Abstract
Signal transducers and activators of transcription (STAT) proteins are latent cytoplasmic transcription factors that affect several cellular processes including cell growth, proliferation, differentiation, and survival. Following phosphorylation, STATs are activated, and their upregulated expressions increase in malignancies with playing a role in the development of leukemia. In this study, transfection of K–562 cells with either unmodified or chemically modified anti-STAT3, -STAT5A, -STAT5B siRNAs for duration of 12 days, determining gene silencing at mRNA and protein levels, evaluating apoptosis rate, and detecting JAK/STAT pathway members’ gene expression profiles via array method were aimed. Quantitative RT-PCR and Western blot assays indicated that STAT expressions were downregulated both at mRNA and protein levels, and TUNEL assay showed that leukemic cell apoptosis was induced due to inhibition of STATs. Array analysis resulted with decreases in signal transducer, phosphorylation inducer, and oncogene expressions, whereas increased expressions in STAT inhibitor and apoptosis inducer genes were observed. These results point out that siRNA application could constitute a new and alternative curative method for supporting therapy of CML-diagnosed patients in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative hematopoietic stem cell disorder and characterized by excess proliferation of myeloid progenitor cells. Over 90 % of the cases, a Philadelphia chromosome is seen which is formed as a result of reciprocal translocation between 9th and 22th chromosomes’ q arms that is characteristic for CML [t(9,22) (q34; q11)]. Over this rearrangement, a highly tyrosine kinase-activated chimeric oncoprotein named bcr-abl is encoded. Actually, tyrosine kinase activity of Abl protein is strictly checked out till Bcr sequence is added to it. Deregulated tyrosine kinase activity of bcr-abl protein stimulates cell proliferation and inhibits apoptosis that is thought to be responsible for the changes in normal cellular functions such as growth, differentiation, adhesion, and migration. Finally, malignant cell transformation is induced that is responsible for CML phenotype [1, 2].
In CML, because the uncontrolled activity of the fusion protein is not under the regulatory control mechanisms of ABL, this results with an increase in malignant disease by activating many cytoplasmic and nuclear signal transduction pathways that affect the proliferation and survival of hematopoietic cells [3]. One of the most important signaling cascades that regulate cell proliferation and apoptosis giving rise to the development of leukemia is the Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathway [4].
A variety of ligands including cytokines, hormones, growth factors, and their receptors stimulate the JAK/STAT pathway. While JAK belongs to a family of non-receptor protein tyrosine kinase, STATs are known as latent cytoplasmic transcription factors. Seven mammalian STAT proteins have been identified from STAT1 to STAT6, including STAT5a and STAT5b, which are encoded by distinct genes. STATs carry a conserved tyrosine residue near the C-terminus of SH-2 domain that is phosphorylated by the activated JAKs. This tyrosine phosphorylation leads the dimerization of STATs and migration of them to the nucleus. In the nucleus, dimerized STATs bind to specific regulatory DNA sequences of target genes to activate or repress their transcription. Thus, the JAK/STAT cascade provides an efficient mechanism to translate an extracellular signal that is resulted with post-transcriptional responses especially seen in hematological malignancies [5, 6]. STAT proteins that usually take part in the activation or suppression of malignant transformations are STAT1, STAT3, and STAT5. While STAT1 exerts an apoptosis inducer gene function [7, 8], STAT3 and STAT5 are constitutively activated in various types of hematooncologic malignancies, and they trigger the formation of leukemia which is p210- and p190-transformed leukemic cells [9, 10]. STAT5 proteins have been demonstrated that they are required for the development of myeloproliferative diseases [7] with playing an important role in anti-apoptotic activity mediated by Bcr-Abl [11, 12]. STAT3 has also been implicated in malignant transformation, and its inhibition triggers tumor cells’ apoptosis [13]. After it has been found out that suppression of STAT gene expression results in inhibition of oncogenesis and leukemogenesis [14] also with induction of tumor cell apoptosis, STATs have become the initial target of emerging anticancer agents [15]. The remarkable point here is that, targeting STATs causes preferential cancer cell death with minimal effects on normal cells. Of late years, intriguing siRNAs are small interfering RNAs that have the ability to silence a target gene by degrading its mRNA, and their usability for the therapy of diseases is exhibited day after day. Thus, varying expression profiles affect cellular fateful events; siRNAs have become good therapeutic targets thereafter [16]. Chemically modified siRNAs can operate RNA interference (RNAi) mechanism stably and efficiently for longer periods rather than unmodified ones because of being more stable to endonucleases [17].
In the current study, we initially aimed to transfect K–562 chronic myeloid leukemia cells efficiently with anti-STAT3, -5A, and -5B specific siRNAs that were either unmodified or chemically modified for duration of 12 days. Secondarily, we focused on to determine expressional changes of STATs at mRNA and protein levels, apoptotic response of the cells, and expression profiles of all JAK/STAT pathway members after RNAi-mediated silencing of STATs.
Materials and methods
Culture conditions of leukemia cells
A human chronic myelogenous leukemia suspension cell line K–562 was purchased from the European Collection of Cell Cultures (morphology, lymphoblast; catalog no., 89121407; lot no., 06/A/015). Cells were cultured in RPMI 1640 medium containing 10 % (v/v) heat-inactivated fetal calf serum, 100 units of penicillin–streptomycin/ml, 1 % l-glutamine at 37 °C in humidified air containing 5 % CO2.
