,2,3,4]. GATA2 gene, located on chromosome 3q21.3, encodes a zing-finger transcription factor which is crucial for hematopoiesis, especially in regulating the development and proliferation of early pluripotent hematopoietic precursors [5].
Previous studies have reported several mutations in GATA2 gene including dominant-negative mutations and loss-of-function mutations that lead to haploinsufficiency. These mutations such as p.Thr354Met have been found related to several hematological malignancies specifically in familial myelodysplastic syndrome (MDS)/acute myeloid leukemia (AML) aggregations [6]. Moreover, epidemiological analysis showed that individuals carrying germline GATA2 mutation have an increased risk (~70%) of developing MDS/AML or chronic myeloid leukemia (CML) syndromes [1, 6,7,8].
Besides germline mutations in GATA2 gene, somatic mutations have been also described in CEBPA gene in several hematological malignancies including basically myeloid lineage such as AML and CML syndromes [9, 10].
Since numerous mutations have been found in GATA2 gene in association with mostly the familial aggregations of MDS/AML, this gene is highly suspected to be involved in the predisposition to familial hematological malignancies. In this study, we evaluate the mutational status of GATA2 gene in familial aggregations of hematological malignancies and sporadic acute leukemia belonging to Tunisian and French populations.
Materials and methods
Patients
A total of 119 patients diagnosed with hematological malignancies, including 98 familial cases and 21 sporadic cases, were recruited via a French national cooperative network focusing on FMH and through the GenHe-mINSERM/DGRS French-Tunisian project.
The hematological malignancies cohort of patients includes chronic or acute, lymphoid or myeloid leukemia, Hodgkin’s or non Hogdkin’s lymphomas, and myeloproliferative or myelodysplastic syndromes.
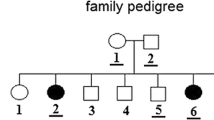
The study cohort consists of 80 patients belonging to 71 familial forms of hematological malignancies (at least two cases of hematological malignancies with or without solid tumors in first, second, or third degree relatives); 17 patients from 17 families with aggregation of tumors including one case of hematological malignancy in first, second, or third degree relatives and 1 patient who had a multiple primitive tumor with hematological malignancy but without familial history. Thirteen sporadic acute lymphoblastic leukemia (ALL) and 8 sporadic acute myeloblastic leukemia (AML) cases from Tunisian population were included in this study. Informed consent was obtained from the patients, relevant family members (healthy relatives) or their legal guardian as required by the Helsinki Declaration.
DNA extraction
Genomic DNA extraction was performed on peripheral blood cells obtained during complete remission as defined by standard protocols of treatment. The EZ1 DNA tissue kit (Qiagen, Hilden, Germany) was used for DNA extraction according to the manufacturer’s instructions. DNA was extracted from paraffin-embedded sections when no peripheral blood was available.
GATA2 sequencing
The entire coding regions of GATA2 gene were amplified and sequenced. The primers used were exon 2F: CGGGACTGGTGCTCTTTCT; exon 2R: TTTTCAGCAGCTCGATTCCT; exon 3-1F: GGGTTCCCTGTAGGGTCTGT; exon 3-1R: GTACTTGACGCCGTCCTTGT; exon 3-2F: ACTCTGGCTCCCACCTTTTC; exon 3-2R: GAGTCACTTCCCTCGCTTCA; exon 4F: GACTCCCTCCCGAGAACTTG; exon 4R: GCGTCTGCATTTGAAGGAGT; exon 5F: TTAGCCCTCCTTGACTGAGC; exon 5R: AGCCAAGCTGGATATTGTGG; exon 6F: CTATGAAGGTCGGGCACAAT; and exon 6R: GGTGGGGAACATTCACAGTA. Purified PCR products were sequenced using the BigDye Terminator cycle sequencing ready reaction Kit v1.1 (Applied Biosystems—Foster City, USA) and loaded onto an ABI Prism 3500 sequencer (Applied Biosystems). The SeqScape software program v2.5 (Applied Biosystems) was used to search for new variants in sequencing products.
