Abstract
Purpose
Deep learning is an emerging reconstruction method for positron emission tomography (PET), which can tackle complex PET corrections in an integrated procedure. This paper optimizes the direct PET reconstruction from sinogram on a long axial field of view (LAFOV) PET.
Methods
This paper proposes a novel deep learning architecture to reduce the biases during direct reconstruction from sinograms to images. This architecture is based on an encoder-decoder network, where the perceptual loss is used with pre-trained convolutional layers. It is trained and tested on data of 80 patients acquired from recent Siemens Biograph Vision Quadra long axial FOV (LAFOV) PET/CT. The patients are randomly split into a training dataset of 60 patients, a validation dataset of 10 patients, and a test dataset of 10 patients. The 3D sinograms are converted into 2D sinogram slices and used as input to the network. In addition, the vendor reconstructed images are considered as ground truths. Finally, the proposed method is compared with DeepPET, a benchmark deep learning method for PET reconstruction.
Results
Compared with DeepPET, the proposed network significantly reduces the root-mean-squared error (NRMSE) from 0.63 to 0.6 (p < 0.01) and increases the structural similarity index (SSIM) and peak signal-to-noise ratio (PSNR) from 0.93 to 0.95 (p < 0.01) and from 82.02 to 82.36 (p < 0.01), respectively. The reconstruction time is approximately 10 s per patient, which is shortened by 23 times compared with the conventional method. The errors of mean standardized uptake values (SUVmean) for lesions between ground truth and the predicted result are reduced from 33.5 to 18.7% (p = 0.03). In addition, the error of max SUV is reduced from 32.7 to 21.8% (p = 0.02).
Conclusion
The results demonstrate the feasibility of using deep learning to reconstruct images with acceptable image quality and short reconstruction time. It is shown that the proposed method can improve the quality of deep learning-based reconstructed images without additional CT images for attenuation and scattering corrections. This study demonstrated the feasibility of deep learning to rapidly reconstruct images without additional CT images for complex corrections from actual clinical measurements on LAFOV PET. Despite improving the current development, AI-based reconstruction does not work appropriately for untrained scenarios due to limited extrapolation capability and cannot completely replace conventional reconstruction currently.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
I Introduction
With the development of positron emission tomography (PET) instrumentation, the axial field of view continually increases, leading to the new area of long axial field of view (LAFOV) PET or total-body PET. Compared with the current clinically standard of care axial field of view (FOV), PET system has a range of 26 cm, and the long-axial FOV-PET systems have larger solid angle coverage and longer axial FOV. In addition, a large anatomical region can be covered with one single bed position. Consequently, a large factor can significantly reduce the total PET acquisition time due to the increased sensitivity [1,2,3].
Several image reconstruction algorithms have been proposed to reconstruct tomographic images using projection data. For instance, conventional methods can solve a mapping function from measurement space to image space based on physical principles. The clinically developed methods include the analytical and iterative methods. The analytical methods, such as the filtered back projection (FBP) [4], can achieve fast image reconstruction. However, the obtained images have a high level of noise. The iterative methods, such as the maximum-likelihood expectation–maximization (MLEM) [5] and ordered-subset expectation maximization (OSEM) [6] with iteratively back-projecting and forward-projecting, are clinically accepted standards. The iterative methods can give reconstructed images with the low noise level and satisfactory contrast. However, the iterative process is time-consuming. Moreover, in order to correctly perform attenuation, a computed tomography (CT) or magnetic resonance (MR) image is used to estimate the attenuation. In recent years, neural networks have been used for tomographic image reconstruction to achieve higher quality results with sparse information and short reconstruction time [7, 8]. Deep learning-based methods have also been applied. For instance, a deep learning method is proposed for noise reduction in order to allow low-dose PET imaging protocols [9,10,11]. A neural network is integrated into the iterative process to speed up the convergence speed and improve the reconstruction quality [12, 13]. A neural network is trained to convert directly from projection to image data. For the direct method, an automated method that uses the transform by manifold approximation (AUTOMAP) to learn the relationship between sensor domain and image domain is proposed [14]. This method mainly tackles the MR image reconstruction, while PET system application is also achievable [14]. A deep encoder-decoder network, referred to as DeepPET, is used for the direct reconstruction of PET images. PET images and projection data simulated based on XCAT digital phantom are used to train the network [15]. Kandarpa et al. [16] propose a double U-Net to learn the sinogram-to-image transformation, while the deep-learning pipeline consists of denoising, image reconstruction, and super-resolution segments. William et al. [17] propose a DirectPET network to achieve full-size neural network PET reconstruction from histo-images data. The XCT-based attenuation maps are used as additional input for corrections. The reconstruction from histo-images using a U-net network is also proposed, where a CT-based attenuation is required [18].