Confirmation of siRNA transfection and cytotoxicity assay
siRNA transfection was optimized through a fluorescent-labeled siRNA by evaluating transfection rate under a fluorescent microscope. At this point, after transfection, an apoptosis assay (Annexin-V-EGFP Apoptosis Detection Kit; Milpitas, CA) was also assed according to kit manual in order to confirm that the decided amounts were appropriate for the cells. Next, in order to verify that used siRNA concentration and transfection reagent (TR) amounts were not cytotoxic and gene silencing would be achieved due to RNAi pathway but not cause of cytotoxic concentrations of siRNA, XTT cell proliferation assay (Cell Proliferation Kit II; Roche, Germany) also with lower and higher concentrations was performed, and absorbance of each sample was measured spectrophotometrically with an ELISA reader (Thermo Multiscan).
Gene silencing by siRNA treatments
siRNA transfection was achieved in accordance with “HiPerFect Transfection Reagent” (Qiagen, Valencia, CA, and USA) manual. Unmodified (UM) or chemically modified same siRNA sequences targeting STATs (Dharmacon, Chicago, IL, USA) were used in order to compare their silencing efficiency in suspension leukemia cells, known as “hard to transfect.” The cholesterol conjugation resided at the 5′-end of siRNA sense strand that specifically binds to STAT mRNAs (STAT-CHL). The FluoroUridine substation with FluoroCytidine is applied to RNA’s 2′-ribose region (sugar modification) that increases the binding affinity of siRNA to STAT mRNA with reduced off-target effects (STAT-FU/FC). The reason why we preferred to use chemically modified siRNA was because it is more stable to endonucleases and thus keeping on gene silencing for longer periods without the need of an extra transfection. In every experiment, positive and negative control siRNA-treated cells [anti-GAPDH and non-targeting (NC)] and also untreated (UT) and “mock control cells” (no siRNA added but only transfection reagent) were used as controls (The used siRNA sequences are given in Supplement Table 1]. For reaching the optimal silencing effect, cells were incubated for 12 days at 37 °C and in 5 % CO2. Among them, a particular cell was used in XTT cell proliferation assay, quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) analyses, and apoptosis assessment, and another transfection was revealed again for Western blot and array assays.
Quantitative assessment of STAT mRNA expression levels via real-time qRT-PCR
Following gene-specific siRNA treatment, mRNA expression levels of STAT3, -5A, -5B and GAPDH genes were assessed by real-time qRT-PCR. Relative expression level of each gene was determined as we described before. Briefly, target gene mRNA copy number was divided to G6PDH mRNA copy number, obtained from the standard curve which was derived from reference standards ranging between 5 × 102 and 5 × 106 copy number of G6PDH [18]. Gene silencing rate was determined by comparing siRNA applied group’s relative expression with UT control group, and repression values were identified for each target gene [18]. The primers and probes used for the amplification of STAT3 (NM_003150), STAT5A (NM_003152), STAT5B (NM_012447), and GAPDH (NM_002046) are given in Supplement Table 2.
Determining STAT protein expression levels by Western blot analyses
Protein extraction from siRNA-treated and UT cells was performed according to the “Proteojet Mammalian Cell Lysis Reagent” (Thermo Scientific, Fermentas) instructions. The supplied protein amounts were assessed by Bradford method by using bovine serum albumin standards ranging between 0.25 and 2 mg/ml concentrations. Finally, 9 μg of each protein extract was resolved at 8 % SDS–PAGE gel and transferred to PVDF membranes using a wet transfer system (Bio-Rad). The used primer antibody concentrations were as follows: 1:1,000 diluted polyclonal STAT3 (06–596, Upstate) and β-actin (#4967, Cell Signaling Technology, USA), 1:2,000 diluted STAT5A (06–968, Upstate, USA), STAT5B (06-969, Upstate, USA), and 1:5,000 diluted GAPDH (#5174, Cell Signaling Technology, USA) by overnight incubation at 4 °C. The protein levels of target genes in each fraction were detected by using the enhanced colorimetric detection kit (Amplified Alkaline Phosphatase Goat Anti-Rabbit Immune-Blot Assay Kit, Bio-Rad) according to the manufacturer’s instructions. The Western blot results were evaluated with gel imaging system (Chemi-Smart 2000).
Apoptosis assay
For the siRNA-treated cells and control group cells—composed of untreated cells, NC siRNA-treated cells, and mock-transfected cells—apoptosis rates were determined with terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) method (DeadEnd™ Fluorometric TUNEL System Protocol, Promega) under a fluorescent microscope according to the kit procedure, and the number of alive and apoptotic cells was evaluated.
Expression array
The same total RNAs used in qRT-PCR were transcribed into cDNA according to “RT2 First Strand Kit” (SABiosciences) insert. The expression of 84 genes related to JAK/STAT-mediated signaling at the sixth day of siRNA transfection was evaluated by an array method with “RT2ProfilerTM PCR Array: Human JAK/STAT Signaling Pathway” (Cat #: PAHS–039; SABiosciences) kit manual in real-time qRT-PCR LightCycler 480 instrument. The members of the pathway are given as a supplementary data in Table 3.
