Abstract
Mutations are responsible for familial cancer syndromes which account for approximately 5–10 % of all types of cancers. Familial cancers are often caused by genetic alterations occurring either in tumor suppressor or genomic stability genes such as TP53. In this study, we have analyzed the TP53 gene by direct sequencing approach, in a panel of 18 Tunisian familial hematological malignancies cases including several forms of leukemia, lymphoma and myeloid syndrome and 22 cases of sporadic acute leukemia. In one familial case diagnosed with acute lymphoblastic leukemia, we reported an intronic substitution 559+1 G>A which may disrupt the splice site and impact the normal protein function. Most of the deleterious mutations (Arg158His; Pro282Trp; Thr312Ser) as classified by IARC data base, were commonly reported in ALL cases studied here. The cosegregation of the two variants rs1042522 and rs1642785 was observed in most patients which may be in favor of the presence of linkage disequilibrium. The most defined TP53 mutations found here were identified in acute lymphoblastic leukemia context whereas only 3 % of mutations have been in previous studies. The cosegregation of the two recurrent variant rs1042522 and rs1642785 should be further confirmed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the discovery of TP53 gene in 1979, several studies have defined this gene as a key tumor suppressor. This gene remains the most important factor in human cancer, by ensuring crucial cells process such as maintaining genomic stability, inducing cell-cycle arrest and assuring replicative senescence or apoptosis and DNA repair in response to various types of stress. The human p53 gene is located on chromosome 17p and consists of 11 exons and 10 introns [1, 2].
The TP53 disruption was estimated to 50 % of all human cancers mostly in solid tumors [3, 4]. However lower frequency (10–20 %) was found in hematological malignancies according to IARC data base: 10–20 % of cases of chronic lymphocytic leukemia (CLL) [5], 10–12 % cases of multiple myeloma (MM) [6], 3–8 % of cases of acute myeloid leukemia (AML) [7] and less than 3 % in acute lymphoblastic leukemia (ALL) [8].
The TP53 mutation was evoked in several familial cancer including breast-ovarian cancer, colorectal cancer and especially in Familial Li–Fraumeni syndrome in which 60–80 % carries TP53 mutations. These mutations were often described in many studies targeting hematological malignancies, but not in familial aggregation context. According to the International Agency for Research in Cancer (IARC) TP53 data base, several germline mutations were reported in several hematological malignancies showing highly polymorphic TP53 gene in coding and non coding regions. Epidemiological studies carried out in hematological malignancies revealed a poor prognosis among patients carrying TP53 mutation characterized by worse overall survival and resistance to chemotherapy [9] compared to those with wild-type form.
We hypothesize that patients with familial hematological malignancies might present TP53 gene mutations. We aimed to determine to what extent the TP53 gene is involved in familial context, compared to sporadic acute leukemia. The entire TP53 gene was investigated in a panel of 18 Tunisian family cases with aggregated hematological malignancies by direct sequencing. 22 sporadic acute leukemias (13 ALL and 9 AML) were included in this study.
Patients and methods
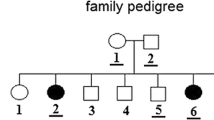
The studied cohort consists of 18 patients belonging to 10 independent Tunisian families in which aggregation of hematological malignancy in first, second or third degree relatives were reported. The familial cases include 10 cases of Hodgkin lymphoma, 1 case of Chronic Myeloid Leukemia (CML), 2 cases of AML, 3 cases of ALL, 1 case of non hodgkin lymphoma (NHL) and 1 case of Myelodysplastic Syndrome (MS). The mutation analysis was performed from blood patients during complete remission according to the stander protocol of treatment of each pathology. Informed consent was obtained from the patients, relevant family members (healthy relatives) or their legal guardian as required by the Helsinki Declaration.
13 ALL and 9 AML sporadic Tunisian cases were included in this study. The mutational analysis was performed from blood patients during the diagnosis.
