Abstract
Background
Computed tomography (CT) has been increasingly used to quantify abdominal muscle and fat in clinical research studies, and multiple studies have shown importance of body composition in predicting clinical outcome. The purpose of study is to compare newly developed semi-automated software (BodyCompSlicer) to commercially available validated software (Slice-O-Matic) for CT body composition analysis.
Methods
CT scans of abdomen at L3 level in 30 patients were analyzed by two reviewers and using two softwares (BodyCompSlicer and Slice-O-Matic). Body composition analysis using BodyCompSlicer was semi-automated. The program automatically segmented subcutaneous fat (SF), skeletal muscle (SM), and visceral fat (VF) areas. Reviewers manually corrected the segmentation using computer–mouse interface as necessary. Body composition analysis using Slice-O-Matic was performed by manually segmenting each area using computer-mouse interface (brush tool). After segmentation, SM, SF, and VF areas were calculated using CT attenuation thresholds. Inter-observer and inter-software variability of measurements were analyzed using intraclass correlation coefficients (ICC) and coefficient of variation (COV).
Results
Inter-observer ICC and COV using BodyCompSlicer were 0.997 and 1.5% for SM, 1.000 and 0.8% for SF, and 1.000 and 1.0% for VF, whereas those using Slice-O-Matic were 0.993 and 2.5% for SM, 0.995 and 3.1% for SF, and 0.999 and 2.3% for VF. Inter-software ICCs and COV were 0.995–0.995 and 2.0–2.1% for SM, 0.991–0.994 and 3.4–3.9% for SF, and 0.998–0.998 and 2.8–3.3% for VF. Time to analyze 30 cases was 70–100 min and 150–180 min using BodyCompSlicer and Slice-O-Matic, respectively.
Conclusion
BodyCompSlicer is comparable to Slice-O-Matic for CT body composition analysis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Computed tomography (CT) has been increasingly used to quantify abdominal muscle and fat in clinical research studies, and multiple studies have shown importance of body composition in predicting clinical outcome. Low skeletal muscle mass, which, in its severe form, is described as sarcopenia has been shown to be associated with adverse oncologic, surgical, and overall survival outcomes in multiple malignancies in the gastrointestinal tract, genitourinary tract, breast, and skin [1,2,3,4,5,6,7,8,9,10] as well as in non-malignant conditions [11]. Increased visceral fat has been associated with numerous metabolic aberrations including insulin resistance, type 2 diabetes mellitus, hypertension, hyperlipidemia, and low-grade inflammatory conditions such as non-alcoholic steatohepatitis and Barrett’s esophagus [12,13,14,15,16].
There are several commercially available software packages that quantify abdominal fat using CT. However, to our knowledge, there are only a few programs available that can quantify skeletal muscle in the abdomen using CT including the Slice-O-Matic [1, 17, 18] and ImageJ-based software [19]. In particular, body composition measurement using Slice-O-Matic has been validated against DEXA scans as well as body impedance absorptiometry, with excellent performance [18]. The software applications segment the muscle and fat from internal organs using human inputs from a mouse–computer interface without use of automated segmentation. This approach overcomes the difficulty in separating muscle and visceral fat from internal organs (such as bowel, liver, kidneys) by automated segmentation, but it introduces the potential for low inter-reader reliability and limits the scalability of the software.
We recently developed software entitled BodyCompSlicer, which automatically segments a single-slice CT image at the third lumbar (L3) level into subcutaneous fat, skeletal muscle, and visceral fat compartments. The software is written in Matlab (Matlab 2015b, MathWorks, Natick, MA) language. The software runs multiple sequential morphological operations including attenuation thresholding with prior knowledge of location, size, and shape of different structures. While the software automatically segments muscle and fat, the boundaries can be manually fine-tuned with modification of the automatically segmented boundaries using a mouse–computer interface.
The purpose of the study is to compare the body composition measurements analyzed using the BodyCompSlicer and the commercially available and validated Slice-O-Matic program.
Materials and methods
Patient population
433 patients with renal cell carcinoma (RCC) or bladder carcinoma (TCC) who underwent preoperative CT were included in the study. These cases were selected from 592 patients (387 with RCC and 205 with TCC) who were previously included in different studies [4, 20] by excluding patients with chronologically most remote CT studies (154 with RCC and 5 with TCC). 400 cases (200 cases each from RCC and TCC cohort) were used as a calibration set, three cases as a training set (RCC cohort), and remaining 30 cases (most recent cases from RCC cohort) as a validation set. The patients in the validation set included 20 males and 10 females with a mean age of 73 years (range 43–86). Mean weight, height, and body mass index were 79 kg (range 42-118), 1.70 m (range 1.38–1.88), and 27.2 (range 17.9–34.5), respectively.
