Abstract
Introduction
The combination of Positron Emission Tomography (PET) with magnetic resonance imaging (MRI) in hybrid PET/MRI scanners offers a number of advantages in investigating brain structure and function. A critical step of PET data reconstruction is attenuation correction (AC). Accounting for bone in attenuation maps (μ-map) was shown to be important in brain PET studies. While there are a number of MRI-based AC methods, no systematic comparison between them has been performed so far. The aim of this work was to study the different performance obtained by some of the recent methods presented in the literature. To perform such a comparison, we focused on [18F]-Fluorodeoxyglucose-PET/MRI neurodegenerative dementing disorders, which are known to exhibit reduced levels of glucose metabolism in certain brain regions.
Methods
Four novel methods were used to calculate μ-maps from MRI data of 15 patients with Alzheimer’s dementia (AD). The methods cover two atlas-based methods, a segmentation method, and a hybrid template/segmentation method. Additionally, the Dixon-based and a UTE-based method, offered by a vendor, were included in the comparison. Performance was assessed at three levels: tissue identification accuracy in the μ-map, quantitative accuracy of reconstructed PET data in specific brain regions, and precision in diagnostic images at identifying hypometabolic areas.
Results
Quantitative regional errors of −20–−10 % were obtained using the vendor’s AC methods, whereas the novel methods produced errors in a margin of ±5 %. The obtained precision at identifying areas with abnormally low levels of glucose uptake, potentially regions affected by AD, were 62.9 and 79.5 % for the two vendor AC methods, the former ignoring bone and the latter including bone information. The precision increased to 87.5–93.3 % in average for the four new methods, exhibiting similar performances.
Conclusion
We confirm that the AC methods based on the Dixon and UTE sequences provided by the vendor are inferior to alternative techniques. As a novel finding, there was no substantial difference between the recently proposed atlas-based, template-based and segmentation-based methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Positron emission tomography (PET) can be used in combination with [18F]-Fluorodeoxyglucose (FDG) to measure regional glucose metabolism. Early stages of Alzheimer’s dementia (AD) exhibit reduced levels of glucose metabolism in the parietotemporal cortex, posterior cingulate, and precuneus regions, spreading to the frontal cortex as the disease evolves. Alternative biomarkers, especially β-amyloid biomarkers such as [11C]-Pittsburgh Compound B or [18F]-Florbetaben, are under investigation to better diagnose AD at early stages. Given the different regional levels of binding in various areas of the brain, and the different sensitivity of particular areas to the aforementioned radioligands, a robust and accurate quantification in PET is of paramount importance.
The combination of PET with magnetic resonance imaging (MRI) can help to differentiate AD from other dementias, which can show glucose metabolism abnormalities in similar ways, and has demonstrated higher accuracy at detecting mild AD than PET or MRI used separately [1, 2]. Simultaneous PET/MRI potentially provides a number of benefits compared to two consecutive PET and MRI scans, ranging from motion correction in PET [3] to anatomically-guided PET image reconstruction [4]. The simultaneous combination of PET and functional MRI can provide information of two mechanisms expressing simultaneously two correlated functions, which would not be observed otherwise. Additionally, for neurological studies, a PET/MRI scan increases patient throughput, patient comfort, and produces perfectly co-registered anatomical/functional images. On the other hand, the use of simultaneous PET/MRI poses a number of challenges that need to be addressed. One of the most important is widely recognized as attenuation correction (AC), mandatory for quantitative PET data. Such is the case that AC in PET/MRI has already been the subject of several reviews [5–8].
MRI data is related to tissue proton density, while computed tomography (CT) data is related to tissue electron density, which is directly related to photon attenuation in the subject. Therefore, to obtain an accurate attenuation map (μ-map) with MRI is not trivial. There exist currently two commercial integrated whole body human PET/MRI systems: the Biograph mMR (Siemens Healthcare GmbH, Erlangen, Germany) and the SIGNA PET/MRI (GE Healthcare, Waukesha WI, USA). AC in the Biograph mMR system is currently performed using a μ-map derived from a 2-point Dixon [9] or a dual Ultra Short Time Echo (UTE) [10] pulse sequence. In the SIGNA PET/MRI system the μ-map is based on a head atlas which takes less than 30 s to calculate for a given patient [11]. With the growing number of simultaneous PET/MRI systems installed worldwide (over 60 systems installed) there is a substantial number of AC methods proposed by different research groups focused mainly in head studies. This activity was partly triggered by the poor performance obtained by the AC methods available in the Biograph mMR system [12, 13], where quantitative errors of ∼20 % in cortical areas and ∼10–15 % in subcortical areas were measured. In contrast, an evaluation of the method in the GE system showed errors of 2.19 ± 1.40 % in eight subjects compared to a μ-map obtained from a CT scan [11]. Most of the published methods reported the accuracy of their performance using different figures of merit, looking into the brain as a whole or in different specific brain areas. Comparing the performance between methods is difficult since different groups used different figures of merit and/or looked into different regions.