The LAFOV of total-body PET increases the probability of LOR detection in order to increase the sensitivity. However, the high obliqueness of the LORs between distant rings suffers from the parallax error [1] and introduces large heterogeneity in the image quality [19, 20]. The increased Compton scattering and ratio between multiple over single scattered photons is another critical bottleneck for the reconstruction of LAFOV PET [21]. The fraction of multiple scatters changes in LAFOV PET [22] heterogeneously. The fractions of random events also depend on the difference of rings in LAFOV PET [20]. The correction of heterogeneity of random and multiple scattered events makes the reconstruction more difficult than in the conventional scanners.
This paper explores the application of the encoder-decoder network to long-axial FOV PET reconstruction using clinical patient data. The study focuses on achieving an end-to-end PET reconstruction directly from the detector to the image domain. In addition, an attenuation correction is integrated into the training process.
II Material and methods
Patients and imaging
Clinical patient list mode data are collected using Biograph Vision Quadra (Siemens Healthineers) at the University of Bern, Switzerland. This system has a FOV of 106 cm. Preliminary assessments of this scanner’s characteristics reveal a sensitivity of 174 cps/kBq and a time of flight (TOF) resolution of 219 ps in ultra-high sensitivity mode [23].
The selected patients are injected with 18F-FDG and undergo a PET/CT examination, including 80 cases (median age, 66 years; age range, 27–83 years; 36 females; BMI, 25.40 ± 4.70 kg/m2) of patients. In all the cases, the subjects fast for more than 4 h and have blood glucose less than 200 mg/dl. All the patients are injected with 18F-FDG with an uptake time of 90 min ± 10%. The patients without complete PET/CT scan images from above head to below thigh, and those with poor image quality because of movement, are excluded from the study. The 80 patients are randomly split into a training dataset of 60 patients (median age, 67 years; age range, 27–83 years; 26 females; BMI, 25.43 ± 4.71 kg/m2), a validation dataset of 10 patients (median age, 62 years; age range, 58–75 years; 5 females; BMI, 26.72 ± 5.96 kg/m2), and a test dataset of 10 patients (median age, 67 years; age range, 40–81 years; 5 females; BMI, 23.92 ± 2.11 kg/m2). Note that this study is performed following the requirements of the respective local ethics committees in Switzerland (Req-2021–00,517).
Data pre-processing
List-mode data obtained from the scanner is reconstructed using a dedicated software prototype (e7-tools, Siemens Healthineers) with CT-based attenuation correction. As performed in our clinical routine, the PET images were reconstructed using PSF-TOF with 4 iterations and 5 subsets [23]. The 3D sinograms are converted into 2-dimensional (2D) slices using single-slice rebinning (SSRB) [4]. A stack of 2D sinograms is created by placing detected events on the plane, perpendicular to the scanner axis (z) and lying in the middle of the line connecting the two detectors of the event. The image matrix size of 2D image volumes is 440 × 440 with dimensions of 1.65 mm × 1.65 mm. A total of 644 2D sinogram slices are obtained for each patient, corresponding to 644 reconstructed images.
The input dataset of the network is the 2D sinogram slices, and the reconstructed images from e7-tools are used as training targets. Several images of the starting and ending positions of each patient’s data have a low count, and therefore they are excluded from the study. Each patient retains 599 sets of data (2D sinogram and reconstructed image). Finally, 60 patients and 10 patients are respectively designated for training validation, while 10 patients are considered for testing.
Deep neural network structure
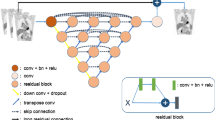
An encoder-decoder network is developed for direct image reconstruction. It comprises two parts: image transform and perceptual loss networks [24] (cf. Figure 1).
The structure of the proposed training network is based on DeepPET [15]. The network consists of the encoder, transformation, and decoder parts (cf. Figure 1, Supplemental Fig. 1, 2) [25]. In addition, 31 convolution blocks and one single convolution layer are involved. Each convolution block includes a convolution layer used to extract features, a batch normalization (BN) layer used to speed up the training and network convergence, and a rectified linear unit (ReLU) activation function. In the decoder and transformation part, the convolution filter for the first two blocks has a size of 7 × 7, that of the following two blocks is of 5 × 5 size, while the others have a size of 3 × 3. The number of extracted features increases from 32 to 1024. The convolution layer decreases the widths and lengths of the feature maps with a kernel stride of 2. In the decoder part, the convolution filter has a size of 3 × 3, and the feature maps are enlarged by upsampling layers. The output layer is a convolution layer with one feature. The 2D sinogram slices are resized to 288 × 269 and used as inputs of the network. The outputs of the image transform network are reconstructed images that are put in the perceptual loss network. Finally, this network comprises 64,544,865 parameters.