Genomic DNA was extracted from whole blood with the EZ1 DNA tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The entire TP53 gene was amplified using standard PCR methods. All primers and PCR conditions are available under request. The amplified PCR products were column-purified and both strands were sequenced using the BigDye Terminator Cycle Sequencing Ready Reaction Kit v1.1 (Applied Biosystems-Foster City, USA) and loaded onto an ABI Prism 3500 sequencer (Applied Biosystems). The sequence chromatograms obtained were compared to the published human TP53 gene sequence (Genebank accession number NC_000017.10) using the SeqScape software program v2.5 (Applied Biosystems).
TP53 variants found were checked in TP53 data base of IARC. The impacts of each variant were evaluated using bioinformatics tools provided in IARC database as SIFT, Align AGVGD and Human Splicing Finder, PolyPhen-2 (Adzhubei et al. 2010) and MEGA-MD (Molecular Evolutionary Genetics Analysis software) http://www.megasoftware.net/mega-md/mega-md.php [10].
Results
The TP53 mutations were screened in 10 independent Tunisian families with aggregation of hematological malignancy and 22 sporadic acute leukemia cases. Among the 40 DNA analyzed, sequences alignment allowed us to identify 10 DNA variations: four mutations were intronic while the six other ones were exonic. Most of them was previously reported and listed in IARC data base. The results are summarized in Table 1.
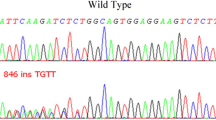
We have identified four intronic variants in TP53 gene: c.74+38 C>G; c.559+1 G>A; c.933+12 T>C; c.1100+30 A>T located respectively in intron: 2, 5, 9 and 10. The most recurrent intronic variant was the c.74+38 C>G found in 37/40 of cases including familial and sporadic AML or ALL cases in which 22 were homozygous. The c.559+1 G>A variant was found in only one familial case, diagnosed with B-ALL. This variant was classified as intronic mutation since it impacts the splice site of intron 5 of TP53 gene.
Two synonymous variants of TP53 gene were found in this study: Pro36 Pro and Arg213Arg. The first substitution Pro36Pro was found in only one case of sporadic ALL, whereas the second one Arg213Arg was more recurrent and was detected in 9 patients including 6 familial cases among them 3 cases of hodgkin lymphoma (HL), 2 cases of ALL and one case of MS and 3 sporadic cases: 1 ALL and 2 AML in which one was homozygous.
We have detected four nonsynonymous substitutions: Pro72Arg, Arg158His, Pro282Trp and Thr312 Ser. The Arg158His and Pro282Trp were found in two patients diagnosed with sporadic ALL. The Pro72Arg variant was the most recurrent substitution found in 36/40 analyzed cases, either in sporadic acute leukemia or familial hematological malignancies. Among the 36,19 patients were homozygous for Arginine amino acid. We notice that each patient carrying Pro72Arg substitution except one, carrys also the intronic variant c.74+38 C>G, with the same degree of homozygosis in most cases. The Thr312Ser substitution (rs145151284) was found in only one case of sporadic ALL. The Sift analysis predicts a neutral effect on protein function.
Discussion
Among the driver Mutations described in several cancers, the TP53 gene has the highest prevalence. Several somatic and germline inherited mutations were described referring to IARC TP53 Data Base and literature. A large study screening 525 patients with several cancer family history, has identified 17.3 % of patients carrying TP53 germline mutations [11].
According to IARC data base, 3.37 % of hematological malignancies were associated with germline TP53 mutations. This data insight us to establish the mutational screening of TP53 gene in familial hematological malignancies since to our knowledge no study was carried out in familial context. For this we sequenced the entire TP53 gene in 22 Tunisian sporadic leukemia cases including AML and ALL and 18 Tunisian familial hematological cases including 10 cases of HL, 1 case of CML, 2 cases of AML, 3 cases of ALL, 1 case of NHL and 1 case of MS.
We report here 10 TP53 variants, 4 of them are located in introns and 6 in exons. According to IARC data base, 90 % of known polymorphisms are identified in TP53 introns including all type of cancer. In our investigation conducted on hematological malignancies context, we found more variations in exons than in introns.