Statistical analyses
The experimental setup studies for gene silencing were triplicated, and the average of relative expression values was taken at the end of qRT-PCR analyses. Statistical analyses were performed by using Student’s t test with a value p < 0.05 defined as significant. Array results analyses were performed with ∆∆CT method that is available at http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php website. In this programmer, the groups to be compared are defined before analyses, and the expression values were normalized according to housekeeping genes; log2 transformation was gained, and fold change and p value results were accesible then. While more than fourfold expressional decreases are given in green spots, >4× increases are supplied in red spots to the user.
Results
Verification of siRNA transfection and cytotoxicity assay
All of the cells were efficiently transfected with siRNA without corruption of cell morphology (Fig. 1). At this point, an apoptosis assay was revealed in order to define the number of apoptotic cells due to siRNA transfection when compared with untreated control group cells under a fluorescent microscope. As a result, we have detected that while UT control group (neither siRNA nor transfection reagent added) had 1 % apoptotic cells, the mock (just transfection reagent added) control group had 2 % ratio. siGLO siRNA-transfected group with 9 μl of transfection reagent showed an apoptosis rate of 15 % when compared to control groups. Besides, 3–6 μl of transfection reagent did not induce apoptosis, so 6 μl was accepted as tolerable for the cells since each cell was transfected with siRNA without an increase in apoptotic cell number (Fig. 2). After generating an average siRNA and TR amount for K–562 cells, cell proliferation assay XTT was carried out. Also, the lower and upper siRNA concentrations (50–100–150 nM) and TR amounts (3–6–9 μl) were assessed. XTT assay revealed that, the used siRNA concentration and TR amount in each experiment were not cytotoxic, thereby 100 nM siRNA and 6 μl TR doses were found to be appropriate for the cells (Fig. 3).
Confirmation of siRNA transfection and annexin V apoptosis assay for cytotoxicity control in siRNA-transfected and control cells: a fluorescein microscopy image captured during annexin V assay, ×40 magnification. The red spots indicate that siRNA was uptaken by each cell. b After fluorescent-labelled siRNA siGLO was transfected into the cells, apoptosis was evaluated, and it was observed that the 6-μl transfection reagent did not induce apoptosis with each cell transfected with siRNA
Cytotoxicity assay for varying transfection reagent and siRNA concentrations. A: Defining unmodified and sugar modified siRNA concentrations’ cytotoxicity targeting STATs. B: Revealing transfection reagent (siRNA was established as 100 nM) and CHL modified siRNA concentrations’ cytotoxicity. Untreated control group’s OD ratio was defined as 1; whereas the treated groups’ absorbance was measured spectrophotometrically according to the control. Finally, 100 nM siRNA and 6 μl transfection reagent concentrations were optimal for K-562 cells
Effect of positive control siRNA uptake and inhibition of GAPDH expression
During 12 days of the study, qRT-PCR results indicated that GAPDH relative expression was at least inhibited by 98.5 % and more with anti-GAPDH siRNA when compared with UT control group. The changing relative expression profiles of GAPDH following siRNA treatment are given as supplement in Table 4. As for protein levels, again GAPDH was downregulated during the 72th and 96th hours (Fig. 4). Because PC gene was knocked down with a very high suppression rate, it was concluded that the transfection protocol was sufficient for K–562, cells and it could be used for silencing of STATs.
Effects of RNAi-mediated silencing of STATs at mRNA and protein expression levels
Gene-specific siRNA-mediated STAT3, -5A, and -5B expressional downregulations were evaluated by comparison of treated group expressions with UT groups. Each gene’s expressional downregulations were assessed by an unmodified and two modified siRNAs having the same sequences, specifically designed for that target gene. The day that a target STAT maximum suppressed was determined via qRT-PCR as “suppression percentage” or “fold reduction” at mRNA level and by Western blot analysis at protein level. The siRNA type for every gene that caused the highest suppression was given below for STAT3, -5A, and -5B. The differential expression values and fold changes of STAT mRNAs for each siRNA compared to control groups for 12 days can be found in Supplement Table 5.
*STAT3 expression levels
While unmodified siRNA caused maximum 85.9 % inhibition (7.09-fold, p = 0.013) of STAT3 at 48th hour, –CHL and –FU/FC-modified siRNAs caused 81.6 and 88.03 % (5.44-fold, p = 0.046 and 7.09-fold, p = 0.043) downregulations, respectively, for the same period of time. STAT3 was suppressed maximally by 86.3 % (7.28-fold, p = 0.013) with CHL-modified siRNA at 24th hour, whereas –UM and –FU/FC-modified siRNAs led to 65.3 and 90.2 % downregulations (2.89-fold, p = 0.039 and 10.2-fold, p = 0.016), respectively, at this hour. With anti-S3-FU/FC siRNA (sugar modified), STAT3 expression was maximally 93.8 % suppressed (16.2-fold, p = 0.004) at sixth day of treatment, in which –UM and –CHL-modified siRNAs caused 63.1 and 79.1 % downregulations (2.72-fold, p = 0.023 and 4.8-fold, p = 0.007), respectively, for that day. For longer periods, STAT3 was inhibited by 79.4 % (4.85-fold, p = 0.001) and 83.5 % (6.06-fold, p = 0.041) with anti-S3-FU/FC siRNA at 10th–12th days (Fig. 5a). As for protein levels, STAT3 expression was mostly downregulated with sugar-modified siRNA and anti-S3 CHL siRNA followed it even at the 10th–12th days, whereas unmodified siRNA lost its silencing efficiency after the sixth day of treatment (Fig. 5b).