The perceptual loss network uses the first 3 convolution blocks of VGG19 [26] (cf. Supplemental Fig. 2). The VGG network uses the accumulation of multiple small-scale convolution kernels (3 × 3) rather than large-scale convolution kernels. This establishment can form multiple non-linear layers to increase the depth of the network and achieve complex feature learning. The convolution blocks in VGG19 include a convolution layer followed by a ReLU activation function. The sizes of the feature maps are reduced by pooling layers. The first 3 convolutional blocks of VGG19 are shallow networks in convolutional neural networks. The features extracted by the first three convolutional blocks are similar to the input and contain more information, such as color, texture, and edge. It can be sufficient to capture the features while remaining robust. In addition, the depth of the three convolutional blocks ensures a sufficient area of receptive field for better reconstruction of structural details. Another consideration of the choice of the first three blocks follows the study of perceptual loss [24]. It has shown that the reconstruction with more than three layers can preserve image content and overall spatial structure while losing color, texture, and exact shape. Another study for PET image fine-tuning found that the features extracted from deeper layers could reduce the quality of the prediction images [9]. The weights of the VGG19 network pre-trained on the ImageNet database (image-net.org) are used. The outputs of the first 3 pooling layers are extracted and used as feature reconstruction loss:
where VGG(x)i represents the output of the i-th pooling layer in VGG19 with the input of ground truth, VGG(y)i denotes the output of the i-th pooling layer in VGG19 with the input of predict image from image transform network.
The other two parts, which are the mean square error (MSE) loss (cf. Equation (2)) and structural similarity (SSIM) loss [27] (cf. Equation (3)), are also involved in the loss function.
where x is the ground truth, y represents the predicted image of the image transform network, and n denotes the total number of image pixels.
where \({u}_{x}\) and \({\sigma }_{x}^{2}\) are respectively the mean and variance of the ground truth image pixels, \({u}_{y}\) and \({\sigma }_{y}^{2}\) are respectively the mean and variance of the predicted image pixels, \({\sigma }_{xy}\) crepresents the covariance of the ground truth and predict images, and \(\mathit{max}\left(x\right)\) denotes the maximum of ground truth image value.
The total loss function is expressed as:
Network training and test procedure
The network is implemented using TensorFlow [28]; the training of the network is performed using a GPU (Tesla V100-PCIe-16 GB, NVIDIA) and tested using GeForce RTX 2080 Ti (NVIDIA). The Adam optimization method [29] is used as an optimizer with a learning rate of 10−4. All the images in the training dataset (35,940 2D sinogram and reconstructed images) are used as input to the network with a batch size of 50 and trained with 300 epochs. The trained network is tested on 10 patients’ data (5590 2D sinograms and reconstructed images).
The network is tested with three noise levels for the input sinograms. Frames of shorter width (1/10 and 1/20) are used to generate sinograms of different noise levels. The sinograms are processed by SSRB and used as the input to the network. The outputs are compared with the reconstruction results of sinograms histogrammed using complete list-mode PET data to evaluate the influence of the noise level of the input sinograms on the AI-based reconstruction results.
Image quality evaluation
The image quality evaluation is processed with the structural similarity index (SSIM), normalized root-mean-squared error (NRMSE), and peak signal-to-noise ratio (PSNR) [27], computed on the regions of the body. SSIM is an index used to measure the similarity of two images. The mean, standard deviation, and covariance estimate the brightness, contrast, and structural similarity, respectively. The values range between 0 and 1. More precisely, a value closer to 1 indicates that the output image is more similar to the target image. The SSIMs are computed as
NRMSE is calculated based on the mean square error (MSE):
where \(\overline{x }\) is the average value of all the pixels in the ground truth image, x represents the ground truth image, and y denotes the predicted image of the network.
The PSNR is computed as
where \({MAX}_{I}\) is the maximum value of the reconstructed image.
Clinical evaluation
The results on the test dataset are further evaluated by 2 nuclear medicine physicians. For each patient, a typical lesion is selected and manually delineated. Among the 10 patients, 1 patient is proved to have no lesion. The mean standardized uptake values (SUVmean) and max standardized uptake values (SUVmax) are measured of the tracer in the selected lesions. The relative errors between the ground truth and DeepPET results and relative errors between the proposed method are calculated and compared. The 3D sinogram data are also reconstructed using the FBP method for comparison, and example visualization and statistics of the comparison are processed.