Intronic variants
Among the 4 intronic variants, one interesting mutation was found: the c.559 + 1 G>A located in the fifth intron. It might interrupt the RNA splicing process. For this reason, it was classified as TP53 splicing mutation [12]. The c.559 + 1 G>A mutation was found in one single familial case diagnosed with B-ALL at the age of 39. This patient showed a worse outcome due to bone marrow infiltration and chemical treatment resistance and died. His 20-years old cousin (first degree relative) was also diagnosed with ALL and also showed a worst outcome with drug resistance. After bone marrow transplantation failed, she relapsed and died. She was not included in this study since the DNA was not available. This mutation may be associated with poor prognoses.
We have found the c.74+38 C>G (rs1642785) variant localized in intron 2 in 92 % of cases including familial cases and sporadic AML/ALL. A recent new role was attributed to rs1642785 on mRNA splicing and stability, and thus on the differential expression of isoform-specific transcripts of the TP53 gene [13]. In only one study, this variant was associated with the Pro72Arg (rs1042522) one to glioma in indian population [14].
The c.933+12 T>C (rs1800899) variant located in the ninth intron was found in two patients diagnosed with sporadic ALL. It was not assigned in IARC database. It was previously described in patients with early onset of Colorectal Cancer [15] and in three breast cancer patients [16]. No significance of this change was described.
In one familial case diagnosed with T-ALL, we have found other intronic variant: the c.1100+30 A>T (rs17880847) which was already described in one study in one patient with malignant salivary gland neoplasms [17].
Exonic variants
We have identified in 40 cases of hematological malignancies analyzed here 6 exonic variants: 2 were Synonymous and 4 non synonymous mutations. The first synonymous substitution was c.108 G>A p.Pro36Pro (rs1800370), located in exon 1, was found in one single case of ALL. According to IARC data base, this variant was classified as a silent mutation. It was detected in 2.4 % of analyzed cases of glioblastoma but was totally absent in Indian controls population [14]. It was previously shown that the rs1800370 silent mutation reduces the ability of TP53 to activate apoptosis by decreasing the affinity of TP53 mRNA for MDM2, consequently reducing TP53 levels [18].
The second synonymous mutation c.639 A>G p.Arg213Arg (rs1800372) located in exon 6, was found in 9 patients: 1 case of sporadic ALL and 2 cases of sporadic AML in which one was found at homozygous level, and 6 familial cases with several hematological malignancies. This variant was classified as silent polymorphism according to IARC data base with medium frequency.
Four missense substitutions were found in our investigation. The first one was c.473 G>A Arg158His (rs139200646) located in exon 5 described in one sporadic ALL. This variant was classified as deleterious missense substitution. The SIFT analysis is in favor of deleterious effect. Several studies have reported this variant in glioblastoma cases and Li–Fraumeni syndrome [19].
The second variant was c.844 C>T, p.Pro282Trp (rs28934574), located in exon 8and found in one sporadic ALL. This deleterious mutation as classified in IARC data baseis located in the DNA binding TP53 core domain. Functional study have showed that it may reduce the DNA binding ability and leads to the loss of transactivation activity of TP53 gene [20].
The third variant found also in sporadic ALL was c.935 C>Gp.Thr312Ser (rs145151284) located in exon 9. According to the IARC data base this variant is classified as missense substitution with neutral effect according to SIFT analysis.
