Abstract
Purpose
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disorder with on average a 1-year delay between symptom onset and diagnosis. Studies have demonstrated the value of [18F]-FDG PET as a sensitive diagnostic biomarker, but the discriminatory potential to differentiate ALS from patients with symptoms mimicking ALS has not been investigated. We investigated the combination of brain and spine [18F]-FDG PET-CT for differential diagnosis between ALS and ALS mimics in a real-life clinical diagnostic setting.
Methods
Patients with a suspected diagnosis of ALS (n = 98; 64.8 ± 11 years; 61 M) underwent brain and spine [18F]-FDG PET-CT scans. In 62 patients, ALS diagnosis was confirmed (67.8 ± 10 years; 35 M) after longitudinal follow-up (average 18.1 ± 8.4 months). In 23 patients, another disease was diagnosed (ALS mimics, 60.9 ± 12.9 years; 17 M) and 13 had a variant motor neuron disease, primary lateral sclerosis (PLS; n = 4; 53.6 ± 2.5 years; 2 M) and progressive muscular atrophy (PMA; n = 9; 58.4 ± 7.3 years; 7 M). Spine metabolism was determined after manual and automated segmentation. VOI- and voxel-based comparisons were performed. Moreover, a support vector machine (SVM) approach was applied to investigate the discriminative power of regional brain metabolism, spine metabolism and the combination of both.
Results
Brain metabolism was very similar between ALS mimics and ALS, whereas cervical and thoracic spine metabolism was significantly different (in standardised uptake values; cervical: ALS 2.1 ± 0.5, ALS mimics 1.9 ± 0.4; thoracic: ALS 1.8 ± 0.3, ALS mimics 1.5 ± 0.3). As both brain and spine metabolisms were very similar between ALS mimics and PLS/PMA, groups were pooled for accuracy analyses. Mean discrimination accuracy was 65.4%, 80.0% and 81.5%, using only brain metabolism, using spine metabolism and using both, respectively.
Conclusion
The combination of brain and spine FDG PET-CT with SVM classification is useful as discriminative biomarker between ALS and ALS mimics in a real-life clinical setting.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating adult-onset neurodegenerative disorder with both upper (UMN) and lower motor neuron (LMN) involvement. ALS diagnosis is based on a combination of clinical and electrophysiological parameters [1]; however, diagnostic timelines remain long (on average 1 year) before a final consensus diagnosis is reached [2]. Therefore, early diagnostic biomarkers are important to facilitate therapeutic trial enrolment in early disease stages and to limit the emotional burden accompanied by the diagnostic uncertainty.
Several [18F]-FDG brain PET studies have demonstrated [18F]-FDG brain PET allows discrimination between ALS patients and healthy volunteers with an accuracy higher than 90% [3,4,5]. Until now, [18F]-FDG studies investigating the essential discriminatory accuracy between ALS patients and ALS mimics are lacking. ALS mimics are a very heterogeneous group of conditions which resemble the presentation and clinical symptoms and signs of early ALS. They include various disorders such as inclusion body myositis, polyradiculopathy, paraneoplastic neuropathy, spinal stenosis, primary progressive multiple sclerosis, functional disorders and cramp fasciculation syndrome. As diagnosis is mainly based on clinical parameters, differential diagnosis from these mimics can be very challenging early in disease.
Current neuroimaging studies have focused on brain (UMN) involvement, as spine (LMN) imaging analysis is more challenging [6]. A limited number of PET and MR studies which included spine data showed promising results for improved staging and disease characterisation. Spinal cord atrophy and diffusion tensor imaging metrics correlated with disease progression, respiratory dysfunction and severity; spinal N-acetyl-aspartate/choline and N-acetyl-aspartate/creatinine measured by spectroscopy correlated with functional vital capacity; and spinal glucose hypermetabolism above the fifth decile resulted in a worse prognosis [7,8,9,10,11]. To allow clinical use of spinal cord data, an automated spinal cord segmentation is needed [7].
Altogether, these results illustrate that a combined approach using brain and spine metabolism together may provide novel insights and strengthen the biomarker potential of [18F]-FDG PET.