III Results
We have trained the networks 3 times with random initialization weights. The obtained final loss curves are shown in Fig. 2. The MSE is used as loss of DeepPET. The perspective loss, shown as Eq. (6), is used for the proposed network. We can see that the losses drop significantly at the first 50 epochs of training, and the loss curves of the validation set stop decreasing after the network has been trained for 300 epochs. Therefore, we stopped network training at that point to prevent overfitting.
The average time cost of this work and DeepPET for predicting 1 patient (644 images) are both 14 s, including 7 s for the data preparation process (SSRB) and 7 s for network prediction NVIDIA GeForce RTX 2080 Ti. The research prototype software (e7-tools, Siemens Healthineers) reconstructed a single patient’s images using approximately 200 s for PSFTOF and 320 s for FBPTOF. The time costs of the methods are then compared (cf. Figure 3).
Network test results
As network selection criterion, the MSEs of 10 validation sets for the DeepPET network and the network proposed in this study are 9.21 and 7.63. For 10 test cases, the predicted images of the network proposed in this work and DeepPET are shown in Fig. 4, along with the ground truth and input sinogram. Figure 6 presents the image quality evaluation results (NRMSE, PSNR, and SSIM) obtained by the proposed network for ten test patients. The same patients’ data are also tested using DeepPET. The average results of ten patients are shown in Table 1.
The input images and reconstruction results from different body areas are shown in Fig. 4. It can be seen that a strong similarity exists between the results obtained by the proposed method and the ground truth. Especially in the regions where the tracer uptake is high such as the head, chest, and heart, the results obtained by the proposed method are coherent with the reference values. Point-like high uptake positions exist in the pelvic cavity, legs, and other areas. However, the proposed network can also accurately restore them. In addition, it can be observed that there are some slight structural differences at the edge regions of some structures, such as the details of the brain and the edge of the heart, that are manifested in the blur of the edges. This is mainly due to the fact that the true value image is directly reconstructed from the 3D sinogram, and the 2D sinograms are the input used for prediction in the proposed method. Some information is lost when the 3D sinogram is converted to a 2D sinogram, and errors are introduced. Compared with the test results obtained by the original DeepPET structure network, the image structure restoration and detail restoration are improved, which demonstrates the efficiency of the perceptual loss network introduction.
The quantitative results statistics are shown in Fig. 6, and the uncertainties are listed in supplemental Table 2. It is shown that the SSIM of the original DeepPET structure prediction result compared with the true value is 0.95 ± 0.02, and the network has a 2% improvement in SSIM (which is close to 1) after the perceptual loss structure is introduced. In addition, it can be observed that the proposed network increases the signal-to-noise ratio from 82.02 ± 0.90 to 82.36 ± 0.87, which represents a slight improvement. Moreover, the NRMSE decreases from 0.63 ± 0.06 to 0.60 ± 0.06, which indicates that the reconstructed image is closer to the true value. The quantitative results obtained by the two networks are statically analyzed with paired t test, and they all show a significant improvement.
It can be seen from Fig. 5 that the AI-based reconstruction is robust with noises in the sinograms. The imaging quality matrices (green cross and red point in Fig. 6) of our results from noisy sinograms are at the same level as the original sinograms. This finding is consistent with the existing studies [17], which demonstrated that the neural network produced images using a half-count sinogram nearly equivalent to full-count data. As a comparison, we reconstructed with sinogram of 1/10 and 1/20 width by FBP methods and compared the results with the AI-based results as shown in Fig. 1 (Supplemental Fig. 3, Table 3). With counts decreasing to 1/10 and 1/20, the SSIM decreases by 1.5% and 2.4% for the AI-based method, NRMSE increases by 2.7% and 3.6%, and PSNR decreases by 0.2% and 0.4%, which are much better than the FBP method. For the FBP method, the SSIM decreases by 10.6% and 14.7%, NRMSE increases by 71.4% and 95.4%, and PSNR decreases by 3.8% and 5.5%. The AI-based reconstruction is less sensitive to the count statistics than FBP reconstruction. The deep learning reconstructed images using a low-count sinogram are very similar to the results of full-count data. This is because the convolutional neural network can extract the features of the input data from a larger space, reducing the noise caused by the low count input. The mechanism is that the convolutional layer can extract the features of the input data from a larger space, reducing the noise caused by the low count input.
