Abstract
Purpose
Pathological complete response (pCR) following neoadjuvant chemoradiotherapy or radiotherapy in locally advanced rectal cancer (LARC) is reached in approximately 15–30% of cases, therefore it would be useful to assess if pretreatment of 18F-FDG PET/CT and/or MRI texture features can reliably predict response to neoadjuvant therapy in LARC.
Methods
Fifty-two patients were dichotomized as responder (pR+) or non-responder (pR-) according to their pathological tumor regression grade (TRG) as follows: 22 as pR+ (nine with TRG = 1, 13 with TRG = 2) and 30 as pR- (16 with TRG = 3, 13 with TRG = 4 and 1 with TRG = 5). First-order parameters and 21 second-order texture parameters derived from the Gray-Level Co-Occurrence matrix were extracted from semi-automatically segmented tumors on T2w MRI, ADC maps, and PET/CT acquisitions. The role of each texture feature in predicting pR+ was assessed with monoparametric and multiparametric models.
Results
In the mono-parametric approach, PET homogeneity reached the maximum AUC (0.77; sensitivity = 72.7% and specificity = 76.7%), while PET glycolytic volume and ADC dissimilarity reached the highest sensitivity (both 90.9%). In the multiparametric analysis, a logistic regression model containing six second-order texture features (five from PET and one from T2w MRI) yields the highest predictivity in distinguish between pR+ and pR- patients (AUC = 0.86; sensitivity = 86%, and specificity = 83% at the Youden index).
Conclusions
If preliminary results of this study are confirmed, pretreatment PET and MRI could be useful to personalize patient treatment, e.g., avoiding toxicity of neoadjuvant therapy in patients predicted pR-.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer is the third leading cause of cancer-related mortality in Western countries and in approximately one-third of cases the tumor is localized in the rectum [1]. The standard therapeutic scheme for locally advanced rectal cancer (LARC) involves surgical resection, preceded by neoadjuvant chemoradiotherapy (CRT) or radiotherapy only (RT) [2, 3]. Neoadjuvant treatment can reduce the risk of local recurrence, downsize the tumor, and facilitate subsequent successful R0 resection and sphincter-preserving surgery [4]. Pathological complete response (pCR) is reached in approximately 15–30% of subjects and in these cases a wait-and-see strategy is becoming a viable therapeutic option [5]. To improve patient management, it could be advantageous to determine the likelihood of pCR or near pCR before treatment to allow clinicians to tailor therapy. Importantly, patients predicted as non-responders could benefit from alternative treatments or up-front surgery, avoiding toxicity and side effects of CRT/RT.
Recently, the idea has emerged that medical images are like the “dark matter in space” since only a small percentage of image “data” is actually used by the radiologist for interpretation whilst the vast majority is locked up within the images themselves. Radiomics analysis can extract hidden data and process large amounts of information from routinely acquired medical images with the scope of providing a comprehensive quantification of tumor phenotype. Radiomics uses advanced quantitative feature analysis, including analysis of the spatial layout of images and of their geometric shape [6]. Improvement in image analysis through the understanding of its texture proprieties has revealed important prognostic information on disease course [7, 8] and on the understanding of underlying genomic patterns [9].
Texture analysis has been applied to MRI to predict long-term survival of patients with LARC [10], to discriminate different stages of rectal cancer [11] and also to predict response to CRT [12, 13]. The role of texture analysis in predicting response to CRT has also been investigated with 18F-FDG PET/CT [14, 15]. To our knowledge, combining 18F-FDG-PET and MRI texture features with the aim of predicting which patients with LARC will respond to neoadjuvant therapy has never been attempted before. However, this approach has shown promising results in other tumor models. For example, Vallières et al. [16] have shown that the predictive value of baseline 18F-FDG-PET texture features in the risk evaluation of lung metastasis at the time of diagnosis of primary soft-tissue sarcomas was significantly enhanced by the addition of MRI parameters.
The aim of this study is to assess if a combination of MRI and 18F-FDG PET/CT texture features can reliably predict response to neoadjuvant therapy (CRT/RT) in LARC and provide clues that could ultimately improve patient management.