CT Protocol and image analysis
CT scans of the abdomen were obtained as part of preoperative evaluation of renal cell carcinoma or bladder carcinoma. Slice thickness ranged from 2.5 to 7.0 mm with a median of 5 mm. One radiologist (MRM) selected a CT image at the L3 level (transverse process) on a workstation, and the image was saved in the Digital Imaging and Communications in Medicine (DICOM) format. The DICOM images were transferred to a personal computer for image analysis. Two reviewers (SPP, MS) analyzed the CT images using BodyCompSlicer and Slice-O-Matic (version 5.0; TomoVision, Quebec, Canada) at separate time-points, blinded to the results of the analysis by the alternate methodologies [18, 21, 22].
Each reviewer had extensive experience using one of the two software packages analyzing over 400 cases (including all cases in the calibration set), but had not used the other software (reviewer 1 had used Slice-O-Matic, reviewer 2 had used BodyCompSlicer). First, each reviewer received training from the other reviewer using the training set cases (n = 3). The training session was approximately 1 h long in total. During the training session, each reviewer agreed upon the definition of the boundaries of the compartments.
Next, calibration set cases (n = 400) were evaluated using each software. Reviewer 1 used Slice-O-Matic and reviewer 2 used BodyCompSlicer. The data were used to calibrate the BodyCompSlicer to minimize the systematic difference in the measurements between the two software packages. Finally, CT images of 30 cases (validation set) were evaluated by each reviewer first using Slice-O-Matic and then using BodyCompSlicer. Each session evaluating validation set was completed in one sitting. The time to complete 30 cases was recorded for each session.
BodyCompSlicer protocol
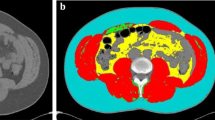
Once the CT image was loaded into the software, the software automatically calculated and placed three boundary lines between external air and subcutaneous fat (boundary 1), between subcutaneous fat and abdominal wall/ paraspinal muscles (boundary 2), and between abdominal wall/paraspinal muscles and visceral fat (boundary 3) (Fig. 1A). The reviewers carefully inspected the boundaries, and manually corrected the boundaries using the mouse–computer interface as necessary (Fig. 1B). When the correction of all boundaries was complete, the software calculated the skeletal muscle area, subcutaneous fat area, and visceral fat area (cm2). Skeletal muscle area was calculated as area containing pixels between boundary 2 and 3, and having CT attenuation value of −30 to 150 HU, but excluding the spine. Subcutaneous fat area was calculated as area containing pixels between the boundary 1 and 3, and having CT attenuation value of −190 to −30 HU. For the purpose of the study, fat in the skeletal muscle compartment was considered as part of subcutaneous fat. Visceral fat area was calculated as area containing pixels within the boundary 3, and having CT attenuation value of −150 to −30 HU, but excluding bowel content. The software automatically created masks for bone and colonic content; these masks were used to exclude bone and colonic content from being included as muscle or fat (Table 1). Once segmentation of all patients is completed, body composition data are exported in a tabular format (Fig. 2).
Axial CT image of abdomen at third lumbar level. The BodyCompSlicer automatically segmented CT image into subcutaneous fat, skeletal muscle, and visceral fat compartments, and the boundaries are displayed (lines are thickened for ease of visualization) (A). The boundaries after manual correction (arrows) (B)
Slice-O-Matic Protocol
Once the CT image was loaded into the software, the reviewer manually segmented subcutaneous fat area, visceral fat area, and skeletal muscle area using a mouse-operated paint brush tool method. Threshold for pixel selection was first set to −190 to −30 HU for subcutaneous and intramuscular fat, −150 to −30 HU for visceral fat, and −29 to 150 HU for skeletal muscle. Using a mouse-operated paint brush tool, the areas of interest (fat or skeletal muscle) were selected. The software automatically selected the pixels that meet the attenuation threshold along the path of the paint brush tool. The procedure was repeated until the structure of interest was selected completely. If the segmented area extended beyond the structure of interest, manual correction using mouse-operated eraser tool was performed. Once the segmentation is complete, the software calculated the skeletal muscle area, subcutaneous fat area, and visceral fat area (cm2).