The fourth variant c.215 C>G p.Pro72Arg (rs1042522) located in exon 4was the most frequent and found in 90 % of cases including sporadic and familial cases. 19 of them were homozygous Arg/Arg.TP53 rs1042522 is one of the most commonly investigated variant in cancer genetic epidemiology [21]. Several association studies have been conducted with many controversies about the pathogenicity effect of this variant. Meta-analysis have suggested that this polymorphism might be a risk factor for breast, colorectal, ovarian, or endometrial cancer and that the Pro allele might cause a small increase in the risk of lung cancer [22]. Among analysis targeting this variant in Tunisian population, an association study of rs1042522 variant conducted by Mabrouk et al. [23] has found that there is no significant difference in frequency distribution of genotyping alleles between 2 groups of Tunisian patients with bladder cancer and breast cancer in comparison to a control population. In a larger group of Tunisian breast cancer, a recent association study showed that TP53 codon 72 polymorphism is involved in susceptibility to develop breast cancer. However, there is no evidence indicating that Arg72Pro SNP may influence response to anthracycline-based chemotherapy [24].
We report in this study that all patients except one, carrying the Pro72Arg variant present also the intronic variant c.74+38 C>G (rs1642785). Most of them were homozygous for these 2 variants, which may suppose a linkage disequilibrium which has to be furtherer confirmed.
TP53 mutations were not much investigated in ALL compared to other cancers, but worst outcome were reported in patients carrying TP53 mutation. This poor outcome was observed in all patients diagnosed with ALL and carrying TP53 mutation [25].
Despite the low frequency 3 % of TP53 mutation reported by previous epidemiological data in sporadic ALL [8], in our investigation most of TP53 mutations were found commonly in patient diagnosed with sporadic ALL compared to sporadic AML and Familial hematological malignancies. Further TP53 investigations targeting sporadic Tunisian ALL are needed.
Conclusion
Germline mutations are responsible for familial cancer syndromes which account for approximately 5–10 % of all types of cancers. These mutations mainly occur within tumor suppressor genes as TP53. Somatic TP53 mutations were included in almost every type of cancer whereas gremline mutations were described in familial cancer context like Li-Fraumeni syndrome. The TP53 gene was screened in several hematological malignancies but not in familial aggregation context. In this study we have target for the first time the TP53 gene in 10 independent families. We reported only one intronic deleterious mutation c.559+1 G>A. The most defined TP53 mutations found here were identified in sporadic ALL whereas only 3 % of mutations have been reported in literature. This finding will encourage us to investigate the TP53 gene in a larger Tunisian ALL cases. We aimed also to focus on the cosegregation of the two most frequent variants rs1042522 and rs1642785 through a linkage study.
References
Moll UM, Wolff S, Speidel D, Deppert W (2005) Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol 17:631–635
Vousden KH, Prives C (2009) Blinded by the light: the growing complexity of p53. Cell 137:413–418
Soussi T, Dehouche K, Beroud C (2000) P53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum Mutat 15:105–113
Olivier M, Hollstein M, Hainaut P (2010) TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2:1–17
Pekova S, Mazal O, Cmejla R, Hardekopf DW, Plachy R, Zejskova L et al (2011) A comprehensive study of TP53 mutations in chronic lymphocytic leukemia: analysis of 1287 diagnostic and 1148 follow-up CLL samples. Leuk Res 35:889–898
Chng WJ, Price-Troska T, Gonzalez-Paz N, Van-Wier S, Jacobus S, Blood E et al (2007) Clinical significance of TP53 mutation in myeloma. Leukemia 21:582–584
Nahi H, Selivanova G, Lehmann S, Möllgard L, Bengtzen S, Concha H et al (2008) Mutated and non-mutated TP53 as targets in the treatment of leukaemia. Br J Haematol 141:445–448