Materials and methods
Patients
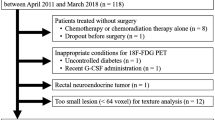
Subjects with LARC that underwent neoadjuvant CRT or only RT followed by surgical resection at our institution were retrospectively enrolled in a single institution study between July 2010 and October 2016. Inclusion criteria were the following: (a) biopsy-confirmed stage II/III LARC (any T, positive N); (b) absence of distant metastasis (M0); (c) axial MRI examination, including T2-weighted (T2w) and diffusion-weighted imaging (DWI), and fluoro-d-glucose (FDG) PET performed at our institute prior to neoadjuvant treatment. Exclusion criteria were: (a) significant image artifacts at MR and/or FDG-PET examinations, (b) absence of tumor regression grading (TRG) evaluation [17], (c) age < to 18 years, (d) pregnancy, (e) mentally incompetent subjects. The study design was approved by the local Ethics Committee, in accordance with the Helsinki Declaration; signed informed consent to use and analyze imaging data was obtained from all participants before entering the study. All accrued patients were evaluated by our internal tumor board before and after the completion of the neoadjuvant treatment.
Treatment
Thirty-two of the 57 patients enrolled in this study were part of a protocol (RectumSIB: ClinicalTrials.gov identifier: NCT01224392) comparing the standard neoadjuvant radiation treatment (CRT arm; 17 patients) to an exclusive radiotherapy treatment protocol with a simultaneous integrated boost (RT arm; 15 patients), as previously reported [18]. The remaining 25 patients performed standard CRT treatment with the same technical specification of the RectumSIB protocol. Six to eight weeks after the end of radiotherapy, all patients repeated the MRI examination of the rectum and FDG-PET and subsequently underwent total mesorectal excision (TME).
Reference standard
Resected tumors were evaluated by an experienced pathologist. All surgical specimens were received under vacuum sealing and stored at 4 °C within 2 h from the resection. The specimens were then opened and the tumor bed was macroscopically identified and extensively sampled at 5-mm intervals. Tissue slices were then fixed in 10% buffered formalin at room temperature for 24 h and subsequently paraffin embedded. Semiserial sectioning at 0.5-mm intervals from each tissue slice from the tumor bed was performed and the sections stained with hematoxylin and eosin for microscopic examination. Semi-quantitative pathological evaluation of primary tumor regression was performed, determining the number of residual tumor cells compared with the desmoplastic response, using the Mandard’s five-point assessment scheme [19].
In this system, TRG 1 represents a complete regression (fibrosis without detectable tissue of tumor); TRG 2 represents a partial response (rare residual tumor cells); TRG 3 is defined as fibrosis outgrowing residual tumor; TRG 4 is defined as residual tumor outgrowing fibrosis; TRG 5 represents a complete non-response (absence of regressive changes).
Imaging
MRI was performed with a 1.5-T scanner using an eight-channel phased-array surface coil (HDx Signa Excite, GE HealthCare, Milwaukee, WI, USA). Patients were positioned in the supine position and, unless contraindicated, a 20-mg intramuscular injection of butyl-scopolamine was administered intravenously 10 min before the beginning of the examination to minimize motion artifact induced by bowel peristalsis. The acquisition protocol included a fast spin-echo T2w sequence acquired on the axial plane perpendicular to the longest tumor diameter having the following scanning parameters: repetition/echo time (TR/TE) = 7660/110 ms, acquisition matrix = 416 × 224, slice thickness = 4 mm, pixel size = 0.4297 × 0.4297 mm2, field of view = 22 cm, and flip angle = 90°, as well as an axial EPI-SE diffusion-weighted (DW) sequence with the following scanning characteristics: TR/TE = 2000/87 ms, acquisition matrix = 96 × 128, slice thickness = 4 mm, pixel size = 0.8594 × 0.8594 mm2, field of view = 22 cm, flip angle = 90°, and b-value = 800 s/mm2. Total examination time including a sagittal and a coronal T2w sequence and a dynamic contrast enhanced T1w sequence was 31 min.