Mutational analysis
Allelic frequencies of the GATA2 variants found were compared to those in general population as presented in the Exome aggregation consortium (ExAC), Cambridge, MA (URL: http://exac.broadinstitute.org).
PolyPhen-2 program was used to predict the impact of the obtained variants on the structure and function of GATA2 protein [11].
Results
The entire GATA2 gene was sequenced in 119 hematological malignancies cases including 21 sporadic cases and belonging either to Tunisian or French populations. All identified GATA2 gene variants are reported in (Table 1, Fig. 1). Their frequencies in general population as showed in ExAC database, and their frequencies in the studied cohort are also presented. To predict the impact of each variant on GATA2 protein, in silico analysis was performed by PolyPhen-2 and the scores are shown in (Table 1).
Mutations distribution in GATA2 gene. a Chromosomal location in 3q21.3. b GATA2 gene structure. Exons are illustrated to scale. c GATA2 gene transcript (NM_032638.4). Coding exons are shown in black. d GATA2 protein structure containing 480 amino acids and the two Zing-finger domains (ZF1, ZF2). The mutations identified in our study are localized
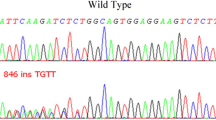
Most of the identified variants in exons 2, 3, 6, and introns 4, 5 are known polymorphisms. However, one p.Arg396Gln variant in exon 6 was found at heterozygous level in one female Tunisian patient diagnosed with sporadic AML at the age of 38 and previously diagnosed with breast cancer. This variant is a known pathogenic mutation located in the second conserved Zinc-finger domain of the GATA2 protein. We also found a new c.1017+49G>A variant in intron 4 with unknown significance, which is carried by another 44-year-old Tunisian male patient diagnosed with sporadic AML.
Discussion
GATA2 gene encodes a zinc-finger transcription factor involved in the regulation of hematopoiesis. Mutations in this gene have been related to various hematological malignancies, particularly in MDS/AML familial aggregations [1, 6,7,8]. Several mutations, involving GATA2 gene, were reported such as p.Thr354Met and p.Thr355del which may hamper the transactivation of target genes and impact the cellular differentiation and apoptosis. Most of GATA2 mutations were associated to myeloid lineage such as AML and MDS which is characterized by a clonal disorder of hematopoietic stem cells and may progress to AML forms [12]. High levels of GATA2 expression were observed in AML and were associated with poor prognosis [13]. Somatic mutations in GATA2 have been reported basically among patients with chronic myeloid leukemia (CML).
Due to the increased incidence of GATA2 mutations in several contexts of sporadic and familial hematological malignancies, we targeted this gene through a mutational analysis. Therefore, we aimed in this study to search for germline mutations by screening GATA2 gene in 119 patients belonging to 98 independents Tunisian and French families diagnosed with hematological malignancies and 21 patients with sporadic acute leukemia.
Among all the identified variants (Table 1), only the p.Arg396Gln variant in exon 6 was found in one patient diagnosed with sporadic AML at the age of 38 and previously diagnosed with breast cancer. This variant has been previously reported in the literature as a disease causing mutation. In fact, this mutation is very common in familial hematological malignancies and particularly in the MonoMac syndrome [14]. The p.Arg396Gln variant is located in the second Zinc-finger domain of the GATA2 protein. This domain is highly conserved through evolution (Fig. 1). Identified mutations have been associated with leukemia and breast cancer [1]. This is observed in a sporadic AML case analyzed through this study. The p.Arg396Gln mutation leads to decreased expression of the protein and loss of protein function which results in haploinsufficiency. In fact, this mutation hampers the DNA binding ability of the Zinc-finger domain of the protein, which impairs the activation of the GATA2 promoter. The p.Arg396Gln mutation can cause disease in the heterozygous state as reported in the literature [14] and found in the patient described through this study.