Agirre X, Novo FJ, Calasanz MJ, Larrayoz MJ, Lahortiga I, Valgañón M et al (2003) TP53 is frequently altered by methylation, mutation, and/or deletion in acute lymphoblastic leukaemia. Mol Carcinog 38:201–207
Peller S, Rotter V (2003) TP53 in hematological cancer: low incidence of mutations with significant clinical relevance. Hum Mutat 21:277–284
Stecher G, Liu L, Sanderford M, Peterson D, Tamura K, Kumar S (2014) MEGA-MD: molecular evolutionary genetics analysis software with mutational diagnosis of amino acid variation. Bioinformatics 30:1305–1307
Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ et al (2009) Beyond Li Fraumeni syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol 27:1250–1256
Nadauld LD, Garcia S, Natsoulis G, Bell JM, Miotke L, Hopmans ES et al (2014) Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol 15:428
Perriaud L, Marcel V, Sagne C, Favaudon V, Guédin A, De-Rache A et al (2014) Impact of G-quadruplex structures and intronic polymorphisms rs17878362 and rs1642785 on basal and ionizing radiation-induced expression of alternative p53 transcripts. Carcinogenesis 35:2706–2709
Jha P, Pathak P, Chosdol K, Suri V, Sharma MC, Kumar G et al (2011) TP53 polymorphisms in gliomas from Indian patients: study of codon 72 genotype, rs1642785, rs1800370 and 16 base pair insertion in intron-3. Exp Mol Pathol 90:167–172
Djansugurova L, Zhunussova G, Khussainova E, Iksan O, Afonin G, Kaidarova D et al (2014) Screening the APC, MLH1, MSH2 and TP53 mutations in patients with early onset of colorectal cancer. J Carcinog Mutagen 5:197–199
Damineni S, Rao VR, Kumar S, Ravuri RR, Kagitha S, Dunna NR et al (2014) Germline mutations of TP53 gene in breast cancer. Tumor Biol 35:9219–9227
Gomes C, Diniz MG, Orsine LA, Duarte AP, Fonseca-Silva T, Brendan-Conn B et al (2012) Assessment of TP53 mutations in benign and malignant salivary gland neoplasms. PLoS ONE 7:1–8
Whibley C, Pharoah PD, Hollstein M (2009) p53 polymorphisms: cancer implications. Nat Rev Cancer 9:95–107
Wong P, Han K (2014) Lack of toxicity in a patient with germline TP53 mutation treated with radiotherapy. Curr Oncol 21:349–353
Calhoun S, Daggett V (2011) Structural effects of the L145Q, V157F, and R282 W cancer-associated mutations in the p53 DNA-binding core domain. Biochemistry 50:5345–5348
Vineis P, Manuguerra M, Kavvoura FK, Guarrera S, Allione A, Rosa F et al (2009) A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst 101:24–36
Dahabreh IJ, Schmid CH, Lau J, Varvarigou V, Murray S, Trikalinos TA (2013) Genotype misclassification in genetic association studies of the rs1042522 TP53 (Arg72Pro) polymorphism: a systematic review of studies of breast, lung, colorectal, ovarian, and endometrial cancer. Am J Epidemiol 177:1317–1318
Mabrouk I, Baccouche S, El-Abed R, Mokdad-Gargouri R, Mosbah A, Saïd S et al (2003) No evidence of correlation between p53 codon 72 polymorphism and risk of bladder or breast carcinoma in Tunisian patients. Ann N Y Acad Sci 1010:764–766
Arfaoui A, Douik H, Kablouti G, Chaaben AB, Handiri N, Zid Z et al (2015) Role of p53 Codon72 SNP in breast cancer risk and anthracycline resistance. Anticancer Res 35:1763–1766
Hof J, Krentz S, van-Schewick C, Körner G, Shalapour S, Rhein P et al (2011) Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol 29:3185–3188
Acknowledgments
This work was supported by the Ministère de l’Enseignement Supérieur, de la Recherche Scientifique et des Technologies de l’Information et de la Communication en Tunisie. It is a part of the GenHem INSERM/DGRS project. We are grateful for English correction provided by Helmi Ben AZIZA.
Authors’ contributions
W.S.H., S.B., and V.B. conceived the study and performed the experiments and wrote the manuscript, Y.B.Y. and M.A.L. provides simple and clinical data, T.N. analyzed the data, A. K., H.S. and Z. S. supervised the project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hamadou, W.S., Besbes, S., Bourdon, V. et al. Mutational analysis of TP53 gene in Tunisian familial hematological malignancies and sporadic acute leukemia cases. Familial Cancer 16, 153–157 (2017). https://doi.org/10.1007/s10689-016-9931-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-016-9931-3