AC methods can be broadly classified into atlas/template-based methods and segmentation-based methods. Alternatively, attenuation information can be estimated using maximum-likelihood reconstruction of activity and attenuation [14], and improved including time-of-flight information in the reconstruction algorithm [15]. However, the feasibility of the latter approach in neurological studies has not been investigated yet. An alternative approach that has shown to provide excellent results is based on MR sequences with special k-space sampling strategies, capable of measuring signal from bone with high quality. Examples of this approach are an improved k-space sampling for UTE sequences [16], the zero-time-echo MR sequence [17], and the pointwise encoding time reduction with radial acquisition MR sequence [18]. Nonetheless, the most common approach for MRI-based AC is to use MR images acquired from well-established sequences without modifying the sampling schemes.
Most of the existing image processing methods are based on atlases created with different types of datasets. Pairs of CT and dual UTE [19–21], or CT and T1 weighted [11, 22] are common choices. Other approaches introduce more information in the atlas like images from Dixon sequences [23, 24] or T2 weighted images and UTE images [25]. In some cases a new patient is compared with the atlas using probabilistic measures [11, 20–23, 25] or pattern recognition approaches [19, 24]. The method presented by [26] contains several similarities compared to the atlas-based methods, using a preliminary atlas based on pairs of CT and T1 weighted images, with the important difference that this method calculates a template out of the atlas averaging the registered data sets. The combination of atlas or template with segmentation of specific tissues was used in [23, 26] to improve the registration process. Most methods target only head AC [19–22, 24–26], although some approaches tackle the more challenging situation of whole body AC [6, 23]. Atlas and template-based methods generally result in high quality μ-maps, with significant similarity with typical CT-based μ-maps (μ-mapCT) in the bone identification and linear attenuation coefficients (LAC). However, these approaches tend to require long computational time due to the large number of datasets to be registered and segmentations to perform. Besides, patients with brain alterations or deformations are likely to fail in the process of identifying the different brain structures with the atlas or template. Additionally, the application of these methods to children requires the acquisition of double scans (MRI and CT) to create a database, which can raise ethical issues due to the radiation exposure.
Alternatively to atlas or template-based methods there is a significant number of studies based on segmentation of MR images. The accuracy of the bone representation in the image to segment is partly the key factor of such approaches. Most methods use dual UTE images, combined by calculating the R2 map [27–29] or similar relations [30, 31], although T1 weighted images have also been used [32]. The accuracy of the R2 map to extract bone structures was thoroughly analysed [33], resulting in a significant correct identification of bone structures but with artefacts in some areas, such as dental implants, folds of the neck fat, and air-bone interfaces.
There is a growing number of AC approaches, but no quantitative comparisons between methods has been yet presented. Therefore, it is not clear whether there is still room for improvement for new AC methods. Moreover, the maximum allowed quantitative error for the different cerebral cortical and subcortical regions is not well known. Test-retest studies suggest that small subcortical structures tend to have larger variability than large cortical structures. Intra- and inter-patient test-retest studies of regional cerebral metabolic rate of glucose consumption (CMRglc) resulted in 5.5 ± 0.5 %, 8.4 ± ,0.7 % and 8.0 ± 0.6 % average variability in the hippocampus, parieta, l and temporal regions, respectively [34, 35].
In this work we present a comparison between four PET/MRI AC methods: two methods based on an atlas [19, 22], one method based on a template [26], and one method based on segmentation [28]. One atlas-based method [22] and the template-based method rely on pairs of CT and T1 weighted images, while the other atlas-based method [19] relies on pairs of CT and dual UTE images. The segmentation-based method relies uniquely on dual UTE images, which are combined using the R2 map. The main reason to compare these methods was their significant quantitative accuracy in reconstructed PET data. Another reason was that these methods were available upon request from their authors. The timing performance was not considered as a critical parameter for the selection of the methods, although this aspect will be discussed in the context of clinical routine work flow.
Comparisons were performed first by measuring the level of agreement between μ-maps for air cavities and bone. Secondly, looking into reconstructed PET data, the focus was on brain regions that can potentially experience functional impairments in patients with suspected AD. An important point considered in this work is the accuracy at identifying regions showing hypometabolism resulting from each μ-map in the AC for each patient. One of the most established analysis tools in clinical routine is the three-dimensional stereotactic surface projection 3D-SSP/NEUROSTAT software toolkit [36], where the reconstructed PET data of each patient is compared with a database comprised of reconstructed PET data from healthy patients. This toolkit produces 3D surfaces showing areas with abnormal levels of FDG uptake compared to the database. We processed, with the 3D-SSP/NEUROSTAT software, the reconstructed PET data produced using all the studied μ-maps. The μ-mapCT and the reconstructed PET data using the μ-mapCT were used as reference for the three comparison levels aforementioned: μ-map, reconstructed PET data, and diagnostic images.