We have identified another variant c.1017+49G>A in AML case without familial history. This variant with unknown significance is located in the intron 4 of GATA2 gene. As indicated in the ExAC database, c.1017+49G>A variant is a very rare in the general population (ExAC frequency = 0.000008257). The sequence of the GATA2 fourth intron is very conserved through evolution and it harbors the GATA-box-E-box composite element which is a regulatory element of transcription. Mutations in this intron result in haploinsufficiency of the protein and have been related to MonoMac Syndrome [2, 7, 15]. Thus, the c.1017+49G>A variant could have a deleterious impact on the GATA2 protein and should be more investigated.
The others p.Pro5Pro (rs1573858) and the p.Ala164Thr (rs2335052) variants found in this study have been identified while associating them with coronary artery disease [16]. The p.Ala164Thr (rs2335052) variant was also associated to reduce disease-free survival in colorectal cancer patients [17]. However, none of these variants were described in hematological malignancies cases. Concerning the p.Ala411Ala (rs34172218) and the p.Pro472Pro (rs376805544) variants identified in this study, both were detected only in familial aggregations of hematological malignancies and were totally absent in sporadic ALL and AML cases in our investigation. These two variants were not assigned to any pathology in literature.
Previous studies have identified other genes while associating them with hematological malignancies. In fact, Gilliland, D. G has suggested a multi-hit model in which many genes should be mutated to result in hematological malignancies [18]. Concomitant mutations to GATA2 were described in literature involving majorly the CEBPA gene in sporadic AML, both genes act as transcription factors that function in the same differentiation pathway in AML and their alteration lead to leukemogenesis [19,20,21,22]. We have already screened several genes in our cohort in order to detect concomitant mutations in TP53, IDH1, IDH2, JAK2, CBL, RUNX1, NPM1, ASXL1, ARL11, and CEBPA [23,24,25,26,27] and AML1, FAS, FASLG, PRF1, CASP8, CASP10, CRAF, BRAF, CBL, and YY1 (data not shown). A new CEBPA mutation p.Gly242Ser was reported in one familial case diagnosed with AML but no concomitant mutation with GATA2 gene was observed [28].
In this study, we targeted for the first time, the entire GATA2 gene in a large series of 119 patients belonging to different ethnic populations; Tunisian and French, consisting of sporadic and familial hematological malignancies. Despite the wide number of analyzed cases, we did not identify a deleterious variant in GATA2 gene. The only p.Arg396Gln mutation found in one sporadic AML case highlights the involvement of GATA2 gene in myeloid lineage malignancies majorly AML as was described in the literature. We did not report any germline GATA2 mutations even in familial aggregations of MDS/AML. These findings underline the fact that GATA2 mutations might be an important predisposing factor, but as a secondary genetic event, required for the development of overt malignant disease. Thus, other investigations should be carried out in order to identify the genetic factors underlying the hematological malignancies in the hole of the cohort. As hematological malignancies are very heterogeneous disorders, a large sequencing panels of the various genes involved in these pathologies are recommended.