Analysis of calibration set data and fine tuning of BodyCompSlicer
The skeletal muscle area, subcutaneous fat area, and visceral fat area of the entire calibration set (n = 400) from both software programs were compared to identify systematic difference in the measurements. The BodyCompSlicer draws boundary lines between two different structures of interest (fat, muscle, or air). The pixels on the boundary lines were included in the compartment inside the line at the default setting. These settings were adjusted for each boundary to minimize the systematic difference in the measurements.
Analysis of validation set data
The body composition measurements for the validation set were calculated using the method determined by the analysis of calibration set. The inter-software and inter-observer variability of skeletal muscle area, subcutaneous fat area, and visceral fat area measurements were analyzed using intraclass correlation coefficients (ICC), coefficient of variation, and Bland–Altman analysis. For the Bland–Altman analysis, the mean difference and mean percentage difference in measurements and 95% limits of agreement (1.96 × SD of mean difference) were calculated. The difference between the two software packages was defined as measurement using Slice-O-Matic subtracted by measurement using BodyCompSlicer. Statistical analysis was performed using Matlab software.
Accuracy of automated segmentation
The Dice similarity indices, level of similarity between the segmented areas and the reference, were calculated for the areas inside the automatically calculated boundaries 1, 2, and 3 by BodyCompSlicer without manual correction with the manually corrected boundaries as Ref. [23]. The Dice similarity index of 1 indicates perfect match of the segmented areas and the reference, whereas 0 indicates no overlap between the two. All data analyses were performed by a single investigator (BC).
Results
Calibration set
The mean percentage difference in the measurements of the skeletal muscle area, subcutaneous fat area, and visceral fat area were −2.7%, 0.6%, and 3.3%, respectively, between the two software packages at the default settings. The BodyCompSlicer had a tendency to underestimate the skeletal muscle area and overestimate visceral fat. To minimize these biases, the boundary 2 was shifted by 1 pixel outward and the boundary 3 was shifted by 1 pixel inward for the calculation of the body composition. After the adjustment, the mean percentage differences in the measurements of the skeletal muscle area, subcutaneous fat area, and visceral fat area were 0.0%, 2.7%, and 0.9%, respectively.
Validation set
The mean skeletal muscle area, subcutaneous fat area, and visceral fat area measurements for each reviewer and software using the validation set are summarized in Table 2. ICC, Bland–Altman analysis, and coefficient of variation between the two software packages and two reviewers are summarized in Table 3.
Inter-observer ICCs using the BodyCompSlicer was 0.997 for skeletal muscle area, 1.000 for subcutaneous fat area, and 1.000 for visceral fat area. The coefficient of variation of two reviewers using the BodyCompSlicer was 1.5% for skeletal muscle area, 0.8% for subcutaneous fat area, and 1.0% for visceral fat area. The 95% limit of agreement of two reviewers using the BodyCompSlicer was 6.2 cm2 and 4.1% for skeletal muscle area, 4.4 cm2 and 2.6% for subcutaneous fat area, and 4.3 cm2 and 6.2% for visceral fat area.
Inter-software ICCs were 0.995 for skeletal muscle area, 0.991–0.994 for subcutaneous fat area, and 0.998 for visceral fat area. The coefficient of variation of two software packages was 2.0%–2.1% for skeletal muscle area, 3.4%–3.9% for subcutaneous fat area, and 2.8%–3.3% for visceral fat area. The 95% limit of agreement of two software packages was 8.1–8.6 cm2 and 6.8%–7.1% for skeletal muscle area, 8.8–15.0 cm2 and 4.2%–6.7% for subcutaneous fat area, and 11.8–14.2 cm2 and 8.7–15.0% for visceral fat area. The systematic differences in the area measurements between the two software packages were negligible for skeletal muscle area and visceral fat area. The mean subcutaneous fat area was 4.9–9.4 cm2 and 3.0%–5.2% higher using BodyCompSlicer compared to Slice-O-Matic.
The time to analyze 30 cases was 100 min (3.3 min/case) and 70 min (2.3 min/case) using the BodyCompSlicer for reviewer 1 and 2, respectively, and 150 min (5.0 min/case) and 180 min (6.0 min/case) using the Slice-O-Matic for reviewer 1 and 2, respectively.
Accuracy of Automated Segmentation
Dice similarity indices for areas within boundary 1, 2, and 3 were 1.000 [1.000–1.000], 0.990 [0.959–1.00], and 0.978 [0.801–0.997] (median, [5 percentile, 95 percentile]), respectively.