The aim of this study was twofold. Firstly, we wanted to validate an automated image analysis pipeline in line with the approach of Marini et al. [7]. Secondly, we investigated the potential of combining brain and spinal cord [18F]-FDG PET data to improve differential diagnosis between ALS and ALS mimics in a real-life clinical setting and examined whether UMN and LMN involvement characterised by cerebellar and spinal hypermetabolism are interrelated.
Materials and methods
Patient characteristics
Patients with a suspected diagnosis of ALS (n = 98; mean age ± SD, 64.8 ± 11 years; 61 M) were recruited from the tertiary neuromuscular clinic at the University Hospital Leuven (Belgium) between November 2016 and May 2018 (Tables 1 and 2). In 62 patients, the diagnosis of ALS was confirmed (mean age ± SD, 67.8 ± 10 years; 35 M) after longitudinal follow-up until death or end of September 2019 (average 18.1 ± 8.4 months). Thirty-six patients were diagnosed with another disease, including motor neuron disease variants primary lateral sclerosis (PLS; n = 4; 53.6 ± 2.5 years; 2 M) or progressive muscular atrophy (PMA; n = 9; 58.4 ± 7.3 years; 7 M), and other disorders mimicking ALS (ALS mimics, n = 23; 60.9 ± 12.9 years; 17 M). Tables 1, 2 and 3 summarise the demographic and disease characteristics of the different groups. The month in which initial symptoms of muscle weakness or dysarthria/dysphagia occurred was used to define disease onset.
All patients were neurologically and electro-diagnostically investigated by an experienced specialist in neuromuscular disorders (PVD) using the revised El Escorial and Awaji-Shima criteria [11, 12]. The clinical stage was determined using ALSFRS-R [13] and the metabolic stage was determined using the corresponding brain FDG scans [14].
This retrospective study was approved by the local University Hospital Ethics Committee (UZ/KU Leuven); because of its retrospective nature, the need for written consent was waived.
PET acquisition and reconstruction
At time of PET imaging, no signs of respiratory distress nor nutritional abnormalities were apparent in any patient. All subjects fasted a minimum of 6 h before [18F]-FDG PET acquisition and had a mean glycaemia of 99 ± 16 mg/dL (range 69–156 mg/dL) before [18F]-FDG administration. [18F]-FDG PET scans were acquired using a Siemens Biograph 16 HiRez PET-CT camera (Siemens Healthcare, Erlangen, Germany). 151 ± 8 MBq [18F]-FDG was injected intravenously in a dimly lit, quiet room with ears and eyes open. Thirty minutes after the [18F]-FDG injection, a low-dose CT of the brain, cervical and thoracic spine was acquired, followed by a static PET scan of 15 min of the brain and a static PET scan of 2 min/bed position of the cervical and thoracic spines (on average 3 bed positions). A vacuum pillow immobilised the subjects’ head during acquisition to limit motion artefacts. On the HiRez PET-CT camera, [18F]-FDG images were reconstructed using iterative ordered-subset expectation maximisation (OSEM). PET data were corrected for dead time, scatter, randoms, decay and CT-based attenuation correction. For the brain scan, 2 iterations and 24 subsets were used with 2 mm post-smoothing (full width at half maximum (FWHM) 8, voxel size 2.1 × 2.1 × 3.0 mm), while for the scan of the spinal cord, 5 iterations and 8 subsets were used with 6 mm Gaussian post-smoothing (FWHM 9, voxel size 2.9 × 2.9 × 3.0 mm). Standardised uptake values (SUV) were calculated by normalising the measured activity to the injected activity and total body weight.
[18F]-FDG brain PET analysis
Volume of interest (VOI)–based analysis of the FDG brain PET data was performed with PMOD PNEURO tool (version 3.4; PMOD Inc. Zürich, Germany) using the Hammers N30R83 maximum probability atlas. Individual VOI activities of the reconstructed individual cortical, brainstem and cerebellar VOIs (in total 30 VOIs) were normalised to average brain activity to obtain relative metabolic activity. Left and right brain VOIs were merged and analysed together.
Voxel-based analysis was performed using statistical parametric mapping (SPM12; Wellcome Trust Centre for Neuroimaging, London, UK). All scans were spatially normalised to Montreal Neurological Institute space using the PMOD FUSION tool (version 3.4; PMOD Inc. Zürich, Switzerland) followed by isotropic Gaussian smoothing with a FWHM of 8 mm in a 2 × 2 × 2 mm matrix. Data were normalised to the average grey matter activity of each image using proportional scaling. Voxel-based group comparison was done using single-way ANOVA using a pheight, uncorr < 0.001 and pcluster, FWE-corr < 0.05. The following comparisons were made: ALS vs all ALS mimics plus PLS/PMA, ALS vs mimics without PLS/PMA.