References
Hahn CN, Chong C-E, Carmichael CL et al (2011) Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet 43(10):1012–1017
Hsu AP, Sampaio EP, Khan J et al (2011) Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 118(10):2653–2655
Ostergaard P, Simpson MA, Connell FC et al (2011) Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet 43(10):929–931
Dickinson RE, Griffin H, Bigley V et al (2011) Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 118(10):2656–2658
Rodrigues NP, Janzen V, Forkert R et al (2005) Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood 106(2):477–484
Kazenwadel J, Secker GA, Liu YJ et al (2012) Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood 119(5):1283–1291
Hsu AP, Johnson KD, Falcone EL et al (2013) GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood 121(19):3830–3837
Micol J-B, Abdel-Wahab O (2014) Collaborating constitutive and somatic genetic events in myeloid malignancies: ASXL1 mutations in patients with germline GATA2 mutations. Haematologica 99(2):201–203
Marceau-Renaut A, Guihard S, Castaigne S, Dombret H, Preudhomme C, Cheok M (2015) Classification of CEBPA mutated acute myeloid leukemia by GATA2 mutations. Am J Hematol 90(5):93–94
Huang Y, Zheng J, Hu JD et al (2014) Discovery of somatic mutations in the progression of chronic myeloid leukemia by whole-exome sequencing. Genet Mol Res 13(1):945–953
Adzhubei I, Jordan DM, Sunyaev SR (2013) Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet, chapter 7:Unit7.20
Barzi A, Sekeres MA (2010) Myelodysplastic syndromes: a practical approach to diagnosis and treatment. Cleve Clin J Med 77(1):37–44
Vicente C, Conchillo A, García-Sánchez MA, Odero MD (2012) The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol 82(1):1–17
Cortés-Lavaud X, Landecho MF, Maicas M et al (2015) GATA2 germline mutations impair GATA2 transcription, causing haploinsufficiency: functional analysis of the p.Arg396Gln mutation. J Immunol 194(5):2190–2198
Johnson KD, Hsu AP, Ryu MJ et al (2012) Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J Clin Invest 122(10):3692–3704
Connelly JJ, Wang T, Cox JE et al (2006) GATA2 is associated with familial early-onset coronary artery disease. PLoS Genet 2(8):e139
Liu X, Jiang B, Wang A et al (2015) GATA2 rs2335052 polymorphism predicts the survival of patients with colorectal cancer. PLoS One 10(8):e0136020
Gilliland DG (2002) Molecular genetics of human leukemias: new insights into therapy. Semin Hematol 39(4 Suppl 3):6–11
Greif PA, Dufour A, Konstandin NP et al (2012) GATA2 zinc finger1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood 120(2):395–403
Fasan A, Eder C, Haferlach C et al (2013) GATA2 mutations are frequent in intermediate-risk karyotype AML with biallelic CEBPA mutations and are associated with favorable prognosis. Leukemia 27(2):482–485
Green CL, Tawana K, Hills RK et al (2013) GATA2 mutations in sporadic and familial acute myeloid leukaemia patients with CEBPA mutations. Br J Haematol 161(5):701–705
Green CL, Koo KK, Hills RK, Burnett AK, Linch DC, Gale RE (2010) Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Oncol 28(16):2739–2747
Hamadou WS, Besbes S, Bourdon V et al (2017) Mutational analysis of TP53 gene in Tunisian familial hematological malignancies and sporadic acute leukemia cases. Familial Cancer 16(1):153–157
Hamadou WS, Bourdon V, Létard S et al (2016) Familial hematological malignancies: new IDH2 mutation. Ann Hematol 95(12):1943–1947
Hamadou WS, Bourdon V, Gaildrat P et al (2016) Mutational analysis of JAK2, CBL, RUNX1, and NPM1 genes in familial aggregation of hematological malignancies. Ann Hematol 95(7):1043–1050
Hamadou WS, El Abed R, Besbes S et al (2016) Familial hematological malignancies: ASXL1 gene investigation. Clin Transl Oncol 18(4):385–390
Hamadou WS, Besbes S, Mani R et al (2017) ARLTS1, potential candidate gene in familial aggregation of hematological malignancies. Bull Cancer 104(2):123–127
El Abed R, Bourdon V, Huiart L et al (2009) Molecular study of CEBPA in familial hematological malignancies. Familial Cancer 8(4):581–585
Acknowledgements
This work was supported by la Société Française d’Hématologie, le groupe Génétique et Cancer and Institut National du Cancer (INCa), and the Ministère de l’Enseignement Supérieur et de la Recherche Scientifique Tunisie. It is a part of the GenHem INSERM/DGRS project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Hamadou, W.S., Mani, R., Besbes, S. et al. GATA2 gene analysis in several forms of hematological malignancies including familial aggregations. Ann Hematol 96, 1635–1639 (2017). https://doi.org/10.1007/s00277-017-3076-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-017-3076-9