FDG-PET was performed using a PET-CT Gemini TF scanner (Philips Medical System, Cleveland, OH) with Time-of-Flight (TOF) technology. All studies were carried out according to the European Association of Nuclear Medicine (EANM) guidelines [19]. Before the examination, patients fasted for a minimum of 6 h and were required to have a serum glucose concentration below 200 ng/dl. A weight-based amount of [18F]FDG, with standard dose of 2.5 MBq/kg was injected. After 60 min of rest, patients were asked to void their bladder and were placed in supine position with arms raised. Image acquisition time was 15–20 min.
Image segmentation
We developed an algorithm using C++ and the ITK libraries to segment tumors on MR images [20]. The semi-automatic segmentation method was applied on both the T2w and the DW images. First, a bounding box enclosing the rectal region (Fig. 1a) was drawn manually on the T2w images. Second, the bounding box was automatically applied to both T2w and DW images to crop the two datasets along the same physical coordinates (Fig. 1b, c). Subsequently, a k-means algorithm was applied on both datasets. The k-means method is an unsupervised learning algorithm that classifies a given dataset through a k number of clusters, in which each observation is associated with the cluster having the closest mean. In our algorithm, we defined k = 3 for the T2w sequence and k = 5 for the DW image. On the T2w images, the cluster having the lowest mean intensity value is more likely to contain voxels belonging to the tumor, while on the DW images the two clusters with the highest intensity values more likely belong to non-necrotic areas of the tumor (Fig. 1d-e). Therefore, the final segmentation was composed of voxels belonging to the tumoral region in both T2w/DW datasets (Fig. 1f, g), i.e., the intersection between the two segmentations (Fig. 1h). Finally, the 2D biggest connected region is kept as the final region of interest, while other non-connected regions (i.e., noise, vessels, and regions outside the tumor) are discarded. Once the automatic segmentation was completed, an experienced radiologist (more than 10 years of experience in interpreting abdominal MRI) manually reviewed the results of segmentation on both T2w and ADC maps to include missing voxels and/or to exclude voxels that were erroneously included by the algorithm (Fig. 1i).
Pipeline of the tumor segmentation on the T2w image. a T2w image; b cropped T2w image: c cropped DW image; d k-means applied on the T2w image; e k-means applied on the DW image; f thresholding to extract tumoral cluster on the T2w image; g thresholding to extract tumoral cluster on the DW image; h intersection between k-means mask f and g; i final mask refined by the radiologist and superimposed to the T2w image
Segmentation of tumors on PET images was obtained using the previously described automatic Adaptive Threshold Algorithm [21]. First, a background area close to the lesion was drawn by a nuclear medicine physician, then the algorithm iteratively determined a threshold value based on the percentage of the maximum intensity in the cross-section area of a sphere containing the tumor. The threshold values were entirely based on the apparent activity concentration in the images and not on known activities. On our dataset, the threshold values ranged from 0.737 to 16.848. Finally, all masks were reviewed by an expert nuclear medicine physician.
Features extraction
The following radiomics features were extracted from voxels belonging to the segmented 2D mask in the T2w, ADC, and PET images: (a) five first-order parameters, i.e., mean intensity, median intensity, 10th, 25th, and 75th percentile; (b) 21 second-order texture parameters derived from the Gray-Level Co-Occurrence matrix (GLCM); (c) the mean standard uptake volume (SUVmean), the metabolic volume, defined as the area of the segmented PET mask, and the glycolytic volume, which is the product between metabolic volume and SUVmean.
The GLCM is a tabulation of how often different combinations of pixel brightness values (i.e., grey levels) occur between neighboring voxels in an image. Therefore, the GLCM allows the calculation of second-order texture features, i.e., describing the relationship between groups of contiguous pixels in the image. To extract the texture parameters, we first equalized the histogram by rescaling the intensities within each ROI between the 1st and the 99th percentile of the ROI over 64 bins. Using 64 equally divided bins has been a common approach for image quantification in radiomics analysis, and at the same time, it makes it possible to explore the whole range of tumor signal intensities [22]. Then, GLCMs were generated for each of the four directions of a 2D image, considering the distance between two neighboring voxels equal to one. Finally, the four matrices were averaged to make the method rotationally invariant to the distribution of texture. Texture features were computed using the MATLAB and Statistics Toolbox Release 2016b (The MathWorks, Inc., Natick, Massachusetts, USA).