Materials and methods
PET/MRI device
The PET/MRI scanner used in this work was the Biograph mMR (Siemens Healthcare GmbH, Erlangen, Germany) with the software version VB20P. The mMR is a fully integrated system with a 3 Tesla MRI magnet and a PET ring based on avalanche photodiode technology. The dimensions of the magnet are a 60 cm inner diameter with an axial field of view of 45 cm, and the dimensions of the PET bore are 59.4 cm diameter per 25.8 cm axially. The spatial resolution in the centre of the scanner is 4.3 mm and the sensitivity is 15 cps/MBq [37]. The gradient coil has a gradient field of 45 mT/m with a switching time of 200 T/m/s. The head/neck coil used in all the studies presented here was the 16 channels Total Imaging Matrix (TIM).
Imaging protocol
Fifteen patients (seven female and eight male) with suspected AD or neurodegenerative disorders were selected for the present retrospective study. The average patient age was 58.8 ± 7.8 years [range 49 to 76 years], and the weight was 76.5 ± 18.5 kg [range 50 to 128 kg]. All subjects gave written informed consent, and all procedures were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The patients were scanned in a PET/CT (Biograph mCT; Siemens Healthcare, Knoxville TN, USA) and in the PET/MRI. The emission data in the PET/MRI were acquired 42.1 ± 17.6 min [range 30 to 84 min] after administering 183.3 ± 3.4 MBq [range 179 to 190 MBq] of 18F-FDG. The data were acquired for 15 min in 3D with an energy window of 430–610 keV, and reconstructed using ordered-subsets expectation-maximization (three iterations, 21 subsets) filtered by a Gaussian mask of 5 mm full-width at half-maximum. Reconstruction was performed with the Siemens off-line e7 tools. Resulting images had 256 × 256 × 127 voxels with 1.40 × 1.40 × 2.03 mm3 dimensions. The PET data reconstructed in every case was always the PET data acquired from the mMR (including the one using the μ-mapCT), to avoid functional discrepancies in the PET data comparison. The MR acquisition took 36 min to acquire several sequences, to obtain anatomical and functional information, the Dixon (19 s) and UTE (100 s) among them.
The MR sequences simultaneously acquired with the PET scan included the T1-weighted 3-dimensional magnetization-prepared rapid gradient echo (MPRAGE – 256 × 240 × 160 voxels with 1 × 1 × 1 mm3), two UTE (192 × 192 × 192 voxels with 1.56 × 1.56 × 1.56 mm3) with different echo times (0.07 and 2.46 ms), and a 2-point Dixon (192 × 126 × 128 voxels with 2.34 × 2.34 × 2.73 mm3). The CT data were acquired in the PET/CT scanner for each patient at 120 kVp and 21 mA for low dose, and contained 512 × 512 × 75 voxels with 0.97 × 0.97 × 3 mm3 per voxel.
Attenuation correction methods
The two methods to calculate the μ-map available in the Siemens mMR scanner are based on a 2-point Dixon sequence (19 s) (μ-mapDX), where bone is ignored, and on a dual UTE sequence (100 s) (μ-mapUTE), where bone is taken into account. The entire MR acquisition took 36 min. All the novel methods studied in this work to create the μ-map for AC are briefly discussed below. In all cases the algorithms produce a continuous spectrum of LACs. For details, their references provide detailed descriptions of the methods.
-
1.
Atlas-based method (μ-mapATL) [22]: This method relies on an atlas database comprised of paired T1-weighted MRI and CT data sets. The T1-weighted data set of each subject is rigidly registered to its corresponding CT data set. Once a new subject is scanned, all the T1-weighted MRI data sets from the database are registered to the target patient T1-weighted data set using a B-splines-based non-rigid registration. The CT data set for the new patient is then calculated from averaged weights, obtained using a similarity measure extracted from the database and the subject T1-weighted data set. The μ-map is calculated from the CT data by converting from Hounsfiled units (HU) to LAC using a bilinear transformation [38].
-
2.
Template-based method (μ-mapTMP) [26]: This method relies on an atlas database created with paired T1-weighted MRI (with a gadolinium-based contrast agent) and CT data sets, in this case to produce a template. All the operations are performed with the SPM8 toolbox (Statistical Parametric Mapping, Wellcome Trust Institute of Neurology, University College London) [39]. To create the template, first all the CT data sets are rigidly registered to their corresponding T1-weighted MRI data sets. Subsequently, all the tissue classes of the MRI data sets are segmented and elastically registered to a common space. Finally, all the CT data sets are transformed to the same common space as the registered MRI data sets. The final CT template is produced by averaging all the registered CT data sets. To calculate the μ-map for a new patient, the patient’s T1-weighted MRI is first registered and segmented using the same procedure as for the atlas. The template CT is then inversely transformed to the subject space, and is converted into a μ-map by applying a bilinear transformation [38] to the CT HU.
-
3.
ANN method (μ-mapANN) [19]: This method relies on a training database, in this case created with paired UTE data sets and CT data sets. The UTE data sets and a template-based AC map (TAC-map) are used as inputs of a feed forward neural network (FFNN). The method is based on a 3 layers FFNN algorithm, the aim of which is to calculate the network weights (training step, TS), in order to directly produce a μ-map with continuous LACs (classification step, CS). Patches of one voxel and six neighbours from both UTE images and the template-based μ-map [19] are the inputs of TS and CS, which use a sigmoid activation function for the middle (hidden) layer and a linear activation function for the output layer. During TS the images of a selected database are compared with the corresponding μ-mapsCT to determine the optimal network weights.