To discriminate between the groups, a support vector machine (SVM) classifier [15] with linear kernel was trained and tested using tenfold cross-validation (CV), with the full brain [18F]-FDG PET data as input. Pre-processing of the data prior to training the SVM classifier included masking, using a full brain mask (Hammers atlas N30R83), demeaning (subject-specific) and unfolding the three-dimensional data as a 1 × N vector, where N is the total amount of voxels. The tenfold CV is applied ten times randomly, and prediction accuracy results are reported with mean and standard deviation with respect to these ten randomisations.
Registration, segmentation and quantification of spinal cord metabolism
The cervical and thoracic spines were first manually delineated in a transversal plane using the low-dose CT in the PMOD FUSION tool (version 3.4; PMOD Inc. Zürich, Germany). As the spine data was acquired at a later time compared with the brain images, a time-correction factor to the beginning of the scan was applied [16].
To overcome the laborious task of manually segmenting the spinal cord in the future, a deep learning convolutional neural network (CNN) based on the UNet architecture [17] was implemented. The network took the 2D CT image slices as input and assigned a class label (out of two classes, spine and background) to each voxel, yielding the segmented spine as output. The CNN was trained using 75% of patient data (n = 11,400, randomly sampled), whereas 25% of the data was used for testing (n = 3800). Patients of whom data were used for training the CNN were excluded from contributing to the test dataset. The 2D images were selected in the transverse plane and were automatically cropped to an image dimension of 256 × 256 to balance the spine/background class information. The CNN architecture is illustrated in Fig. 1. After every convolutional operation, images were padded such that the spatial output dimension stays the same as the input; therefore, cropping was not necessary when feature channels were concatenated. Between consecutive convolution operations, a dropout of 10% was applied [18]. The CNN model was trained using the ADAM optimiser for a maximum of 50 epochs comprising early dropout (5 consecutive epochs without improvement), a batch size of 16, binary cross-entropy as loss function and a learning rate of 0.001. To build and train the CNN architecture, Keras (version 2.3.1, keras.io) and TensorFlow (version 1.15.0, www.tensorflow.org) were used. The accuracy of the predicted spinal cord segmentation was quantified by the Dice similarity coefficient (DSC) between the automatic and manual segmentations. DSC measures the spatial overlap between two segmentations A and B and is defined as DSC(A, B) = 2(A ∩ B)/(A + B), where ∩ is the intersection and + is the sum of both. The DSC value ranges from 0, indicating no spatial overlap, to 1, expressing complete overlap.
Convolutional neural network based on the UNet architecture and consists of a contracting path (left side) and expanding path (right side). The leftmost input yields the 256 × 256 map sampled from the CT image, and the rightmost output the CNN’s binary spine segmentation prediction. Boxes represent feature maps, where each map’s dimension is indicated on its lower left, and the number of channels (feature maps) is indicated above. Operations are represented by arrows; i.e. a blue arrow indicates a convolution operation with 3 × 3 kernel followed by a rectified linear unit (ReLU) activation function [35], a red arrow represents a down-sampling operation using max-pooling with 2 × 2 pool size, a green arrow consists of a 2 × 2 up-sampling operation applying inverse-convolution, a grey arrow means concatenating the feature maps, and an orange arrow represents a convolution operation with 1 × 1 kernel followed by the sigmoid activation function
In order to calculate and compare the metabolic profile along the spine across subjects, spinal cord PET-CT data were spatially aligned with a reference image, which is illustrated in Fig. 2. The first step comprised the straightening of the spinal cord (CT image) using the segmented spine; then, this straightening transformation was applied onto the PET data. Straightening of the spinal cord was performed using the Spinal Cord Toolbox (SCT, version 4.0.2, Montreal, Canada) [19], by solving analytically the straightening equations for each image voxel to compute the forward and inverse deformation fields for straightening [20]. Subsequently, the straightened CT data was non-linearly registered to a reference image using Advanced Normalisation Tools (ANTs; [21]) by applying an initial rigid transformation followed by an affine and non-linear transformation using symmetric image normalisation (SyN; [22]) as diffeomorphic image registration.