Statistical analysis
Patients were dichotomized as responders (pR+), having TRG = 1 or 2, vs. non-responders (pR-) having TRG ≥ 3. The relationship between pR+/pR- and texture features was explored by both the mono-parametric and multi-parametric approach. When using the first approach, we evaluated the predictive value of each feature individually, using the Mann–Whitney test. For those variables that were statistically different between pR+ and pR- patients, we evaluated the area under the receiver operating characteristic (ROC) curve (AUC), sensitivity, and specificity at the best cut-off. Sensitivity was defined as the number of correctly classified pR+ patients over the total number of pR+ patients, while specificity was defined as the number of correctly classified pR- patients over the total number of pR- patients. The best cut-off maximizes the Youden index, which is the cut-point of the ROC curve that optimizes the biomarker’s differentiating ability when equal weight is given to sensitivity and specificity [23]. A p value < 0.05 was considered as indicating a significant AUC greater than 0.5. Analyses were performed with statistical software (MedCalc Statistical Software version 17.4, Ostend, Belgium).
Conversely, in the multiparametric approach, the accuracy in predicting pR+ of different feature subsets was assessed using a logistic classifier. Three feature subsets were created composed of: (a) features from MRI (T2w and ADC maps), (b) features from PET images, and (c) features from both MRI and PET images. However, since the dimensionality of features was high for all three subsets, a feature selection step was necessary to exclude irrelevant or redundant attributes that might cause overfitting and that might be a source of noise for the classifier [24]. To this scope, features were first ranked according to their AUC in discriminating between pR+ and pR-; subsequently, the correlation matrix between features was computed to detect which pairs were highly correlated. When a couple of features showed a Pearson’s linear correlation ≥ 0.8, we discarded the feature with the lower AUC. To improve stability and avoid bias, this selection was repeated 100 times using random training sets composed of 80% of the whole patients’ dataset. Only features chosen more than 60 times were included in the subsequent analysis. Once the three subsets of features were created, they were fed into a logistic classifier, applying the stepwise regression method to further exclude irrelevant variables. Within this procedure, the method searches for terms to add to or remove based on the p value of the F-statistics and iteratively add or remove terms that have p value ≤ 0.05 and/or p value > 0.20, respectively. Sensitivity and specificity, as previously defined, were then estimated.
Results
Patients
From the initial cohort of 57 patients, three were excluded due to MRI artifacts and two because the TRG score was not evaluated. The final dataset included 52 patients, of whom 35 men (68%). Twenty-two patients were classified pR+ (nine with TRG = 1, 13 with TRG = 2), the remaining 30 were classified as pR- (16 with TRG = 3, 13 with TRG = 4, and 1 with TRG = 5). Patient and lesion characteristics are reported in Table 2. Of note, age was statistically lower in the pR+ group, grade 0 and 1 tumors were more represented in the pR+ group, grade 2 and 3 were more represented in the pR- group, pT0 tumors were significantly more represented in the pR+ group while pT3 were significantly more represented in pR- group.
Mono-parametric approach
In total, 12 features derived from PET images and five features computed on the ADC maps accurately predicted response to neoadjuvant therapy (Table 3). The maximum AUC (0.77) was obtained by PET homogeneity, which also showed a good balance between sensitivity (72.7%, 16/22) and specificity (76.7%, 23/30). Lower values of the PET homogeneity feature were measured in responders (< 0.18). The highest sensitivity in the prediction of response was obtained by PET glycolytic volume and ADC dissimilarity (both 90.9%; 20/22) at the cost of a low specificity (56.8% [17/30] and 43.3% [13/30], respectively). Conversely, the highest specificity in response prediction was obtained by metabolic volume (83.3% [25/30]) to the detriment of sensitivity (63.6% [14/22]). Figure 2a shows ROC curves of PET and ADC features having the highest AUC values and reaching the highest sensitivities. T2w features did not differ between pR+ and pR- patients with the mono-parametric approach (Fig. 3).