Effects of RNAi Mediated Silencing of STATs at mRNA and Protein Expression Levels. A-C-E: STAT3/STAT5A /STAT5B mRNA relative expression results following siRNA treatment at days 10 and 12 detected by qRT-PCR. B-D-F: Stat3/Stat5a/Stat5b protein expression results at the 10th and 12th days of siRNA transfection revealed by western blot
*STAT5A expression levels
STAT5A relative expression was maximum decreased by 91.2 % (11.38-fold, p = 0.001) at 96th hour with unmodified siRNA; –CHL and –FU/FC-modified siRNAs caused 61.5 and 77.4 % expressional downregulations (2.60-fold, p = 0.005 and 4.43-fold, p = 0.002), respectively, for this hour. While 81.7 % suppression was detected (5.47-fold, p < 0.001) with FU/FC siRNA at 48th hour, –UM and –CHL-modified siRNAs caused 68.4 and 65.9 % downregulations (3.17-fold, p = 0.043 and 2.93-fold, p = 0.036), respectively, for the same hour. For longer periods, modified siRNAs caused 76.3 % (4.21-fold, p < 0.001) and 82.7 % (5.77-fold, p = 0.038) inhibition of STAT5A on days 10 and 12, respectively, for sugar and cholesterol (Fig. 5c). Unmodified anti-STAT5A siRNA showed 66.6 and 72.3 % suppressions (2.99-fold, p < 0.001 and 3.60-fold, p = 0.048) for 10th and 12th days, respectively. When STAT5A protein expressions were evaluated, it was observed that all of the siRNAs efficiently suppressed STAT5A at early stages of the experiments up to the eighth day of treatment; then, only the modified ones—initially anti-S5A CHL and secondly FU/FC siRNA—still strongly inhibited its expression on days 10 and 12 (Fig. 5d).
*STAT5B expression levels
While STAT5B relative expression was maximum inhibited by 85.8 % (7.06 fold, p = 0.001) at 48th hour with unmodified siRNA, –CHL and –FU/FC-modified siRNAs caused 65.2 and 84.3 % expressional downregulations (2.88-fold, p = 0.081 and 6.38-fold, p = 0.030) for this time period. CHL-modified siRNA exhibited 83.1 % inhibition (5.89-fold, p = 0.001) at 72th hour, whereas –UM and –FU/FC-modified siRNAs caused 69.6 and 78.4 % downregulations (3.30-fold, p = 0.001 and 4.64-fold, p = 0.001), respectively, for this hour. STAT5B mRNA expression was maximally suppressed by 85.9 % (7.11-fold, p = 0.001) with FU/FC siRNA on day 12 (Fig. 5e). For day 12, –UM and –CHL-modified siRNAs showed a bit lower suppression with 47.9 and 68.02 % (1.92-fold, p = 0.198 and 3.13-fold, p = 0.004), respectively. As for Western blot results, STAT5B protein expression was downregulated similarly with three of the siRNAs at the beginning of the experiments till the eighth day of treatment, but at 10th-12th days, modified siRNAs—especially FU/FC—increased their silencing efficiencies and repressed the expression of STAT5B, whereas the unmodified one lost its effect and did not cause any expressional suppression (Fig. 5f).
Triggered apoptosis of K–562 cells in response to STAT knockdown
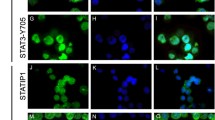
Apoptosis was induced due to siRNA-mediated gene silencing of STATs in leukemic K–562 cells at earlier periods of the study. Apoptosis rate was found to be extremely high especially at 72th and 96th hours of the experimental setup mostly with cholesterol conjugated anti-STAT5A and -S5B siRNAs ,and no apoptotic cell was detected after that day for any of the siRNA-treated groups. A picture of apoptotic and live cells is given in Fig. 6. In brief, 43 and 71 % of the cells were apoptotic in anti-S3 FU/FC-treated groups at 72th and 96th hours, respectively, whereas and 98 and 97 % of the cells were apoptotic at 96th hour with anti-STAT5A, –S5B CHL-modified siRNAs, but unmodified siRNAs triggered differing between 6 and 12 % of leukemic cell apoptosis (Fig. 7a–c). Other control groups’ apoptotic cell rate including UT, non-targeting, mock, and PC was very low and ranged between 0 and 2 %. The apoptosis rate for each day is given in Supplement Table 6. When suppression rates of target STATs were evaluated for 96th day that apoptosis was highly induced, it was observed that STAT3 mRNA was mostly downregulated with sugar-modified siRNA, whereas STAT5A and -5B mRNAs were inhibited with UM siRNAs (Fig. 7d–f). As for protein expression levels, while STAT5A was mostly suppressed via UM siRNA, STAT3 and STAT5 were inhibited by modified ones for triggering high apoptosis rate at 96th hour (Fig. 7g–i).
siRNA applied and untreated K-562 cells going under apoptosis with their downregulations detected at mRNA and protein levels for the same hours. A-C: STAT3/STAT5A/STAT5B apoptosis rate % seen in siRNA applied groups compared to control group for 72th and 96th hours. D-F: STAT3/STAT5A/STAT5B relative mRNA expression values that apoptosis was significantly triggered at hours 72 and 96. G-I: Stat3/Stat5a/Stat5b protein expression results at the 72th and 96th days of siRNA transfection that highest degrees of apoptosis was seen
Changes in gene expression profiles of JAK/STAT pathway following RNAi knockdown of STATs—mRNA array assay
JAK/STAT pathway members’ expression profiles were evaluated by an array method at the sixth day of siRNA transfection. When STAT3 was knocked down with any of the siRNAs, A2M and STAM expressions were downregulated −4.08- and −32.03-fold, respectively, with anti-STAT3 CHL-modified siRNA, and while STAT1 was 2.22-fold upregulated, CEBPB expression was decreased with a −52.78-fold decline with anti-S3 FU/FC siRNA (p < 0.05 all).