Registration of spinal cord PET-CT data to a reference image. The first step consists of straightening the spinal cord CT image by using the segmented spine, and subsequently applying the straightening transformation to the PET image. Thereafter, the straightened CT image is non-linearly registered to a reference image using Advanced Normalisation Tools (ANTs), and the non-linear registration transformation is applied to the PET image
Once the PET-CT data were straightened and registered to a common reference space, the mean [18F]-FDG PET uptake (absolute SUV values) in the plane perpendicular to the spinal axis is calculated, followed by one-dimensional spatial smoothening along the spine with different Gaussian kernels. Subsequently, these spinal cord metabolic profiles, containing spatial information, were used as input for a SVM classifier to differentiate between the groups. Training and testing were performed using tenfold CV, applying ten random selections of CV scheme.
To combine spinal and brain metabolic information for differentiation between both groups, two independent SVM classifiers were trained and tested using a tenfold CV for both the brain and spine [18F]-FDG PET data. The SVM prediction scores, i.e. the distances to the dividing hyperplanes, of the test data obtained for each classifier, were standardised (Z-values) and combined in a tenfold CV of two-dimensional input data for training and testing a second classification step using a linear SVM.
General statistics
General statistics were performed in SPSS (version 26.0; Armonk, USA). Normality was tested using the Shapiro-Wilk test. Kruskal-Wallis tests were performed to compare ALS patients with ALS mimics plus or minus the PLS/PMA group. Additionally, a Spearman correlation coefficient was calculated between cervical and thoracic spines and the cerebellum. Significance was thresholded at p < 0.05.
Results
Metabolic differences in brain between ALS, ALS mimics and PLS/PMA
VOI-based analysis showed a significantly decreased metabolism (< 5%) in the superior frontal (p = 0.01), midfrontal (p = 0.02) and precentral (p = 0.01) cortex, and significantly increased metabolism (< 5%) in the cerebellum (p = 0.02) in ALS compared with ALS mimics plus PLS/PMA. Comparing ALS versus ALS mimics minus PLS/PMA, both the superior frontal gyrus (p = 0.03) and precentral gyrus (p < 0.05) remained significant. However, these regions were not significant after multiple comparison correction. No significant differences were observed between ALS mimics and the PLS/PMA group. Voxel-based analysis showed no significant results between ALS patients, ALS mimics and the PLS/PMA group. As brain metabolism was very similar between ALS mimics and PLS/PMA patients, groups were pooled to increase number of subjects in the non-ALS group for training purposes.
If a full brain mask (all VOIs of Hammers atlas N30R83, except for the ventricles) is applied, training the SVM classifier using ten random tenfold CV schemes resulted in a classifying accuracy of 60.6 ± 2.3% and 59.2 ± 3.7% for ALS versus ALS mimics with PLS/PMA, respectively. An improvement in prediction accuracy of 66.5 ± 4.3% for ALS and 64.6 ± 5.1% for ALS mimics with PLS/PMA was found in case a selective brain mask is used comprising only regions that showed significant VOI-based metabolic differences (without multiple comparison correction) between ALS and ALS mimics, i.e. cerebellum, and superior frontal, midfrontal and precentral gyri.
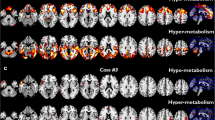
The corresponding discriminate weight vectors represented in the three-dimensional image space are illustrated in Fig. 3. Since a positive class label was a priori assigned to the ALS group and a negative class label to the ALS mimics with PLS/PMA patients, red regions (positive values) in the discriminate brain pattern illustrated in Fig. 3 represent a relative higher glucose metabolic uptake for ALS with respect to ALS mimics with PLS/PMA, while blue regions (negative values) correspond to a relative lower glucose metabolic uptake for ALS compared with ALS mimics with PLS/PMA.
Discriminating weight vectors obtained by training the support vector machine (SVM) classifier differentiating between amyotrophic lateral sclerosis (ALS) and ALS mimics with PLS/PMA, for full brain mask (upper figure) and for a selective brain mask (lower figure) comprising the regions that showed significant volume of interest (VOI)–based metabolic differences, i.e. cerebellum, and superior frontal, midfrontal and precentral gyri. The length of the weight vector is normalised to one, implying arbitrary units (a.u.) are used for the colour bar
Automatic segmentation of the spinal cord
After 18 epochs, the early stopping criterium was reached, i.e. 5 consecutive epochs without improvement in loss function. The average DSC calculated for the test data of the optimal CNN was equal to 0.82.5.