a Receiver operating characteristic (ROC) curves of PET and ADC features having the highest areas under the ROC curve and the highest sensitivities; b radiomics signature score for every patient: the green marks indicate the patients in the responder group, while the red marks represent the patients in the non-responder group. The dotted line is the threshold which optimizes sensitivity over specificity
Figure 4 shows the mean AUC reached by each radiomics feature computed on PET, T2w, and ADC images during the 100 repetitions. Features from PET images obtained higher AUCs compared to features from MRI. The median AUC obtained by texture features from PET was 0.65 (IQR = 0.61–0.73); from T2w image, 0.57 (IQR = 0.56–0.58); and from ADC maps, 0.59 (IQR = 0.57–0.63).
Area under the receiver operating characteristics (ROC) curve reached by each of radiomics features computed on both PET and MRI (T2w and ADC) images. ROCs were computed 100 times using random training sets composed of 80% of the whole patients’ dataset. Means and standard deviations are shown. IMC information measure of correlation; INN inverse difference moment; IDNN inverse difference moment normalized
Multi-parametric approach
The feature selection step returned three features subsets composed of: (a) features from PET images, (b) features from MRI, (c) features from both PET and MRI. Features included in each subset are listed in Table 1 (columns 2–4).
The logistic classifier obtained using only PET features included homogeneity, contrast, metabolic volume, glycolytic volume, and 10th percentile, and reached an AUC of 0.84, a sensitivity and a specificity at the Youden index (0.5) of 77% (17/22) and 83% (25/30), respectively. In order to obtain the highest sensitivity in predicting pR+, i.e., ensuring treatment to the large majority of responder patients, we could lower the cut-off value. With a cut-off value of 0.28 we obtained a sensitivity of 91% (20/22) and a specificity of 57% (17/30) in recognizing pR+ patients. When only features from MRI were used, the logistic regression created a model containing 10th percentile and correlation 1 from T2w images, and cluster prominence and information measure of correlation 2 from the ADC maps. This model reached an AUC of 0.72, and a sensitivity and specificity of 73% (16/22) and 70% (21/30) respectively, at the Youden index (0.37). Finally, when features from PET and MRI were combined, the logistic regression computed a model containing: PET homogeneity, PET contrast, PET 10th percentile, glycolytic volume, metabolic volume, and T2w correlation 1. The AUC obtained by this model was 0.86, while sensitivity and specificity at the Youden index (0.42) were 86% (19/22) and 83% (25/30), respectively. When lowering the cut-off to 0.25, the model will correctly recognize as responder one additional patient (sensitivity 91%; 20/22) but at the expense of a marked reduction in specificity (53%; 16/30) (see Fig. 2b).
Discussion
In this study, we show that a logistic regression model containing five second-order PET texture features and one second-order texture feature from the T2 MRI sequence yields the highest predictivity in determining which patients will or will not respond to neoadjuvant therapy (AUC = 0.86). However, the combined PET-MRI regression model yielded results that were only slightly better than a model including only 5 PET features (AUC = 0.84) and far better than a model including only MRI features (AUC = 0.72). If FDG-PET is confirmed as a reliable predictor of response, then hybrid PET-MRI imaging could be implemented in the future both to stage LARC and to tailor treatment to the individual patient.
In the above-described regression models the Youden index allows determination of the best cut-off value between sensitivity and specificity. However, from a clinical standpoint, a high-sensitivity value would ensure that most patients that will benefit from CRT treatment actually receive it, even if this will come at the cost of a large group of patients having to undergo treatment unnecessarily. In our study, the highest sensitivity was obtained by lowering the cut-off value of the ROC curve of the multiparametric PET model to 0.28. Assuming we were in clinical practice, the PET model would have correctly suggested the use of CRT in over nine out of ten patients (i.e., 91% sensitivity) and avoided unnecessary treatment in almost six of ten patients (i.e., 57% specificity).