When STAT5A was inhibited with any of the siRNAs, A2M, CEBPB, STAM, SOCS4, and SOCS5 expressions were decreased. Among them, the highly expressional downregulations caused by anti-S5A CHL siRNA are CEBPB with −185.59-fold, CRK with −111.63-fold, STAM with −66.22-fold, and A2M −8.22-fold; decreases came forward. STAT1 expression was 3.41- and 2.21-fold increased with respectively anti-STAT5A CHL and sugar-modified siRNA (p < 0.05 all).
Suppression of STAT5B resulted with A2M, STAM, SOCS4, and SOCS5 expressional downregulation and PIAS2 and STAT1 upregulation. Among them, STAM, SOCS4, and SOCS5 expressions were decreased by −33.01-, −11.84-, and −5.50-fold, respectively, with anti-S5B CHL treatment, whereas PIAS2 and STAT1 expressions were 3.33- and 2.94-fold upregulated with CHL-modified anti-S5B siRNA (p < 0.05 all). Up- and downregulated genes are given as hierarchical cluster analysis in Fig. 8.
Discussion
To date, several curative strategies have been developed for CML, but the usage of tyrosine kinase inhibitors was a new generation therapy [19] till gain of resistance. One possible mechanism responsible for CML progression is the activation of transcription factors that should be inhibited some way for giving rise to cell death of leukemic cells [15]. From this point of view, in this study, we aimed to induce leukemic cell apoptosis due to RNAi-mediated gene-specific silencing of STAT3 and STAT5 that function as silent transcription factors but when activated, overexpressed and triggered leukemia development. RNAi is a powerful tool in biological studies, but has also become one of the most challenging therapeutic strategies because siRNA-based applications suffer from their poor delivery and biodistribution and degradation by nucleases inside the cell [20]. Consequently, the obstacles that we have to overcome in this study were: the transfection of suspension K–562 cells with target gene-specific siRNAs and duration of siRNA affectivity avoiding degradation in the cell. Because suspension cells are known as “hard to transfect” due to their fast growing and not attaching to the surface where siRNA is found, initially we have focused on siRNA uptake of cells that seemed to be the key point for a successful gene silencing. In our previous experiences, we had demonstrated that siRNA transfection of leukemic cells was achieved by a lipid-based detergent TR rather than an electroporation method. Besides this, another gene silencing tool we used was “antisense oligonucleotides” by which we observed that, silencing efficiency was much more less than siRNA for suspension cells in an in vitro environment (unpublished data). Regarding our aims, at the beginning of the study, we detected the siRNA uptake of cells after transfection of florescent-labeled siRNA and concluded that siRNA was penetrated into every cell, and this result led us to think that siRNA was the right tool for gene knockdown experiments. As we focused on siRNA, the unmodified ones were also good enough to knock down a target gene for about a maximum duration of 1 week, but to accelerate the silencing efficacy of STATs for longer periods without the need of an extra transfection, eliminating off-target effects and avoiding degradation as possible, we have also used chemically modified siRNAs and revealed their ability of silencing in leukemic cells. Gene silencing efficiencies between unmodified and chemically modified siRNAs targeting STAT3, STAT5A, and STAT5B were compared at mRNA and protein levels for 12 days duration. The reason why we continued with the experiments up to 12 days was the accelerating importance and curiosity about RNAi mechanism response for long-term gene silencing in primary cell lines.
Next, we checked the TR and siRNA concentrations’ effect upon cell viability in order to prove that the used volumes were not cytotoxic for the cells. Eventually, 6 μl TR and 100 nM siRNA concentrations were found to be convenient for K–562 cells. This was a critically important point because if the used doses were already cytotoxic for a cell, then any strategy to overcome a disease such as RNAi mechanism would not serve the purpose of an alternative therapy, owing to effecting healthy cells also. The second step was optimizing the transfection protocol for a specific cell type by making a transfection with positive control siRNA that should be inhibited by at least 75 % at mRNA level. As for our PC siRNA transfection results, we observed that it was suppressed by 98.5 % and more even at the 12th day of transfection and also at protein level at 72th and 96th hours, so the protocol could be reliably used in other target genes for transfection of siRNA into K–562 cells.