Discussion
This present study showed that inter-observer agreement using the BodyCompSlicer was very high and inter-software agreement between the BodyCompSlicer and Slice-O-Matic was comparable to the inter-observer agreement of the Slice-O-Matic. These results suggest that the accuracy and reproducibility of the BodyCompSlicer is at least comparable to the Slice-O-Matic.
The previously reported inter-observer coefficient of variation ranged from 2% to 5% for abdominal skeletal muscle area and 0.4-2.5% for subcutaneous fat or visceral fat areas [24,25,26], and intra-observer coefficient of variation was 0.3% (using same slice) or 1.3-1.6% (using two consecutive slices) for abdominal skeletal muscle area [1, 17, 18]. The inter-observer coefficient of variation in our study is comparable to the previous reports.
High inter-observer agreement does not guarantee the accuracy of measurements. When automated segmentation is used without any manual correction, the inter-observer coefficient of variation and limit of agreement are theoretically zero (perfect agreement). Therefore, we assessed the agreement between inter-software agreement between BodyCompSlicer and the Slice-O-Matic. The inter-software agreement was comparable to the inter-observer agreement using Slice-O-Matic alone.
The BodyCompSlicer has integrated the automated masking of bone and colonic contents. Part of the bone may have soft tissue attenuation (−30 to 150HU), and could be erroneously categorized as skeletal muscle without masking. Similarly, spinal canal has soft tissue attenuation and could be erroneously categorized as skeletal muscle without masking. Intracolonic air may have fat attenuation (−150 to −30HU) when mixed with fluid. A previous study demonstrated that colonic air resulted in overestimation of visceral fat area by 20% when masking was not used [27].
In the calibration set, the skeletal muscle area was underestimated by 2.8% using the default setting of the BodyCompSlicer compared to Slice-O-Matic. This is likely from the difference in segmentation methods. The BodyCompSlicer draws smooth, curved, or straight boundary lines based on CT attenuation and other factors. On the other hand, the segmentation by Slice-O-Matic is strictly based on CT attenuation values. By enlarging the skeletal muscle compartment by 1 pixel both at inner and outer boundaries, the underestimation of skeletal muscle was nearly completely corrected. The inter-software mean percentage difference was 0.2% in the calibration set after adjustment and −0.9% to 0.4% in the validation set after the adjustment.
The major advantage of the BodyCompSlicer is the use of automated segmentation and reduction in time to evaluate cases. The time to analyze 30 cases was 70 and 100 min for experienced and inexperienced reviewer using the BodyCompSlicer compared to 150 and 180 min using the Slice-O-Matic. The automated segmentation is not perfect, and the BodyCompSlicer almost always required some degree of manual correction. This is particularly true for the boundary between skeletal muscle and visceral fat compartments (boundary 3). The presence of internal organs abutting the abdominal wall muscle and complex contour of the paraspinal muscles makes automated segmentation more difficult. The Dice similarity index was lowest for the boundary 3 with a median value of 0.978. The major disadvantage of the BodyCompSlicer is that it cannot handle CT images at different anatomical locations such as thigh. This is because the software automatically segments the different compartments using prior knowledge of location, size, and shape of different structures. In addition, MR images cannot be analyzed.
In conclusion, the body composition measurements obtained using the BodyCompSlicer are comparable to those obtained using the Slice-O-Matic.