-
4.
R2 method (μ-mapR2) [28]: This method relies uniquely on the dual UTE images, where air, bone, and soft tissue are segmented. Air cavities are estimated by calculating the mean and standard deviation from the voxels located outside the head in the UTE scans, and then identifying those voxels inside the head that have similar statistical properties. The bone is estimated using the R2-map [27], derived from the difference of the logarithms between two images obtained from two consecutive UTE sequences acquired at different echo times. After the bone is identified, the intensity values of the voxels corresponding to bone are equalized to match the intensity values measured with CT. Finally, the remaining voxels are set as soft tissue.
Tissues classification evaluation
The first level to evaluate the different methods to estimate the μ-map was focused on studying the accuracy of each method to determine to which tissue (soft, bone, or air) each voxel corresponded in the μ-map. The μ-mapCT was used as reference. Since the μ-mapCT was acquired with the Biograph mCT scanner and all the other μ-maps were derived from MR images from the Biograph mMR, the μ-mapCT was rigidly registered to the MRI-derived μ-maps using the Syngo Multimodality Workplace (Siemens Healthcare GmbH, Erlangen, Germany) application. The registered μ-mapCT was later used for AC correction in the image reconstruction step for further quantitative PET analysis.
The figures of merit used to evaluate the level of agreement between μ-maps were the sensitivity and precision defined as \( \frac{TP}{TP+FN} \) and \( \frac{TP}{TP+FP,} \) respectively, where TPs are true positives, FNs are false negatives, and FPs are false positives. For this purpose all the μ-maps were binarized using the same thresholds: 500 HU (0.1165 cm−1) and −300 HU (0.03 cm−1) for bone and air, respectively. Once thresholded, the sensitivity and precision were calculated.
Regions close to air interfaces, nasopharyngeal cavities, within the folds of the neck fat, and where dental implants were present, were usually problematic for AC based on UTE sequences [33]. Moreover, the neck is a challenging region to analyse, since the rigid registration between the reference μ-mapCT and the MRI-derived μ-maps fails. The tissue classification evaluation was performed for the entire head (including neck) and also only for the part of the head where the brain is enclosed by applying a manually generated mask. Hence, potential mismatches produced in the areas enumerated above were not present in the brain-region tissue analysis.
Quantitative comparison
Each method was additionally compared by using each μ-map for AC and reconstructing the PET data as explained above. The resulting reconstructed PET data were analysed with SPM8. First, the MPRAGE dataset of each patient was rigidly registered to the common coordinate space of the μ-maps and PET images. Then, the T1 (MPRAGE) Montreal Neurological Institute (MNI) template was elastically registered to the MPRAGE data set of each patient. The MNI template contains a voxel atlas holding 116 anatomical predefined regions based on the automated anatomical labeling atlas. Once the MNI template was in the same coordinate space as the MPRAGE data, it was used to extract the quantitative information from the PET data. Finally, the mean was extracted from all the anatomical predefined regions in the template. The figure of merit used for the comparison between methods was the normalized error (En), defined as
using the μ-mapCT corrected PET images as reference. ĀCT and ĀX are the mean activities measured in a given region of interest (RoI) of the PET data reconstructed using the μ-mapCT for AC and the alternative μ-maps. For this study we analysed only those regions related to different stages of AD.
Quantitative evaluation in diagnostic images
For the clinical evaluation we analysed the results obtained after processing each reconstructed PET patient data obtained with the different μ-maps under study, with the 3D-SSP/Neurostat software toolkit, which is a tool used in clinical routine in our institution. The 3D-SSP/Neurostat software calculates statistical Z-scores from a patient, comparing with a database of normal controls that has μ-maps obtained from CT scans. The aim is to determine areas with abnormal levels of CMRglc using FDG normalized to a reference, which can be the global count, pons, cerebellum, or thalamus. Increased and decreased levels of CMRglc are separately produced. In this study we focused on reduced levels of CMRglc compared to the database. The 3D-SSP/Neurostat software extracts information of the metabolic activity projected onto surfaces; hence, it can be displayed from different views: superior, inferior, anterior, posterior, right, left, and medial. For more details about the 3D-SSP/Neurostat software, we refer the reader to the work of S. Minoshima et al. [36].
The amount of information generated for visual inspection for each patient and each different μ-map was substantial (eight views × four references × 15 patients × five μ-maps). To simplify the comparison, to reduce the amount of information to present, and to perform a systematic analysis, we defined two parameters. First, we focused our study on data normalized by the FDG signal in the thalamus, as suggested in [36], where larger differences between methods were obtained. This effect can be attributed to the fact that the reference regions can be under or overestimated in the same range as the analysed regions of interest. Secondly, we analysed the data using a range of thresholds of 1–3σ, to classify whether a cerebral region exhibits abnormal metabolic uptake. By applying the threshold to every view, for each reconstructed PET data from each patient and μ-map, we produced binary maps with a value of ‘0’ assigned to voxels corresponding to healthy tissue and ‘1’ to voxels corresponding to tissue with abnormal glucose uptake. Using the binary map obtained from the reconstructed PET data obtained with the μ-mapCT as ground truth, we calculated the precision at identifying regions with reduced CMRglc obtained with each of the μ-maps under study.