Comparison between the reconstruction results of the sinogram generated with full frames, with frames of 1/10 width and with frames of 1/20 width. Left to right: sinogram generated with full frames, results of sinograms generated with full frames, with frames of 1/10 width and with frames of 1/20 width
The image quality evaluation results of this work for ten test patients: including root-mean-squared error (NRMSE), peak signal-to-noise ratio (PSNR), and structural similarity index (SSIM). (This work—results of sinograms generated with full frames, this work_n1—sinogram generated with frames of 1/10 width, this work_n2—sinogram generated with frames of 1/20 width)
Clinical evaluation results
The mean standardized uptake values (SUVmean) and max standardized uptake values (SUVmax) of the tracer uptake are measured in a region of interest of lesions (cf. Figure 7) for the test sets (median age, 67 years; age range, 40–81 years; 5 females; BMI, 23.85 ± 2.38 kg/m2). The relative errors between the ground truth, DeepPET, and the proposed method are calculated (cf. Table 2). It can be seen from the comparison for smaller lesions, such as lesions 1 and 6, that the reconstruction results obtained by the proposed method are closer to the ground truth value. For larger lesions, such as lesions 3 and 7, the recovery performed by the proposed method and DeepPET on SUVmax is slightly worse. However, by comparing the shape and contour of the lesion, it can be seen that the results obtained by the proposed method are more similar to the ground truth. The two reconstruction results do not contain enough details for some cases, and lesion 8 is not significantly separated. In addition, the proposed method shows superior performance in anatomical structure with non-intensive uptaken. For instance, in the same layer of lesion 7, it better displays the non-uptaken area in the liver, which is not clearly shown by DeepPET results. In general, compared with DeepPET, the SUVmean and SUVmax of lesions obtained by the proposed method are closer to the ground truths. This indicates that the obtained prediction results can provide a better clinical reference at the lesion level. The currently trained network and DeepPET both have a possible degradation of small lesions such as lesion 2. Compared with the results obtained by the AI methods, the FBP method generates more accurate SUVmean and SUVmax for some lesions, such as lesions 6, 7, and 8. However, as expected, the noise level of the FBP reconstruction results is higher.
IV Discussion
This study follows the mainstream of AI development for PET reconstruction. It focuses on the direct reconstruction from sinogram data. In contrast to most of the existing studies on sinogram data from phantom-based simulation [15] or anthropomorphic simulation by projecting real patient data [30], the training and test in this study are directly performed on real PET measurements. In addition, a critical concern for AI development is its reproducibility and extensibility to complexity in real applications [31]. Compared with the development on simulated sinogram data, the development on real measurement data in this study can better tackle the challenges of physical and physiological complexity. It also enhances the translational potential of data-driven methods.
An advanced LAFOV PET scanner is used to develop and test the AI-based direct reconstruction from Sinogram data. Although conversion of 2D sinogram data of LAFOV PET led to loss of information, the preliminary results demonstrate that the deep neural network can reconstruct PET images with corrections of attenuation and scattering directly from sinogram data without requiring the CT input. The ground truth data used for training are reconstructed PET images with corrections of attenuation and scattering. This potential of AI in complex reconstruction with different corrections may benefit the reconstruction of LAFOV PET, considering the increased complexity in its reconstruction [21]. Although the current study does not consider all the challenging issues, such as the larger and heterogenous solid angles in LAFOV PET reconstruction, the advantage of the AI methods may deal with the complexity and heterogeneity, which encourages the development of this technology. At this stage, the AI-based reconstruction may be less advanced and accurate than the physics-based reconstruction. Further improvements of the input sinogram and training data with more accurate corrections may enhance the performance of this data-driven approach in LAFOV PET reconstruction. In addition, it may eventually reach or outperform the physics-based reconstruction.
Due to a large number of LORs received in LAFOV PET, the storage and processing of the acquisition data are daunting [21, 22]. For instance, the 106-cm LAFOV system has roughly 10 times data to process, compared with a SAFOV PET system. However, when using more oblique LORs, there could be a 40-fold increase [22]. In fact, the prompts count rate peaks at 10 million events, a few orders of magnitude larger than for a traditional PET scanner [21]. The conventional PET reconstruction algorithms are inefficient in processing the vast data of LAFOV PET reconstruction. Although the presented test is performed on sinogram data for reconstruction, the results demonstrate that deep learning can significantly shorten (up to 36 times) the reconstruction time for whole-body imaging, compared with a conventional iterative algorithm. This potential in accelerating the computational speed may bring advantages for the practice of LAFOV PET in the clinical routine.