References
Martin L, Birdsell L, Macdonald N, et al. (2013) Cancer cachexia in the age of obesity: skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. J Clin Oncol 31:1539–1547
Del Fabbro E, Parsons H, Warneke CL, et al. (2012) The relationship between body composition and response to neoadjuvant chemotherapy in women with operable breast cancer. Oncologist 17:1240–1245
Mir O, Coriat R, Dhooge M, et al. (2012) Feasibility of gemcitabine and oxaliplatin in patients with advanced biliary tract carcinoma and a performance status of 2. Anticancer Drugs 23:739–744
Psutka SP, Carrasco A, Schmit GD, et al. (2014) Sarcopenia in patients with bladder cancer undergoing radical cystectomy: impact on cancer-specific and all-cause mortality. Cancer 120:2910–2918
Joglekar S, Asghar A, Mott SL, et al. (2014) Sarcopenia is an independent predictor of complications following pancreatectomy for adenocarcinoma. J Surg Oncol 111:771–775
Kirihara Y, Takahashi N, Hashimoto Y, et al. (2013) Prediction of pancreatic anastomotic failure after pancreatoduodenectomy: the use of preoperative, quantitative computed tomography to measure remnant pancreatic volume and body composition. Ann Surg 257:512–519
Tan BH, Birdsell LA, Martin L, Baracos VE, Fearon KC (2009) Sarcopenia in an overweight or obese patient is an adverse prognostic factor in pancreatic cancer. Clin Cancer Res 15:6973–6979
Torres ML, Hartmann LC, Cliby WA, et al. (2013) Nutritional status, CT body composition measures and survival in ovarian cancer. Gynecol Oncol 129:548–553
Sabel MS, Lee J, Cai S, et al. (2011) Sarcopenia as a prognostic factor among patients with stage III melanoma. Ann Surg Oncol 18:3579–3585
Thoresen L, Frykholm G, Lydersen S, et al. (2013) Nutritional status, cachexia and survival in patients with advanced colorectal carcinoma. Different assessment criteria for nutritional status provide unequal results. Clin Nutr 32:65–72
Tsien C, Shah SN, McCullough AJ, Dasarathy S (2013) Reversal of sarcopenia predicts survival after a transjugular intrahepatic portosystemic stent. Eur J Gastroenterol Hepatol 25:85–93
Bray GA (2004) Medical consequences of obesity. J Clin Endocrinol Metab 89:2583–2589
Weltman A, Despres JP, Clasey JL, et al. (2003) Impact of abdominal visceral fat, growth hormone, fitness, and insulin on lipids and lipoproteins in older adults. Metabolism 52:73–80
Sironi AM, Gastaldelli A, Mari A, et al. (2004) Visceral fat in hypertension: influence on insulin resistance and beta-cell function. Hypertension 44:127–133
Thomas EL, Hamilton G, Patel N, et al. (2005) Hepatic triglyceride content and its relation to body adiposity: a magnetic resonance imaging and proton magnetic resonance spectroscopy study. Gut 54:122–127
Cook MB, Greenwood DC, Hardie LJ, Wild CP, Forman D (2008) A systematic review and meta-analysis of the risk of increasing adiposity on Barrett’s esophagus. Am J Gastroenterol 103:292–300
Prado CM, Lieffers JR, McCargar LJ, et al. (2008) Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: a population-based study. Lancet Oncol 9:629–635
Mourtzakis M, Prado CM, Lieffers JR, et al. (2008) A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl Physiol Nutr Metab 33:997–1006
Gomez-Perez SL, Haus JM, Sheean P, et al. (2015) Measuring abdominal circumference and skeletal muscle from a single cross-sectional computed tomography image: A Step-by-Step Guide for Clinicians Using National Institutes of Health ImageJ. JPEN J Parenter Enter Nutr 21:0148607115604149
Psutka SP, Boorjian SA, Moynagh MR, et al. (2015) Decreased skeletal muscle mass is associated with an increased risk of mortality after radical nephrectomy for localized renal cell cancer. J Urol 195:270–276
Mitsiopoulos N, Baumgartner RN, Heymsfield SB, et al. (1985) Cadaver validation of skeletal muscle measurement by magnetic resonance imaging and computerized tomography. J Appl Physiol 1998(85):115–122
Prado CM, Birdsell LA, Baracos VE (2009) The emerging role of computerized tomography in assessing cancer cachexia. Curr Opin Support Palliat Care 3:269–275
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Irving BA, Weltman JY, Brock DW, et al. (2007) NIH ImageJ and Slice-O-Matic computed tomography imaging software to quantify soft tissue. Obesity 15:370–376 ((Silver Spring))
Hicks GE, Simonsick EM, Harris TB, et al. (2005) Cross-sectional associations between trunk muscle composition, back pain, and physical function in the health, aging and body composition study. J Gerontol A Biol Sci Med Sci 60:882–887
Keller A, Gunderson R, Reikeras O, Brox JI (2003) Reliability of computed tomography measurements of paraspinal muscle cross-sectional area and density in patients with chronic low back pain. Spine 28:1455–1460 ((Phila Pa 1976))
Potretzke AM, Schmitz KH, Jensen MD (2004) Preventing overestimation of pixels in computed tomography assessment of visceral fat. Obes Res 12:1698–1701
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funding was received for this study.
Conflict of interest
Mayo Clinic and one author (Naoki Takahashi) have intellectual property rights relevant to the technology studied in this paper and a potential financial interest. The other authors had control over the data and information submitted for publication.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. For this type of study formal consent is not required.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Takahashi, N., Sugimoto, M., Psutka, S.P. et al. Validation study of a new semi-automated software program for CT body composition analysis. Abdom Radiol 42, 2369–2375 (2017). https://doi.org/10.1007/s00261-017-1123-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-017-1123-6