Of note, the AUC was lower for each first- or second-order feature taken individually. Overall, PET homogeneity yielded the best results with an AUC of 0.77 followed by PET inverse difference normalized and ADC entropy. Based on current knowledge, an explanation cannot be given as to why patients with metabolically inhomogeneous LARC respond better to neoadjuvant treatment; however, a similar trend was also reported by Lovinfosse et al. [14]. We might envisage that molecular traits of tumor may be responsible for poor response and that combining molecular and imaging metrics will allow better comprehension of the mechanisms that underlie tumor objective response. One additional finding of this study is that we confirm that dissimilarity and contrast directly correlate with good response, while metabolic volume and glycolytic volume inversely correlate with good response to RT as previously demonstrated by other authors who adopted a different method, i.e., odds ratio to evaluate correlation between individual features and response to CRT instead of ROC analysis [14, 15].
There are some points of strength of this study that deserve consideration. First, unlike most previous studies on the assessment of radiomics features of LARC [12, 28], in this study not only first- but also second-order features were extracted to assess tumor characteristics. Second-order features provide information on the spatial relations between neighboring regions/voxels of the tumor, which first-order features, such as histogram-derived features, cannot explore. Lovinfosse et al. [14] showed that histogram features failed to independently predict outcome measures in multivariate analysis. Contrarily, the same authors identified coarseness, a local textural feature that quantifies granularity of tumor, as a predictor of disease-free survival [14]. Second, multivariate analysis was performed to identify groups of features that were more predictive of response to treatment than individual parameters. Third, the collection of PET and MR images we used were all from the same institution, taken on the same equipment and using the same exam protocols, ensuring a high reproducibility of test. Fourth, to our knowledge, this is the first time that PET and MRI features have been combined in a single classifier with the aim of predicting response to neoadjuvant therapy in LARC, with promising results. For data analysis, we used the original image instead of filtered images as in Dinapoli et al. [28]. Using original images avoids including bias in the original data and allows exploitation of a larger number of texture parameters.
There are also limitations to this work. First, our results should be validated on data originating from different scanners and different acquisition protocols. Second, texture analysis was performed on the largest single-slice mask rather than on the whole tumor. However, the decision to use the whole dataset or only the slice most representative of the tumor to measure radiomics features has long been debated and which is best has not been convincingly determined. Indeed, Ng et al. [29], in a study involving 55 patients with primary colorectal cancer, showed that entropy computed on CT images was higher and uniformity lower for the whole tumor volume compared to the largest cross-sectional area at all filter levels and Kaplan–Meier analysis showed better separation of entropy and uniformity for whole-tumor analysis for 5-year overall survival. Nevertheless, findings of other authors appear to be going in a different direction [30]. Lubner et al. [31], for example, compared 2D and 3D texture features from CT images in a subset of 20 patients with hepatic metastatic colorectal cancer and demonstrated that overall results were fairly similar in Bland–Altman analysis (e.g., for entropy, the limits of agreement were − 0.0182, 0.029, bias 0.005). Also, in a larger study involving 588 patients with non-small cell lung cancer, Shen et al. [32] demonstrated that 2D texture features performed slightly better in discriminating between high- and low-risk tumors, thus suggesting their use in clinical practice since they are less time-consuming and do not require heavy-load computation needed for 3D analysis.
Moreover, 2D analysis has been previously demonstrated to be a robust prognostic tool to provide important information for patient management [33,34,35,36,37,38].
Several advantages of 2D analysis may be worthwhile addressing. First, in a clinical perspective where time is an important issue, 2D analysis is more straightforward than 3D analysis, whether performed manually or via semi-automatic segmentation. Second, in the specific setting of rectal cancer, 3D segmentation of 2D MRI images may not be accurate, in particular on the cranial and caudal margins of the lesion due to low tissue contrast on T2w images and low spatial resolution of DWI.