Based on the lesion demarcation, overall image quality, and visually assessed signal-to-noise ratio, the proposed method improves image quality more than the traditional DeepPET approach. In addition, the semiquantitative measurement method is used. The obtained results are shown in Table 2. This paper estimates a series of lesions located in different organs such as the rib, muscle, mediastinum, and retroperitoneal space soft tissue (cf. Figure 7). Lesions 1 and 2 both present a lesion located in the rib, where the image obtained by the proposed method shows a better-outlined shape than the traditional DeepPET, which is easily misdiagnosed to be located in the sternum. Considering the purpose of optimizing the reconstruction, the outputs show a satisfying performance in presenting the morphological character of the primary lesion with elevator uptake values. The lack of structure details leads to misdiagnosis in the location and conceals some small lesions in worse cases. This may be due to the limited training cases. The use of more varied and larger capacity training sets can improve the prediction’s accuracy. Compared with the actual reconstruction, the currently developed AI-based reconstruction can generally recover the primary anatomy of patients similar to the trained ones. It can generally maintain contrasts and quantitative relations. As we can see from Fig. 7, no artifacts of attenuation or scatter have been observed in the AI-reconstructed images. The AI-reconstructed images look generally smoother than the actual reconstruction. It may miss the lesions in complex anatomy contexts, such as ribs, intercostal spaces, and supra-/sub-clavicular area (e.g., lesion 2, 8). For the quantitative analysis of the lesions, our network led to missing 2 lesions, reduced SUV values for 4 lesions, and increased SUV values for 3 lesions, while the DeepPET led to missing 2 lesions, reduced SUV values for 5, and increased SUV values for 2 lesions. Although our network has lower biases than the DeepPET, the AI-based reconstruction methods are still suboptimal and remain in the early research stage. They are not able to replace conventional reconstruction. Nevertheless, with the proof of concept in this study, it is expected that further development of AI-based reconstruction with a more extensive and diverse training dataset may overcome the limitations and improve the performance. Eventually, the AI-derived results may recapitulate or outperform the conventional reconstruction.
The proportion of female cases is 43%, 50%, and 50% in the training, test, and validation datasets. We compared the test results of the 10 test cases, including 5 males and 5 females. The NRMSEs, PSNRs, and SSIMs of different gender are calculated, and the one-way analysis of variance is processed. It can be observed from Table 3 that all the p values of NRMSE, PSNR, and SSIM were over 0.05, which indicates no significant gender bias for the trained network in this study.
One limitation of this study is the conversion of 3D sinograms to 2D sinograms, where certain noises are introduced, and part of the spatial information in the axial direction is lost [32, 33]. This results in information loss for LAFOV PET and hampers the throughput of image reconstruction. This was due to the large memory requirements of 3D sinograms and our limited GPU memory. The memory required for the processing of one patient can reach 19 Gb, and it is almost impossible to train a 3D network on hundreds of patients with the current GPU capacities in most research institutes. Consequently, a compromise is made to focus on reconstructing 2D sinograms after transformation. With the anticipated increase of computational capacity, exploring 3D AI-based reconstruction can be feasible in the future. Nevertheless, our results demonstrate that AI-based reconstruction can somehow overcome the limitation of 2D sinograms and is relatively robust with information loss. We believe that the current development of 2D AI-based reconstruction on real clinical data can move an important step forward toward the breakthrough of AI technology in PET image reconstruction.
We tested the trained network with NEMA International Electrotechnical Commission (IEC) body phantom [34] and patients with extreme anatomies and found that the network trained on regular clinical data failed in extreme situations. It is known that deep learning is a data-driven method, and the performance of deep learning models depends heavily on the knowledge established from the training data [35,36,37]. Although the AI-based reconstruction methods have several advantages over conventional reconstruction methods, they have limited extrapolation capability and cannot be suitable for untrained scenarios such as physical phantom and extreme anatomy cases in this study. It is a long way to replace traditional reconstruction methods with AI-based methods. Although this study focused on developing and evaluating regular clinical data, it is expected that the developed methodology will work for these extreme situations provided sufficient relevant training data can be prepared in future works.
This study uses sinograms and reconstructed images with attenuation and scatter corrections as train data sets. Given the appropriate supervision of these reconstructed images with attenuation and scatter corrections, it is assumed that the deep neural network can learn the complex principles of reconstruction with attenuation and scatter correction, but the underlying mechanism is not clear. The black-box nature of the AI-based reconstruction methods is a critical limitation [38], and further interpretation of AI-based methodology can be an important direction for future research.
V Conclusion
This paper proposes a network structure combining the encoding–decoding and perceptual loss structure to improve the direct PET image reconstruction from projection data. This is the first AI-based reconstruction method tested on real clinical data from a LAFOV PET to the best of our knowledge. The preliminary results demonstrate that the improvement of deep learning architecture can improve the performance of AI-based reconstruction. In response to the challenge of real data training, the perceptual loss network structure is used to optimize the neural network. The pre-trained VGG network extracts the feature map from the predicted images and ground truth. The perceptual loss is added to the loss function calculation, improving the training efficiency and network effect. The comparison of the prediction results demonstrates that the similarity of the reconstructed image structure and the signal-to-noise ratio is improved. This is because the perceptual loss function can calculate the distance between the predicted image and the target image from the feature level but not from the pixel level. Therefore, the structure of the image can be better reconstructed in a larger area [24].