Metabolic differences in spinal cord between ALS, ALS mimics and PLS/PMA
No significant differences in spinal cord metabolism were observed between ALS mimics and PLS/PMA patients. We observed a significantly higher metabolism of 18% in the thoracic spinal cord (p < 0.001) and a trend to higher metabolism of 10% in the cervical spinal cord (p = 0.07), in ALS compared with ALS mimics. If we pooled the ALS mimics with the PLS/PMA group, we obtained similar findings, namely a significant higher metabolism in the thoracic spinal cord (p < 0.001) and in the cervical spinal cord (p = 0.04). Similarly, spinal cord metabolism was very similar between ALS mimics and PLS/PMA patients so they were pooled to compare against ALS patients for training purposes.
The spatial normalisation of the individual spinal cord data to the spine template could not be done in an automated way for 24 out of 98 subjects due to a relatively low CT resolution. Misregistration was obvious by visual assessment of the results. Therefore, we only included 74 subjects with a good, fully automated normalisation to template space to create the spinal metabolic profiles. There were no differences in patient characteristics between the different subgroups of these 74 subjects versus those characteristics of all 98 subjects (Tables 1 and 2). The corresponding mean spinal metabolism and standard deviation across the ALS and ALS mimics with PLS/PMA are illustrated in Fig. 4. Using these spinal metabolic profiles as input for the SVM classifier, a prediction accuracy of 80.6 ± 2.7% and 79.7 ± 4.1% was obtained for ALS and ALS mimics with PLS/PMA, respectively. Varying the FWHM of the smoothing kernel along the spinal axis (range from 0 to 1 cm) did not influence the accuracy of the classification results.
Mean and standard deviation (STD) of the spinal metabolism across amyotrophic lateral sclerosis (ALS) in red and ALS mimics with PLS/PMA in blue. Spinal metabolism is quantified by calculating the mean [18F]-FDG PET activity (standard uptake value (SUV)) in the plane perpendicular to the spine, after straightening and registration of the PET-CT data
Combined analysis of spinal cord and brain metabolism
We did not observe any significant correlations between cerebellar and spine subsection/whole metabolism.
The combination of standardised (Z-values) SVM prediction scores for both brain and spine data is illustrated in Fig. 5. Figure 5 also demonstrates PLS/PMA patients have very similar brain and spinal metabolism compared with ALS mimics. Training a classifier (SVM, tenfold CV) in this two-dimensional space resulted in a prediction accuracy of 82.2 ± 1.6% for ALS and 79.7 ± 1.6% for ALS mimics with PLS/PMA.
Hyperplane differentiating between amyotrophic lateral sclerosis (ALS, red), ALS mimics (blue), progressive lateral sclerosis (PLS, cyan) and primary muscular atrophy (PMA, orange) in the two-dimensional feature space (Z-values), by combining support vector machine (SVM) prediction scores obtained by classification using brain and spinal metabolic data
Discussion
This is the first study to explore differences in brain and spinal metabolism between ALS and ALS mimics and investigate the possibility for differential diagnosis. We observed a remarkably similar pattern of brain glucose metabolism between ALS patients, ALS mimics and PLS/PMA patients, demonstrated by the low prediction accuracy, the very small VOI-based differences which were not statistically significant after multiple comparison correction and the lack of cluster-wise significant differences on a voxel level. This is concordant with previous studies which investigated brain metabolism in PLS, PMA and an ALS mimic, namely Kennedy disease [4, 5, 23]. One possible hypothesis is the brain-muscle crosstalk: physical activity contributes to cognitive function and metabolic control [24], and vice versa mental imagination of strong muscle contractions may reduce muscle-disuse weakness [25]. In line with this hypothesis, previous studies demonstrated a relative glucose increase in the frontal, temporoparietal, occipital, premotor and cerebellar cortex after physical activity [26, 27] and likewise, Watson et al. observed that lack of exercise reduces neurogenesis [28]. Therefore, we may hypothesise that muscle disuse/loss, present in ALS mimics and PLS/PMA patients, causes widespread brain hypometabolism.