The main step of the study was transfection of siRNAs targeting STATs and revealing silencing rates both at mRNA and protein levels also revealing the apoptotic case of siRNA applied and untreated control leukemia cells. According to our qRT-PCR results, FU/FC-modified siRNA caused the highest suppression of STAT3 with 16.2-fold expressional decrease at the sixth day of treatment, and STAT3 protein expression was also inhibited greatly for that day with 71 % of apoptosis in K–562 cells. We have revealed STAT5A and STAT5B expressions independently and evaluated the results for each gene to expose their exact effect in the development of CML. Our results indicated that STAT5A was the highest silenced by 11.38-fold decline at mRNA level, and also, protein expression was downregulated at 96th hour by UM siRNA with 98 % apoptotic cells. As for STAT5B, the expression was knocked down highest by 7.11-fold inhibition at mRNA and protein levels at 12th day with anti-S5B CHL siRNA, and apoptosis was induced at very high levels like 97 %. No apoptotic cell was detected for any of the siRNA applied groups after 96th hour of the study. We think that this situation was related with the short doubling time of K–562 cells (~24 h) because after siRNA transfection, the cells continued to divide, and sometime later, the cells gained their proliferative phase again with generating rapid division of cells that escaped from apoptosis, and the number of alive cells was arisen in the population.
Taken all qRT-PCR and Western blot results together, we concluded that STAT5A and -5B seemed to play more efficient roles in the pathogenesis and development of CML. The reason why we attributed this conclusion could be explained with our apoptosis assay results indicating that, STAT5 regulated an anti-apoptotic program in CML, and that suppression of STAT5 expression led to a rapid induction of apoptosis. While sugar-modified siRNAs targeting STAT5A and STAT5B triggered by 78 and 75 % of leukemic cell apoptosis, respectively, the rates were 98 and 97 % with cholesterol-modified siRNA at 96th day. These results indicated two outcomes for us: One of them is the efficiency comparison of sugar- and cholesterol-modified siRNAs; and we concluded that because the cell membrane is composed of phospholipids, CHL-conjugated siRNA could be more easily uptaken by the cells and exhibit more impact upon leukemic cells owing to higher apoptosis rates. The second outcome was the importance of STAT5A and -5B and the fact that they came forward as therapeutic novel genes in CML. There are some studies that are in the same line with ours by supporting that CML indicator bcr/abl oncoprotein induces the activation of STAT5 making it a remarkable target for anticancer therapy [9, 18, 21, 22]. Besides this, in our previous study when we compared differing expression patterns of STATs in imatinib-resistant (IMA-3) cells to sensitive K–562 cells, we had observed that while STAT3 mRNA expressional increase was by 4 %, STAT5A and 5B expressions were most significantly changed (by 67 and 56 % upregulation, respectively) giving rise to a conclusion that STAT3 expression did not differ much in the gain of resistance. Besides this, CHL-modified siRNA silenced STAT5A mRNA expression and increased the rate of already induced apoptosis with different doses of imatinib in resistant cells. So in drug resistance, silencing STAT5A seemed to have more importance in response to therapy [23]. In the current study, as we concluded that STAT3 had the least impact among STATs upon CML, there can be two different explanations for that: One possible reason was owing to moderate apoptotic leukemic cell rate by 64 and 71 % with modified siRNAs at the 96th hour of treatment. The second reason could be the loss of post-transcriptional gene silencing mechanism on coming days of the transfection because STAT3 expression was maintained to be inhibited at only mRNA level, and protein suppression was lost even at the sixth day of the experiments. The reason why STAT3 was less likely to have an impact on leukemia could be explained with IL-6-induced STAT3 activation having more effect upon lymphomas and myelomas rather than CML [13].
The last step of our study was the array assay, and we performed it at the sixth day of transfection because this was the middle of experiments, and all target STATs were downregulated both at the mRNA and protein levels. So, we wanted to evaluate the JAK/STAT pathway members changing mRNA expression levels due to siRNA treatment and profiling significant fold changes that might have cellular response when STATs were sufficiently knocked down. According to these results, suppression of STAT3 with any of the anti-STAT3 siRNA resulted with downregulations in the expressions of A2M from “cytokines,” STAT3 from “transcription factors,” STAM from “signal transducers,” and CEBPB from “transcription factors.” Because STATs are activated upon cytokines and signal transducers, the expressional decreases seen in A2M and STAM gave rise to a conclusion that STAT3 signaling could be inhibited upon them. As STAT3 augments the transcriptional activity of CEBPB, we think that as response to gene silencing, CEBPB expression was also downregulated. In a study, 15 CML-diagnosed patients’ and 7 normal donors’ blood materials were analyzed using microarray assay, and they revealed that positive regulators of hematopoietic cell differentiator CEBPB expression were downregulated [24] similar to ours. In another one, when STAT3 signaling was inhibited by specific short hairpin RNAs, they reported that CEBPB expression was also downregulated [25]. As for pro-apoptotic gene STAT1; its expression was upregulated in only modified anti-STAT3 siRNA applied groups which might be the reason of the high apoptosis rate seen at the beginning of the experiments with modified siRNA targeting STAT3.