Despite the limitations of the AI-based methods, the end-to-end reconstruction process from the sinogram data demonstrates the potential of deep learning to learn complex reconstruction principles such as projection, normalization, attenuation correction, and scattering correction, for example. The current research results reveal the possibility and advantages of AI methods for PET image reconstruction, but it should be acknowledged that reconstruction accuracy cannot fully meet the clinical requirements. In future work, further optimization and development of AI-based reconstruction may provide an efficient solution for complex PET reconstruction such as LAFOV PET.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Vandenberghe S, Moskal P, Karp JS. State of the art in total body PET. EJNMMI Phys. 2020;7:35. https://doi.org/10.1186/s40658-020-00290-2.
Cherry SR, Jones T, Karp JS, Qi J, Moses WW, Badawi RD. Total-body PET: maximizing sensitivity to create new opportunities for clinical research and patient care. J Nucl Med : Off Publ Soc Nucl Med. 2018;59:3–12. https://doi.org/10.2967/jnumed.116.184028.
Cherry SR, Badawi RD, Karp JS, Moses WW, Price P, Jones T. Total-body imaging: transforming the role of positron emission tomography. Sci Transl Med. 2017;9:eaaf6169. https://doi.org/10.1126/scitranslmed.aaf6169.
Defrise M, Kinahan PE, Michel CJ. Image reconstruction algorithms in PET. In: Bailey DL, Townsend DW, Valk PE, Maisey MN, editors. Positron emission tomography: basic sciences. London: Springer, London; 2005. p. 63–91.
Shepp LA, Vardi Y. Maximum likelihood reconstruction for emission tomography. IEEE Trans Med Imaging. 1982;1:113–22. https://doi.org/10.1109/TMI.1982.4307558.
Hudson HM, Larkin RS. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans Med Imaging. 1994;13:601–9. https://doi.org/10.1109/42.363108.
Uribe CF, Mathotaarachchi S, Gaudet V, Smith KC, Rosa-Neto P, Benard F, et al. Machine learning in nuclear medicine: part 1—introduction. J Nucl Med. 2019;60:451–8. https://doi.org/10.2967/jnumed.118.223495.
Reader AJ, Corda G, Mehranian A, da Costa-Luis C, Ellis S, Schnabel JA. Deep learning for PET image reconstruction. IEEE Trans Radiat Plasma Med Sci. 2020;1-. https://doi.org/10.1109/trpms.2020.3014786.
Gong K, Guan J, Liu CC, Qi J. PET image denoising using a deep neural network through fine tuning. IEEE Trans Radiat Plasma Med Sci. 2019;3:153–61. https://doi.org/10.1109/TRPMS.2018.2877644.
Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, et al. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging. 2019;46:2780–9.
Katsari K, Penna D, Arena V, Polverari G, Ianniello A, Italiano D, et al. Artificial intelligence for reduced dose 18F-FDG PET examinations: a real-world deployment through a standardized framework and business case assessment. EJNMMI Physics. 2021;8:25. https://doi.org/10.1186/s40658-021-00374-7.
Gong K, Guan J, Kim K, Zhang X, Fakhri G, Qi J, et al. Iterative PET image reconstruction using convolutional neural network representation. IEEE Trans Med Imaging. 2017;38. https://doi.org/10.1109/TMI.2018.2869871.
Gong K, Wu D, Kim K, Yang J, Sun T, El Fakhri G, et al. MAPEM-Net: an unrolled neural network for fully 3D PET image reconstruction. 15th International Meeting on Fully Three-Dimensional Image Reconstruction in Radiology and Nuclear Medicine: Int Soc Opt Photon. 2019; p. 110720O.
Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature. 2018;555:487–92. https://doi.org/10.1038/nature25988.
Haggstrom I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: a deep encoder-decoder network for directly solving the PET image reconstruction inverse problem. Med Image Anal. 2019;54:253–62. https://doi.org/10.1016/j.media.2019.03.013.
Kandarpa VSS, Bousse A, Benoit D, Visvikis D. DUG-RECON: a framework for direct image reconstruction using convolutional generative networks. IEEE Trans Radiat Plasma Med Sci. 2021;5:44–53. https://doi.org/10.1109/trpms.2020.3033172.
Whiteley W, Luk WK, Gregor J. DirectPET: full-size neural network PET reconstruction from sinogram data. J Med Imaging (Bellingham). 2020;7:032503. https://doi.org/10.1117/1.JMI.7.3.032503.
Whiteley W, Panin V, Zhou C, Cabello J, Bharkhada D, Gregor J. FastPET: near real-time PET reconstruction from histo-images using a neural network. arXiv preprint arXiv:200204665. 2020.