On the other hand, ALS patients have a significantly higher spinal cord glucose metabolism compared with ALS mimics with and without PLS/PMA, with a mean discriminative accuracy of 80% including PLS/PMA patients. These findings are in line with the higher spinal cord metabolism in ALS patients compared with healthy volunteers as observed by Marini et al. [7, 29]. Interestingly, although the spinal cord metabolism is very heterogeneous across subjects (illustrated by the high standard deviation, see Fig. 4), the average ALS spinal cord metabolic profile is roughly only shifted upwards (higher SUV) compared with ALS mimics with an almost identical spatial gradient along the spine.
Hypermetabolism has been observed both in the spinal cord and cerebellum in ALS which may suggest a similar disease progression [4, 5, 7, 29, 30]. In this study, no significant correlation was observed between both hypermetabolic regions, concordant with the clinical findings of Marini et al. [30]. The current hypothesis about the onset of ALS disease is that it starts in the motor cortex and spinal cord and from there migrates to other cortical brain regions. In support of the latter, Brettschneider et al. reported pathological spine involvement and motor cortex involvement from an early stage in ALS [31]. The lack of correlation might reflect an independent disease progression and therefore may suggest the necessity of a dual therapeutic strategy. In agreement with this, mean prediction accuracy increased to 82% by combining both brain and spinal metabolic information. Therefore, even though not discriminative on its own, our study demonstrated that adding brain metabolic information to spinal metabolism resulted in a slightly higher predictive power to differentiate between ALS and ALS mimics. We agree that a prediction accuracy of 82% is lower than the discrimination accuracy of 90% of neurofilaments; nevertheless, it might certainly contribute to approach a prediction accuracy of 100% early in disease [32].
This study took place in a real-life clinical work-up of patients with suspected ALS of which 2/3 received a definitive diagnosis of ALS. Early final diagnosis is of critical importance to enrol patients in therapeutic trials before widespread irreversible neuronal damage occurs. As [18F]-FDG PET-CT scans are part of standard clinical practice in our centre, no additional scans were needed to perform [18F]-FDG PET-CT scans of the spine. Spinal metabolism can be investigated using a 6–8-min additional spine scan as part of a routine [18F]-FDG PET-CT brain scan protocol. Van der Burgh et al. demonstrated atrophy of the spine is present in all groups of the motor neuron disease spectrum, including PLS/PMA patients [33]. Thus, although cervical atrophy is a common feature of all groups in the motor neuron disease spectrum, our study confirms that a discriminant metabolic pattern is present. Future studies are necessary to confirm this finding as we could only include 4 PMA and 9 PLS patients in our cohort.
Software, such as the SCT [19], is freely available to segment, register and analyse spinal cord data based on anatomical MR imaging; however, no standard approach is available to quantify the spinal cord based on CT data. Recently, Marini et al. proposed an automated approach based on the Hough transform which is able to adequately delineate the spinal cord [7]. As an alternative, we proposed a deep learning approach which took advantage of the extensive set of available manual segmentations for training and testing and does not require any pre-processing in terms of denoising or edge detection except for automatic cropping. For this purpose, we have implemented a convolutional neural network based on the UNet architecture to automatically segment the spinal cord using the CT imaging data as input. The high DSC of 0.84 based on an independent test dataset proved that our trained CNN can be applied successfully to new unseen data for very fast automatic spinal cord segmentation without the need of manual delineations, which are very time-consuming. These small differences in spine segmentation between prediction and manual segmentation (ground truth) did not result in differences in quantification of spinal cord, since averaging and smoothing operations were performed on the [18F]-FDG PET activity within the segmented spine.
Limitations
Some limitations have to be mentioned. Firstly, no control group was available to compare our results for brain and spine data. However, our results are in line with previous findings describing spinal hypermetabolism in ALS compared with healthy volunteers [7, 29]. Secondly, we acknowledge that the average follow-up was only 1.5 year and that PLS/PMA patients may still convert to ALS until 10 years after symptom onset. Nevertheless, PLS/PMA patients represented only 13 subjects of the ALS mimics with PLS/PMA group. Therefore, it is unlikely this will alter our findings. Finally, spinal template normalisation was only feasible in 74 out of 98 subjects due to a relatively low CT resolution and signal to noise ratio for the low-intensity whole body CT (30 mAs). Optimising the integrated current for the CT may overcome this problem.