Results
Attenuation maps evaluation
Figure 1 shows a 3D rendering of the bone structure from the μ-map of one of the patients considered in this study, obtained with CT and the new four methods under study: μ-mapANN, μ-mapR2, μ-mapTMP, and μ-mapATL. All sagittal, coronal, and transversal views for the μ-mapCT, μ-mapDX, μ-mapUTE, μ-mapANN, μ-mapR2, μ-mapTMP, and μ-mapATL are shown in supplemental Fig. 1. The En (defined as Eq. (1) but applied to the μ-maps) obtained with the μ-mapUTE, μ-mapANN, μ-mapR2, μ-mapTMP, and μ-mapATL, compared to the μ-mapCT, is shown in supplemental Fig. 2. Relatively high errors are visible, mainly where bone and air cavities are present, and around the head, which were partly attributed to a mild mismatch between the μ-mapCT and the rest of the μ-maps due to small inaccuracies in the rigid registration performed with the Syngo Multimodality Workplace.
The sensitivity and precision for bone and air cavities, for the entire head and the brain area, are shown in Fig. 2. The results represent the mean calculated among the 15 patients. The error bars are the standard deviation computed between all patients and hemispheres. The numerical values for the true positives, false negatives, and false positives, from which sensitivity and precision were calculated, are shown in supplemental Tables 1 and 2.
In the case of the sensitivity analysis in the brain, comparing the different methods, the bone sensitivity for μ-mapUTE was 57.8 ± 9.2 %, while for the other four methods, it was 78.5–86.0 ± 4.3–11.3 %. For air cavities, the μ-mapUTE obtained the best sensitivity with 92.0 ± 4.2 %, whereas the other four methods obtained a sensitivity of 77.9–85.3 ± 6.6–14.8 %. The four novel methods performed similarly, while the μ-mapUTE showed better results for air cavities and worse results for bone. Similar conclusions were extracted in general from the analysis in the head but with lower sensitivity, especially in the case of the μ-mapUTE.
Regarding precision in bone, μ-mapANN slightly underperformed (62.1 ± 10.1 %) compared to the other methods, especially in the entire head analysis. The other four methods obtained similar performance (74.0–76.9 ± 8.2–9.7 %). In the air cavities, the μ-mapUTE obtained a precision of 40.7 ± 8.7 %, worse than the other four methods that resulted in a precision of 53.8–60.3 ± 9.3–12.3 %. Comparing the analysis in head and brain, as with sensitivity, the analysis in the entire head reduced the precision compared to the brain analysis, also reducing the differences between methods. For completeness we also calculated the Jaccard index for bone and air at different thresholds between the μ-mapCT and the μ-maps under evaluation (supplemental Fig. 3). Results consistently showed a higher Jaccard index obtained with the μ-maps obtained with the four assessed methods compared to μ-mapUTE, especially for bone.
In summary, μ-mapUTE produced high precision but with low sensitivity (less bone is identified but it is correctly identified), while the other four methods performed similarly, with μ-mapANN producing slightly lower precision, especially in bone tissue. Results obtained considering only the brain area showed that the sensitivity increased compared to the entire head, especially for the air cavities. Interestingly, the differences between methods were reduced considering only the brain region, which meant that the main differences were due to the nasopharyngeal cavities and neck. The precision also slightly increased for the brain analysis but still showing the same differences between methods as those observed for the head analysis.
Evaluation of reconstructed PET data
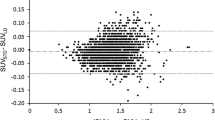
The PET data from each of the 15 patients were reconstructed using the μ-mapCT, μ-mapDX, μ-mapUTE, μ-mapANN, μ-mapR2, μ-mapTMP, and μ-mapATL for AC. The reconstructed PET data using μ-mapCT for AC was used as reference to calculate the En. The average En measured in those regions related to AD is shown in Fig. 3, together with the regions that were analysed. The variability was calculated as the standard deviation between patients and brain hemispheres. The area between ±5 % error is indicated in Fig. 3. The numerical values are shown in supplemental Table 3.
The analysis showed that μ-mapDX produced the most inaccurate results, compared to all the other methods, including the μ-mapUTE also available in the scanner [7, 8, 28], with an average En of -14.9 %, compared to −8.9 % measured with the μ-mapUTE. Among the four new methods compared in this study, the average errors in the analysed regions were 2.13, 1.04, −1.36, and −0.21 % for the μ-mapANN, μ-mapR2, μ-mapTMP and μ-mapATL, respectively, which means that the average FDG uptake was slightly overestimated with μ-mapANN and μ-mapR2 and slightly underestimated with mapTMP and μ-mapATL. The positive sign of the first two methods can be interpreted as an overestimation of the amount or density of the bone structure. The ANN-based method produced slightly higher errors than the other methods in the parietal region and the precuneus regions, in particular the parietal superior showed an En>5 %, which was over the limit considered as acceptable in this study. For completeness we show in supplemental Fig. 4 the absolute error measured in the same AD-related regions, and their numerical values in supplemental Table 4. The average absolute error with μ-mapDX and μ-mapUTE was below −1.0 kBq/mL, while the four new methods resulted in average errors of −0.04–0.24 kBq/mL.