Schmall JP, Karp JS, Werner M, Surti S. Parallax error in long-axial field-of-view PET scanners—a simulation study. Phys Med Biol. 2016;61:5443–55. https://doi.org/10.1088/0031-9155/61/14/5443.
Zhang X, Badawi RD, Cherry SR, Qi J. Theoretical study of the benefit of long axial field-of-view PET on region of interest quantification. Phys Med Biol. 2018;63:135010. https://doi.org/10.1088/1361-6560/aac815.
Efthimiou N. New challenges for PET image reconstruction for total-body imaging. PET Clin. 2020;15:453–61. https://doi.org/10.1016/j.cpet.2020.06.002.
Zhang X, Zhou J, Cherry SR, Badawi RD, Qi J. Quantitative image reconstruction for total-body PET imaging using the 2-meter long EXPLORER scanner. Phys Med Biol. 2017;62:2465–85. https://doi.org/10.1088/1361-6560/aa5e46.
Alberts I, Hünermund JN, Prenosil G, Mingels C, Bohn KP, Viscione M, et al. Clinical performance of long axial field of view PET/CT: a head-to-head intra-individual comparison of the Biograph Vision Quadra with the Biograph Vision PET/CT. Eur J Nucl Med Mol Imaging. 2021;48:2395–404. https://doi.org/10.1007/s00259-021-05282-7.
Johnson J, Alahi A, Fei-Fei L. Perceptual losses for real-time style transfer and super-resolution. European conference on computer vision: Springer; 2016. p. 694–711.
Shan H, Zhang Y, Yang Q, Kruger U, Kalra MK, Sun L, et al. 3-D convolutional encoder-decoder network for low-dose CT via transfer learning from a 2-D trained network. IEEE Trans Med Imaging. 2018;37:1522–34.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Comput Sci. 2014.
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13:600–12.
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. Tensorflow: large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:160304467. 2016.
Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014.
Hu Z, Xue H, Zhang Q, Gao J, Zhang N, Zou S, et al. DPIR-Net: direct PET image reconstruction based on the Wasserstein generative adversarial network. IEEE Trans Radiation X Plasma Med Sci. 2021;5:35–43. https://doi.org/10.1109/trpms.2020.2995717.
Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021;3:199–217.
Daube-Witherspoon ME, Muehllehner G. Treatment of axial data in three-dimensional PET. J Nucl Med. 1987;28:1717–24.
Gundlich B, Musmann P, Weber S, Nix O, Semmler W. From 2D PET to 3D PET: issues of data representation and image reconstruction. Z Med Phys. 2006;16:31–46. https://doi.org/10.1078/0939-3889-00290.
Prenosil GA, Sari H, Fürstner M, Afshar-Oromieh A, Shi K, Rominger A, et al. Performance characteristics of the Biograph Vision Quadra PET/CT system with a long axial field of view using the NEMA NU 2–2018 standard. J Nucl Med. 2022;63:476–84. https://doi.org/10.2967/jnumed.121.261972.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. https://doi.org/10.1038/nature14539.
Webb S. Deep learning for biology. Nature. 2018;554:555–7. https://doi.org/10.1038/d41586-018-02174-z.
Towards trustable machine learning. Nat Biomed Eng. 2018;2:709–10. https://doi.org/10.1038/s41551-018-0315-x.
Wang S, Cao G, Wang Y, Liao S, Wang Q, Shi J, et al. Review and prospect: artificial intelligence in advanced medical imaging. Front Radiol. 2021;1. https://doi.org/10.3389/fradi.2021.781868.
Acknowledgements
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided.
Funding
This project received funding from China’s National Natural Science Foundation [Grant No. 11875036], Tsinghua University Initiative Scientific Research Program, and the Swiss National Science Foundation [SNSF Grant No. 188350 and 188914].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by RM, JH, HS, and SX. The first draft of the manuscript was written by RM, JH, and KS, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. WL, RQ, AR, JL, and KS supported this work with funding acquisition.
Corresponding authors
Ethics declarations
Ethics approval
The study was conducted in accordance with the requirements of the respective local ethics committees in Switzerland (Req-2021–00517).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
The authors affirm that human research participants provided informed consent for publication of Figs. 4, 5, 6, 7.
Conflict of interests
Hasan Sari is a full-time employee of Siemens Healthcare AG, Switzerland. No other potential conflict of interest relevant to this article was reported.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, R., Hu, J., Sari, H. et al. An encoder-decoder network for direct image reconstruction on sinograms of a long axial field of view PET. Eur J Nucl Med Mol Imaging 49, 4464–4477 (2022). https://doi.org/10.1007/s00259-022-05861-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-022-05861-2