Conclusion
The combination of brain and spine FDG PET-CT yielded an accuracy of 82% as discriminative biomarker between ALS and ALS mimics in a real-life clinical setting. Moreover, we have implemented a convolutional neural network based on the UNet architecture to automatically segment the spinal cord using the CT imaging data as input, facilitating future spinal cord PET-CT studies.
References
Salameh JS, Brown RH Jr, Berry JD. Amyotrophic lateral sclerosis: review. Semin Neurol. 2015;35(4):469–76.
Paganoni S, Macklin EA, Lee A, Murphy A, Chang J, Zipf A, et al. Diagnostic timelines and delays in diagnosing amyotrophic lateral sclerosis (ALS). Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(5–6):453–6.
Pagani M, Oberg J, De Carli F, Calvo A, Moglia C, Canosa A, et al. Metabolic spatial connectivity in amyotrophic lateral sclerosis as revealed by independent component analysis. Hum Brain Mapp. 2016;37(3):942–53.
Van Laere K, Vanhee A, Verschueren J, De Coster L, Driesen A, Dupont P, et al. Value of 18fluorodeoxyglucose-positron-emission tomography in amyotrophic lateral sclerosis: a prospective study. JAMA Neurol. 2014;71(5):553–61.
Van Weehaeghe D, Ceccarini J, Delva A, Robberecht W, Van Damme P, Van Laere K. Prospective validation of 18F-FDG brain PET discriminant analysis methods in the diagnosis of amyotrophic lateral sclerosis. J Nucl Med. 2016;57(8):1238–43.
Wheeler-Kingshott CA, Stroman PW, Schwab JM, Bacon M, Bosma R, Brooks J, et al. The current state-of-the-art of spinal cord imaging: applications. Neuroimage. 2014;84:1082–93.
Marini C, Cistaro A, Campi C, Calvo A, Caponnetto C, Nobili FM, et al. A PET/CT approach to spinal cord metabolism in amyotrophic lateral sclerosis. Eur J Nucl Med Mol Imaging. 2016;43(11):2061–71.
Branco LM, De Albuquerque M, De Andrade HM, Bergo FP, Nucci A, Franca MC Jr. Spinal cord atrophy correlates with disease duration and severity in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(1–2):93–7.
El Mendili MM, Cohen-Adad J, Pelegrini-Issac M, Rossignol S, Morizot-Koutlidis R, Marchand-Pauvert V, et al. Multi-parametric spinal cord MRI as potential progression marker in amyotrophic lateral sclerosis. PLoS One. 2014;9(4):e95516.
Grolez G, Kyheng M, Lopes R, Moreau C, Timmerman K, Auger F, et al. MRI of the cervical spinal cord predicts respiratory dysfunction in ALS. Sci Rep. 2018;8(1):1828.
Costa J, Swash M, de Carvalho M. Awaji criteria for the diagnosis of amyotrophic lateral sclerosis: a systematic review. Arch Neurol. 2012;69(11):1410–6.
Schrooten M, Smetcoren C, Robberecht W, Van Damme P. Benefit of the Awaji diagnostic algorithm for amyotrophic lateral sclerosis: a prospective study. Ann Neurol. 2011;70(1):79–83.
Balendra R, Jones A, Jivraj N, Knights C, Ellis CM, Burman R, et al. Estimating clinical stage of amyotrophic lateral sclerosis from the ALS Functional Rating Scale. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(3–4):279–84.
van Weehaeghe D, Ceccarini J, Willekens SM, de Vocht J, van Damme P, van Laere K. Is there a glucose metabolic signature of spreading TDP-43 pathology in amyotrophic lateral sclerosis? Q J Nucl Med Mol Imaging. 2017.
Hearst MA. Support vector machines. IEEE Intell Syst App. 1998;13(4):18–21.
van den Hoff J, Lougovski A, Schramm G, Maus J, Oehme L, Petr J, et al. Correction of scan time dependence of standard uptake values in oncological PET. EJNMMI Res. 2014;4(1):18.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. Lect Notes Comput Sc. 2015;9351:234–41.
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–58.