Quantitative comparison in diagnostic images
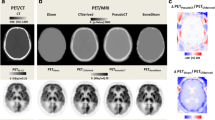
The reconstructed PET data of each patient were analysed with the 3D-SSP/Neurostat software toolkit. The Z-scores of the right lateral, left lateral, right medial, and left medial views normalized by the FDG signal in the thalamus, of the reconstructed PET data obtained with all the μ-maps for one exemplar patient are shown in supplemental Fig. 5. The En (defined as in (1) but used for the Z-score views) in Z-score calculated using the reconstructed PET data obtained with the μmapCT as reference for one patient is shown in Fig. 4.
En of the Z-scores obtained with the 3D-SSP/Neurostat for the right lateral (first column), left lateral (second column), right medial (third column), and left medial (fourth column) views, for the reconstructed PET data obtained with the μ-mapCT as reference compared to the μ-mapDX (a), μ-mapUTE (b), μ-mapANN (c), μ-mapR2 (d), μ-mapTMP (e), and μ-mapATL (f) for an exemplar patient
The mean precision and standard deviation (uncertainty) at identifying hypometbolic regions, for the 15 patients and the eight views analysed by the 3D-SSP/Neurostat toolkit are shown in Fig. 5. Figure 5a shows the different precisions obtained for a range of 1–3σ thresholds, demonstrating the precision drop as the σ threshold increases for μ-mapDX, while the other methods show similar performance for all the thresholds. Figure 5b shows the precision at a threshold of 2σ with the different background colors indicating different ranges of precision (0–50 % in red, 50–80 % in blue and 80–100 % in green). Visually, results obtained with μ-mapANN, μ-mapR2, μ-mapTMP, and μ-mapATL are in the green area, and μ-mapUTE and μ-mapDX are in the blue area. From Fig. 5b, the most precise approach was the method based on the template to calculate the μ-map (93.3 % in average), closely followed by the R2-based method (91.9 % in average), the atlas-based method (91.3 % in average), and the ANN-based method (87.5 % in average). The method based on the UTE resulted in a precision of 79.5 % in average. Finally, the precision obtained with the Dixon-based method was 62.9 % in average. The uncertainty measured with the template-based, atlas-based, and R2-based methods resulted in an uncertainty of 5.5–5.7 %, whereas the uncertainty measured with the ANN-based, UTE-based, and Dixon-based methods was in a range of 9.3–15.4 %. The numerical values of Fig. 5b are in supplemental Table 5.
Discussion
Different methods to calculate the μ-map derived from MRI data were analysed. Out of all the studied methods, the atlas/template methods, the most extended approach in the literature, produced visually the most CT-like μ-maps. However, looking at the quantitative analysis and the precision obtained at identifying hypometabolic regions, the results obtained with the atlas, template, and R2 approaches were similar, and slightly better compared to the ANN method.
For the quantitative analysis, the four analysed methods produced errors in the range of −3.2–4.1 %, with the exception of the parietal inferior region for the ANN-based method that resulted in an error of 6.4 %. In contrast, the two vendor’s AC methods produced errors between −5.9 and −10 %, when bone was included in the μ-map, and −10.2–−18.8 % when bone was completely ignored. The atlas-based and template-based methods slightly obtained underestimated results, while the ANN-based and R2-based methods obtained slightly overestimated PET uptake, in agreement with their original works (19, 22, 26, 28). This observations indicated that the former methods slightly underestimated, and the latter overestimated, the amount or density of bone tissue, while the consistently negative normalized error obtained with the vendor methods indicated that bone was underestimated. Insignificant differences were observed in the uncertainty between methods.
In the case of diagnostic images, the precision at identifying hypometabolic regions was 87.5–93.3 % in average, among views for the four AC methods under investigation. These results demonstrate that there are small differences between the four methods, and reveal that sophisticated (and computationally demanding) methods are not strictly required to obtain high precision diagnostic images. Among the analysed methods, negligible differences were observed between the atlas-based, template-based, and R2-based methods regarding PET quantitative and precision performance, while the ANN-based method performed marginally worse. The uncertainty obtained with the atlas/template-based and the R2-based method was within a range of 4.5–5.7 %, increasing to 9.3 % for the ANN-based method, and finally 12 % and 15.4 % for the UTE-based and Dixon-based methods, respectively.
It is worth noting that atlas/template methods can fail for patients with significant alterations in the brain that deviate too much from the atlas/template, while patient specific μ-maps are less likely to fail in these cases. On the contrary, methods based on the dual UTE sequence are susceptible to produce artefacts where there are dental implants, as shown in supplemental Fig. 6, where the μ-mapCT, μ-mapR2, and μ-mapTMP of a dementing patient with a dental implant are shown. However, there are methods to correct for such artefacts [40].