Inhibition of STAT5A with any of the siRNA targeting STAT5A resulted with PIAS2 and SOCS2 expressional upregulation from “STAT inhibitors” and downregulations of A2M cytokine, STAT5A, and CEBPB from “transcription factors,” and STAM, SOC4, and SOCS5 from “STAT inhibitors” were observed. PIAS2 and SOCS2 were actually responsible for inhibiting the activation of STATs; because STAT5A expression was downregulated following siRNA treatment, we think that the need for the overexpression of these genes was diminished, but due to their upregulation, we concluded that STAT signaling was inhibited. Similar to this result but by doing just the opposite of our experimental setup, Quentmeier et al. have reported that, when SOCS2 was knocked down via RNAi mechanism in cytokine-dependent JAK2V617F mutation positive cell line MB–02, STAT5 phosphorylation was increased, whereas they could not define the underlying mechanism of it [26]. Upon the downregulated genes, CEBPB came forward due to the −185.60-fold decrease of expression in anti-S5A CHL group and led us to think whether activation of this gene might be a new therapeutic target for CML. Because Guerzoni C et al. have reported that CEBPB expression was downregulated in BCR-ABL-positive cells and when they treated leukemia cells with imatinib for 24 h, CEBPB expression was increased about fourfold, and proliferation of leukemic cells was inhibited; thus, it could be a new target gene for CML [27]. Besides this, while CRK expression was highly downregulated by −111.63-fold decrease, MYC was upregulated from “oncogenes” with 2.33-fold with anti-STAT5A CHL siRNA. These results indicated that leukemic cell proliferation was balanced, cancer cells had tendency to survive, and maybe due to the upregulation of MYC, the cells were protected from apoptosis and passed to proliferative phase again. Similar to our results, Ulger et al. have used microarray technology for identification of candidate cancer-related genes in the promyelocytic cell line HL-60 and reported that after genomewide analyses, while MYC was upregulated, CRK was downregulated [28].
Silencing STAT5B with either modified or unmodified siRNAs resulted with upregulation of PIAS2 from “STAT inhibitors” and downregulation of STAM, STAT5B, SOCS4, and SOCS5 expressions. As PIAS2 is a negative regulator of STATs that binds to activated STAT dimers and prevent them from binding to DNA, upregulation of PIAS2 after silencing both STAT5A and -5B revealed that, cytokine/growth factors’ signaling that was responsible for activating JAK/STAT pathway was switched off. The SOCS4 and SOCS5 genes are discovered recently as inducible feedback inhibitors, and they are labile and undergo proteasome-dependent degradation in a few hours within the cell due to their 1–2 h long half-life [29] we speculated that, the extra need for such a negative control mechanism was reduced due to STAT5 knockdown, and their endogenous levels might already be very low in cells to make a considerable impact upon leukemic cells at the sixth day of analyses.
Finally, there are two other important data seen at the end of array analyses. Anti-apoptotic gene BCL2L1 expression was upregulated in each siRNA application group. The upregulation of BCL2L1 could explain why any apoptotis was not promoted in leukemic cells after the 96th hour of the siRNA treatments which caused tumor cells to go under proliferative phase again and escape from apoptosis. Eventually, STAT1 transcription factor expression was upregulated in each siRNA application group, but this increase was significant especially in STAT5 groups. Because STAT1 exerts a pro-apoptotic effect, it is a desired and inevitable result and the cause of high apoptotic leukemic cell rate seen after siRNA treatments. The expressional increase in STAT1 was balanced with BCL2L1 and MYC expressional upregulation due to the inhibition of STAT5, and this resulted with regulated uncontrolled proliferation and apoptosis of leukemia cells.
Taken all together, since RNAi has been regarded as a pioneering tool by targeting biological processes and we have proved its utility in an in vitro environment, these results might have contribution to clinics, and we think that especially modified siRNA could be an alternative therapeutic strategy in leukemia that gained resistance to drugs in the future with targeted delivery of siRNA in vivo with different choices like nonviral carrier Trojan horses, cholesterol–siRNA conjugates, liposomes and lipoplexes, folate-conjugated siRNA carriers, or antibody-conjugated siRNA carriers [30].
Conclusion
This study showed that by the application of specific siRNAs, expressions of target STATs was suppressed both at gene and protein levels, and apoptosis of leukemic cells was induced. Modified siRNAs were highly efficient and more effective than unmodified siRNAs in an in vitro environment for duration of 12 days. After array analysis, a decrease in the expressions of signal transducer and STAT phosphorylation inducer genes, cytokines, transcription factors, and oncogenes; and increase in the expressions of STAT inhibitors could be observed. In conclusion, we think that in the future, siRNA applications could constitute a new and alternative curative method for the supporting therapy especially of CML-diagnosed patients who gained resistance to treatment.