In our study, we tried to reduce reader variability by automatically detecting the largest slice of the tumor and semi-automatically segmenting it. The final segmentation required a minimal user intervention and might be easily integrated in clinical practice, providing a straightforward tool for better management of patients. Currently, we are working on the implementation of a deep-learning algorithm to automatically segment rectal cancers on MR images, and, if successful, in the future we will test this algorithm to extract 3D texture features, comparing the results with our current findings of 2D analysis.
Third, we used a semi-automatic method for segmentation of the tumor, which cannot completely avoid inter-reader variability. However, this is the first study attempting to perform a semi-automatic segmentation, which is a very challenging task due to the low contrast between tumor and healthy regions.
In conclusion, in this study we explore the potential role of texture parameters derived from pretreatment MRI and PET images in predicting the response to CRT/RT in patients with LARC. These preliminary results, if confirmed, could be useful to personalize patient treatment, for example to avoid toxicity of neoadjuvant therapy in patients predicted non-responders.
References
Siegel R, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30.
NCCN Clinical Practice Guidelines in Oncology: Rectal Cancer, Version 1.2016. (2016). NCCN.org. Accessed 11 Jan 2019.
Sauer R, Becker H, Hohenberger W, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351(17):1731–40.
Li Y, Wang J, Ma X, Tan L, Yan Y, Xue C, et al. A review of neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Int J Biol Sci. 2016;12(8):1022–31.
Maas M, Nelemans PJ, Valentini V, et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2010;11:835–44.
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced. Eur J Cancer. 2012;48(4):441–6.
Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105(13):5213–8.
Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative. Nat Commun. 2014;5:4006.
Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RT, Hermann G, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114(3):345–50.
Jalil O, Afaq A, Ganeshan B, Patel UB, Boone D, Endozo R, et al. Magnetic resonance based texture parameters as potential imaging biomarkers for predicting long-term survival in locally advanced rectal cancer treated by chemoradiotherapy. Color Dis. 2017;19(4):349–62.
Liu L, Liu Y, Xu L, Li Z, Lv H, Dong N, et al. Application of texture analysis based on apparent diffusion coefficient maps in discriminating different stages of rectal cancer. J Magn Reson Imaging. 2017a;45(6):1798–808.
De Cecco CN, Ganeshan B, Ciolina M, Rengo M, Meinel FG, Musio D, et al. Texture analysis as imaging biomarker of tumoral response to neoadjuvant chemoradiotherapy in rectal cancer patients studied with 3-T magnetic resonance. Investig Radiol. 2015;50(4):239–45.
Cusumano D, Dinapoli N, Boldrini L, Chiloiro G, Gatta R, Masciocchi C, et al. Fractal-based radiomic approach to predict complete pathological response after chemo-radiotherapy in rectal cancer. Radiol Med. 2017;123:286.
Lovinfosse P, Polus M, Van Daele D, Martinive P, Daenen F, Hatt M, et al. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur J Nucl Med Mol Imaging. 2018;45(3):365–75.
Bundschuh R, Dinges J, Neumann L, Seyfried M, Zsótér N, Papp L, et al. Textural parameters of tumor heterogeneity in 18F-FDG PET/CT for therapy response assessment and prognosis in patients with locally advanced rectal cancer. J Nucl Med. 2014;55(6):891–7.
Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60(14):5471–96.
Mandard A, Dalibard F, Mandard JC, Marnay J, Henry-Amar M, Petiot JF, et al. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathol Correl Cancer. 1994;73(11):2680–6.
Engels B, De Paoli A, Cattari G, Munoz F, Vagge S, Norkus D, et al. Preoperative radiotherapy with a simultaneous integrated boost compared to chemoradiation therapy for T3-4 rectal cancer: interim analysis of a multicentric randomized trial. Int J Radiat Oncol Biol Phys. 2014;90(1):S22–3.
Boellaard R, O’Doherty MJ, Weber WA, Mottaghy FM, Lonsdale MN, Stroobants SG, et al. FDG PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0. Eur J Nucl Med Mol Imaging. 2010;37(1):181–200.
Johnson HJ, McCormick M, Ibanez L. The ITK software guide. 3rd ed. New York: Kitware Inc.; 2013.