De Leener B, Levy S, Dupont SM, Fonov VS, Stikov N, Collins DL, et al. SCT: spinal cord toolbox, an open-source software for processing spinal cord MRI data. Neuroimage. 2017;145:24–43.
De Leener B, Mangeat G, Dupont S, Martin AR, Callot V, Stikov N, et al. Topologically preserving straightening of spinal cord MRI. J Magn Reson Imaging. 2017;46(4):1209–19.
Tustison NJ, Cook PA, Klein A, Song G, Das SR, Duda JT, et al. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage. 2014;99:166–79.
Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41.
Lai TH, Liu RS, Yang BH, Wang PS, Lin KP, Lee YC, et al. Cerebral involvement in spinal and bulbar muscular atrophy (Kennedy’s disease): a pilot study of PET. J Neurol Sci. 2013;335(1–2):139–44.
Pedersen BK. Physical activity and muscle-brain crosstalk. Nat Rev Endocrinol. 2019;15(7):383–92.
Clark BC, Mahato NK, Nakazawa M, Law TD, Thomas JS. The power of the mind: the cortex as a critical determinant of muscle strength/weakness. J Neurophysiol. 2014;112(12):3219–26.
Shimada H, Ishii K, Ishiwata K, Oda K, Suzukawa M, Makizako H, et al. Gait adaptability and brain activity during unaccustomed treadmill walking in healthy elderly females. Gait Posture. 2013;38(2):203–8.
Tashiro M, Itoh M, Fujimoto T, Fujiwara T, Ota H, Kubota K, et al. 18F-FDG PET mapping of regional brain activity in runners. J Sports Med Phys Fitness. 2001;41(1):11–7.
Watson N, Ji X, Yasuhara T, Date I, Kaneko Y, Tajiri N, et al. No pain, no gain: lack of exercise obstructs neurogenesis. Cell Transplant. 2015;24(4):591–7.
Marini C, Morbelli S, Cistaro A, Campi C, Caponnetto C, Bauckneht M, et al. Interplay between spinal cord and cerebral cortex metabolism in amyotrophic lateral sclerosis. Brain. 2018;141(8):2272–9.
Cistaro A, Valentini MC, Chio A, Nobili F, Calvo A, Moglia C, et al. Brain hypermetabolism in amyotrophic lateral sclerosis: a FDG PET study in ALS of spinal and bulbar onset. Eur J Nucl Med Mol Imaging. 2012;39(2):251–9.
Brettschneider J, Del Tredici K, Toledo JB, Robinson JL, Irwin DJ, Grossman M, et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol. 2013;74(1):20–38.
Feneberg E, Oeckl P, Steinacker P, Verde F, Barro C, Van Damme P, et al. Multicenter evaluation of neurofilaments in early symptom onset amyotrophic lateral sclerosis. Neurology. 2018;90(1):E22–30.
van der Burgh HK, Westeneng HJ, Meier JM, van Es MA, Veldink JH, Hendrikse J, et al. Cross-sectional and longitudinal assessment of the upper cervical spinal cord in motor neuron disease. Neuroimage Clin. 2019;24:101984.
Acknowledgements
The authors want to thank the nuclear medicine team for their contributions to the scanning and data handling, the PET radiopharmacy team and the medical physics team of UZ Leuven for their skilled contributions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
PVD, KVL and MK are senior clinical investigators of the research foundation of Flandres (FWO). PVD is supported by the E. von Behring Chair for Neuromuscular and Neurodegenerative Disorders, the ALS Liga België and the KU Leuven funds ‘Een Hart voor ALS’, ‘Laeversfonds voor ALS Onderzoek’ and ‘the Valéry Perrier Race against ALS fund’. MD and DVW and PhD fellows from FWO. JDV is funded by the Klinische onderzoeks- en opleidingsraad (KOOR) of the University Hospitals Leuven. No other conflicts of interest were present.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
This retrospective study was approved by the local University Hospital Ethics Committee; because of its retrospective nature, the need for written consent was waived.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Neurology
Rights and permissions
About this article
Cite this article
Van Weehaeghe, D., Devrome, M., Schramm, G. et al. Combined brain and spinal FDG PET allows differentiation between ALS and ALS mimics. Eur J Nucl Med Mol Imaging 47, 2681–2690 (2020). https://doi.org/10.1007/s00259-020-04786-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-020-04786-y