References
Melo JV, Hughes TP, Apperley JF (2003) Chronic myeloid leukemia. Hematol Am Soc Hematol Educ Program 1:132–152
Kantarjian H, Melo JV, Tura S, Giralt S, Talpaz M (2000) Chronic myelogenous leukemia: disease biology and current and future therapeutic strategies. Hematol Am Soc Hematol Educ Program 1:90–109
Kantarjian H, Talpaz M, Giles F, O'Brien S, Cortes J (2006) New insights into the pathophysiology of chronic myeloid leukemia and imatinib resistance. Ann Intern Med 145(12):913–923
Vaidya S, Ghosh K, Vundinti BR (2011) Recent developments in drug resistance mechanism in chronic myeloid leukemia: a review. Eur J Haematol 87(5):381–393
Valentino L, Pierre J (2006) JAK/STAT signal transduction: regulators and implication in hematological malignancies. Biochem Pharmacol 71(6):713–721
Rawlings JS, Rosler KM, Harrison DA (2004) The JAK/STAT signaling pathway. J Cell Sci 117(Pt 8):1281–1283
Levy DE, Gilliland DG (2000) Divergent roles of STAT1 and STAT5 in malignancy as revealed by gene disruptions in mice. Oncogene 19(21):2505–2510
Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE Jr (1996) Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A 93(15):7673–7678
Yu H, Jove R (2004) The STATs of cancer—new molecular targets come of age. Nat Rev Cancer 4(2):97–105
Ilaria RL Jr, Van Etten RA (1996) P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem 271(49):31704–31710
Donato NJ, Wu JY, Zhang L, Kantarjian H, Talpaz M (2001) Down-regulation of interleukin-3/granulocyte-macrophage colony-stimulating factor receptor beta-chain in BCR-ABL(+) human leukemic cells: association with loss of cytokine-mediated Stat-5 activation and protection from apoptosis after BCR-ABL inhibition. Blood 97(9):2846–2853
Nieborowska-Skorska M, Wasik MA, Slupianek A, Salomoni P, Kitamura T, Calabretta B, Skorski T (1999) Signal transducer and activator of transcription (STAT)5 activation by BCR/ABL is dependent on intact Src homology (SH)3 and SH2 domains of BCR/ABL and is required for leukemogenesis. J Exp Med 189(8):1229–1242
Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernández-Luna JL, Nuñez G, Dalton WS, Jove R (1999) Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10(1):105–115
Jing N, Tweardy DJ (2005) Targeting Stat3 in cancer therapy. Anticancer Drugs 16(6):601–607
Benekli M, Baumann H, Wetzler M (2009) Targeting signal transducer and activator of transcription signaling pathway in leukemias. J Clin Oncol 27(26):4422–4432
Whitehead KA, Langer R, Anderson DG (2009) Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov 8(2):129–138
Chiu YL, Rana TM (2003) siRNA function in RNAi: a chemical modification analysis. RNA 9(9):1034–1048
Bowman T, Garcia R, Turkson J, Jove R (2000) STATs in oncogenesis. Oncogene 19(21):2474–2488
Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J (2000) Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 289(5486):1938–1942
Crombez L, Aldrian-Herrada G, Konate K, Nguyen QN, McMaster GK, Brasseur R, Heitz F, Divita G (2009) A new potent secondary amphipathic cell-penetrating peptide for siRNA delivery into mammalian cells. Mol Ther 17(1):95–103
Buettner R, Mora LB, Jove R (2002) Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res 8(4):945–954
Chai SK, Nichols GL, Rothman P (1997) Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol 159(10):4720–4728
Kosova B, Tezcanli B, Ekiz HA, Cakir Z, Selvi N, Dalmizrak A, Kartal M, Gunduz U, Baran Y (2010) Suppression of STAT5A increases chemotherapeutic sensitivity in imatinib-resistant and imatinib-sensitive K562 cells. Leuk Lymphoma 51(10):1895–1901
Nowicki MO, Pawlowski P, Fischer T, Hess G, Pawlowski T, Skorski T (2003) Chronic myelogenous leukemia molecular signature. Oncogene 22(25):3952–3963
Piva R, Pellegrino E, Mattioli M, Agnelli L, Lombardi L, Boccalatte F, Costa G, Ruggeri BA, Cheng M, Chiarle R, Palestro G, Neri A, Inghirami G (2006) Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and BCL2A1 as critical target genes. J Clin Invest 116(12):3171–3182
Quentmeier H, Geffers R, Jost E, Macleod RA, Nagel S, Röhrs S, Romani J, Scherr M, Zaborski M, Drexler HG (2008) SOCS2: inhibitor of JAK2V617F-mediated signal transduction. Leukemia 22(12):2169–2175
Guerzoni C, Bardini M, Mariani SA, Ferrari-Amorotti G, Neviani P, Panno ML, Zhang Y, Martinez R, Perrotti D, Calabretta B (2006) Inducible activation of CEBPB, a gene negatively regulated by BCR/ABL, inhibits proliferation and promotes differentiation of BCR/ABL-expressing cells. Blood 107(10):4080–4089
Ulger C, Toruner GA, Alkan M, Mohammed M, Damani S, Kang J, Galante A, Aviv H, Soteropoulos P, Tolias PP, Schwalb MN, Dermody JJ (2003) Comprehensive genome-wide comparison of DNA and RNA level scan using microarray technology for identification of candidate cancer-related genes in the HL-60 cell line. Cancer Genet Cytogenet 147(1):28–35
Segatto O, Anastasi S, Alemà S (2011) Regulation of epidermal growth factor receptor signalling by inducible feedback inhibitors. J Cell Sci 124(11):1785–1793
Shim MS, Kwon YJ (2010) Efficient and targeted delivery of siRNA in vivo. FEBS J 277(23):4814–4827
Acknowledgments
We would like to thank Dr. Vildan Bozok ÇETİNTAŞ, Phd (Ege University Medical School, of Medical Biology Department) for the great help in creating figures of the manuscript. This work was supported by The Scientific and Technological Research Council of Turkey, by the Turkish Society of Hematology, and Ege University Medical School Scientific Research Projects (TUBITAK 105S459, THD 200–02/08 and APAK 2010-TIP–004 respectively, to BK).
Conflict of interest
The authors declare no potential conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 273 kb)
Rights and permissions
About this article
Cite this article
Kaymaz, B.T., Selvi, N., Gündüz, C. et al. Repression of STAT3, STAT5A, and STAT5B expressions in chronic myelogenous leukemia cell line K–562 with unmodified or chemically modified siRNAs and induction of apoptosis. Ann Hematol 92, 151–162 (2013). https://doi.org/10.1007/s00277-012-1575-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-012-1575-2