Regarding the computing time required to calculate each μ-map, the μ-mapATL required 2 h, the μ-mapTMP required 30 min, the μ-mapANN required 5 min and the μ-mapR2 required 5 s on a standard computer. The implementation of these methods could be accelerated, but that has not been the purpose of these projects so far. It is important to highlight that there are currently no hard constraints about the time required to calculate the μ-map. However, if high throughput is required, and given that patient scans are read by physicians the same day they are taken, the computational burden to calculate the μ-map can represent a bottle-neck which may potentially hinder a smooth work-flow in clinical routine. Therefore, the time required to calculate the μ-map is dependent on the number of patients and nature of studies performed.
Conclusion
Four different AC methods were evaluated, and compared with the methods provided by the vendor. Results showed that the four new methods performed in a range of ±5 % quantitative En in AD-related brain regions, and showed a precision in the range of 87.5–93.3 % at identifying hypometabolic regions, compared to the 3D-SSP/Neurostat database. This study showed that a range of methods for AC with different levels of sophistication produced similar quantitative and diagnostic images, as opposed to methods that ignore the bone structure or that calculate the bone structure in too simplistic ways.
References
Kawachi T, Ishii K, Sakamoto S, et al. Comparison of the diagnostic performance of FDG-PET and VBM-MRI in very mild Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2006;33(7):801–9.
Teipel S, Drzezga A, Grothe M, et al. Multimodal imaging in Alzheimer’s disease: validity and usefulness for early detection. Lancet Neurol. 2015;14(10):1037–53.
Grimm R, Fürst S, Souvatzoglou M, et al. Self-gated MRI motion modelling for respiratory motion compensation in integrated PET/MRI. Med Image Anal. 2015;19(1):110–20.
Bai B, Li Q, Leahy RM. MR guided PET image reconstruction. Semin Nucl Med. 2013;43(1):30–44.
Hofmann M, Pichler B, Schölkopf B, Beyer T. Towards quantitative PET/MRI: a review of MR-based attenuation correction techniques. Eur J Nucl Med Mol Imaging. 2009;36:S93–104.
Wagenknecht G, Kaiser HJ, Mottaghy FM, Herzog H. MRI for attenuation correction in PET: methods and challenges. MAGMA. 2013;26(1):99–113.
Keereman V, Mollet P, Berker Y, Schulz V, Vandenberghe S. Challenges and current methods for attenuation correction in PET/MR. MAGMA. 2013;26(1):81–98.
Visvikis D, Monnier F, Bert J, Hatt M, Fayad H. PET/MR attenuation correction: where have we come from and where are we going? Eur J Nucl Med Mol Imaging. 2014;41(6):1172–5.
Dixon WT. Simple proton spectroscopic imaging. Radiology. 1984;153(1):189–94.
Robson MD, Gatehouse PD, Bydder M, Bydder GM. Magnetic resonance: an introduction to ultrashort TE (UTE) imaging. J Comput Assist Tomogr. 2003;27(6):825–46.
Sekine T, Buck A, Delso G, et al. Evaluation of atlas-based attenuation correction for integrated PET/MR in human brain - application of a head atlas and comparison to true CT-based attenuation correction. J Nucl Med. 2016;57(2):215–20.
Dickson JC, O’Meara C, Barnes A. A comparison of CT- and MR-based attenuation correction in neurological PET. Eur J Nucl Med Mol Imaging. 2014;41:1176–89.
Andersen FL, Ladefoged CN, Beyer T, et al. Combined PET/MR imaging in neurology: MR-based attenuation correction implies a strong spatial bias when ignoring bone. NeuroImage. 2014;84:206–16.
Nuyts J, Dupont P, Stroobants S, Benninck R, Mortelmans L, Suetens P. Simultaneous maximum a posteriori reconstruction of attenuation and activity distributions from emission sonograms. IEEE Trans Med Imaging. 1999;18:393–403.
Rezaei A, Defrise M, Bal G, Michel C, Conti M, Watson C, et al. Simultaneous reconstruction of activity and attenuation in time-of-flight PET. IEEE Trans Med Imaging. 2012;31:2224–33.
Aitken AP, Giese D, Tsoumpas C, et al. Improved UTE-based attenuation correction for cranial PET-MR using dynamic magnetic field monitoring. Med Phys. 2014;41(1):012302.
Delso G, Wiesinger F, Sacolick LI, et al. Clinical evaluation of zero-echo-time MR imaging for the segmentation of the skull. J Nucl Med. 2015;56(3):417–22.
Grodzki DM, Jakob PM, Heismann B. Ultrashort echo time imaging using pointwise encoding time reduction with radial acquisition (PETRA). Magn Reson Med. 2012;67(2):510–8.
Santos Ribeiro A, Rota-Kops E, Herzog H, Almeida P. Hybrid approach for attenuation correction in PET/MR scanners. Nucl Inst Methods Phys Res A. 2014;A734:166–70.