Brambilla M, Matheoud R, Basile C, Bracco C, Castiglioni I, Cavedon C, et al. An adaptive thresholding method for BTV estimation incorporating PET reconstruction parameters: a multicenter study of the robustness and the reliability. Comput Math Methods Med. 2015;2015:571473.
Cook GJR, Azad G, Owczarczyk K, Siddique M, Goh V. Challenges and promises of PET radiomics. Int J Radiat Oncol Biol Phys. 2018;102(4):1083–9.
Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5.
Chizi B, Maimon O. Dimension reduction and feature selection. In: Maimon O, Rokach L, editors. Data mining and knowledge discovery handbook. Boston: Springer; 2005. p. 93–111.
Soh L, Tsatsoulis C. Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Trans Geosci Remote Sens. 1999;37(2):780–95.
Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;SMC-3:610–21.
Clausi DA. An analysis of co-occurrence texture statistics as a function of grey level quantization. Can J Remote Sens. 2002;28(1):45–62.
Dinapoli N, Barbaro B, Gatta R, Chiloiro G, Casà C, Masciochi C, et al. Magnetic resonance, vendor-independent, intensity histogram analysis predicting. Int J Radiat Oncol Biol Phys. 2018;102:765.
Ng F, Kozarski R, Ganeshan B, Goh V. Assessment of tumor heterogeneity by CT texture analysis: can the largest cross-sectional area be used as an alternative to whole tumor analysis? Eur J Radiol. 2013;82(2):342–8.
Liu Y, Liu S, Qu F, Li Q, Cheng R, Ye Z. Tumor heterogeneity assessed by texture analysis on contrast-enhanced CT in lung adenocarcinoma: association with pathologic grade. Oncotarget. 2017b;8(32):53664–74.
Lubner MG, Stabo N, Lubner SJ, del Rio AM, Song C, Halberg RB, et al. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40(7):2331–7.
Shen C, Liu Z, Guan M, Song J, Lian Y, Wang S, et al. 2D and 3D CT radiomics features prognostic performance comparison in non-small cell lung cancer. Transl Oncol. 2017;10(6):886–94.
Henderson S, Purdie C, Michie C, Evans A, Lerski R, Johnston M, et al. Interim heterogeneity changes measured using entropy texture features on T2-weighted MRI at 3.0 T are associated with pathological response to neoadjuvant chemotherapy in primary breast cancer. Eur Radiol. 2017;27(11):4602–11.
Giannini V, Mazzetti S, Marmo A, Montemurro F, Regge D, Martincich L. A computer-aided diagnosis (CAD) scheme for pretreatment prediction of pathological response to neoadjuvant therapy using dynamic contrast-enhanced MRI texture features. Br J Radiol. 2017;90(1077):20170269.
Vignati A, Mazzetti S, Giannini V, Russo F, Bollito E, Porpiglia F, et al. Texture features on T2-weighted magnetic resonance imaging: new potential biomarkers for prostate cancer aggressiveness. Phys Med Biol. 2015;60(7):2685–701.
Skogen K, Schulz A, Dormagen JB, Ganeshan B, Helseth E, Server A. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol. 2016;85(4):824–9.
Ganeshan B, Skogen K, Pressney I, Coutroubis D, Miles K. Tumour heterogeneity in oesophageal cancer assessed by CT texture analysis: preliminary evidence of an association with tumour metabolism, stage, and survival. Clin Radiol. 2012;67(2):157–64.
Ganeshan B, Burnand K, Young R, Chatwin C, Miles K. Dynamic contrast-enhanced texture analysis of the liver: initial assessment in colorectal cancer. Investig Radiol. 2011;46(3):160–8.
Funding
This work was funded by “AIRC 5xmille Special Program Molecular Clinical Oncology - Ref. 9970” and “FPRC 5xmille 2013 Ministero Salute”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Giannini, V., Mazzetti, S., Bertotto, I. et al. Predicting locally advanced rectal cancer response to neoadjuvant therapy with 18F-FDG PET and MRI radiomics features. Eur J Nucl Med Mol Imaging 46, 878–888 (2019). https://doi.org/10.1007/s00259-018-4250-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-018-4250-6