Delso G, Zeimpekis K, Carl F, Wiesinger F, Hüllner M, Veit-Haibach P. Cluster-based segmentation of dual-echo ultra-short echo time images for PET/MR bone localization. Eur J Nucl Med Mol Imaging. 2014;1:7.
Roy S, Wang WT, Carass A, Prince JL, Butman JA, Pham DL. PET attenuation correction using synthetic CT from ultrashort echo-time MR imaging. J Nucl Med. 2014;55(12):2071–7.
Burgos N, Cardoso MJ, Thieleman K, et al. Attenuation correction synthesis for hybrid PET-MR scanners: application to brain studies. IEEE Trans Med Imaging. 2014;33:2332–41.
Hofmann M, Steinke F, Scheel V, et al. MRI-based attenuation correction for PET/MRI: a novel approach combining pattern recognition and atlas registration. J Nucl Med. 2008;49:1875–83.
Navalpakkam BK, Braun H, Kuwert T, Quick HH. Magnetic resonance-based attenuation correction for PET/MR hybrid imaging using continuous valued attenuation maps. Investig Radiol. 2013;48(5):323–32.
Johansson A, Karlsson M, Nyholm T. CT substitute derived from MRI sequences with ultrashort echo time. Med Phys. 2011;38(5):2708–14.
Izquierdo-Garcia D, Hansen AE, Förster S, et al. An SPM8-based approach for attenuation correction combining segmentation and non-rigid template formation: application to simultaneous PET/MR brain imaging. J Nucl Med. 2014;55:1825–30.
Keereman V, Fierens Y, Broux T, De Deene Y, Lonneux M, Vandenberghe S. MRI-based attenuation correction for PET/MRI using ultrashort echo time sequences. J Nucl Med. 2010;51:812–8.
Cabello J, Lukas M, Förster S, et al. MR-based attenuation correction using ultrashort-echo-time pulse sequences in dementia patients. J Nucl Med. 2015;56(3):423–9.
Choi H, Cheon GJ, Kin HJ, et al. Segmentation-based MR attenuation correction including bones also affects quantitation in brain studies: an initial result of 18F-FP-CIT PET/MR for patients with parkinsonism. J Nucl Med. 2014;55(10):1617–22.
Catana C, van der Kouwe A, Benner T, et al. Toward implementing an MRI based PET attenuation-correction method for neurologic studies on the MR-PET brain prototype. J Nucl Med. 2010;51:1431–8.
Berker Y, Franke J, Salomon A, et al. MRI-based attenuation correction for hybrid PET/MRI systems: a 4-class tissue segmentation technique using a combined ultrashort-echo-time/Dixon MRI sequence. J Nucl Med. 2012;53:796–804.
Schulz V, Torres-Espallardo I, Renisch S, et al. Automatic, three-segment, MR-based attenuation correction for whole-body PET/MR data. Eur J Nucl Med Mol Imaging. 2011;38(1):138–52.
Delso G, Carl M, Wiesinger F, et al. Anatomic evaluation of 3-dimensional ultrashort-echo-time bone maps for PET/MR attenuation correction. J Nucl Med. 2014;55:780–5.
Schaefer SM, Abercrombie HC, Lindgren KA, et al. Six-month test–retest reliability of MRI-defined PET measures of regional cerebral glucose metabolic rate in selected subcortical structures. Hum Brain Mapp. 2000;19(1):1–9.
Camargo EE, Szabo Z, Links JM, Sostre S, Dannals RF, Wagner JRHN. The influence of biological and technical factors on the variability of global and regional brain metabolism of 2-[18F]Fluoro-2-deoxy-D-glucose. J Cereb Blood Flow Metab. 1992;12:281–90.
Minoshima S, Frey KA, Koeppe RA, Foster NL, Kuhl DE. A diagnostic approach in Alzheimer’s disease using three-dimensional stereotactic surface projections of fluorine-18-FDG PET. J Nucl Med. 1995;36:1238–48.
Delso G, Fürst S, Jakoby B, et al. Performance measurements of the Siemens mMR integrated whole-body PET/MR scanner. J Nucl Med. 2011;52:1914–22.
Watson CC, Rappoport V, Faul D, Townsend DW, Carney JP. A method for calibrating the CT-based attenuation correction of PET in human tissue. IEEE Trans Nucl Sci. 2006;53:102–7.
Friston KJ, Ashburner J, Frith CD, Poline J-B, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Hum Brain Mapp. 1995;3:165–89.
Ladefoged CN, Hansen AE, Kelelr SH, et al. Dental artifacts in the head and neck region: implications for Dixon-based attenuation correction in PET/MR. Eur J Nucl Med Mol Imaging. 2015;2:8.
Acknowledgments
The PET/MRI facility at the Technische Universität München was funded by the Großgeräteinitiative from the Deutsche Forschungsgemeinschaft, (DFG). The research leading to these results has received funding from the European Union Seventh Framework Program (FP7) under Grant Agreement n° 602621- Trimage, n° 294582- MUMI, and from the DFG grant no. FO 886/1–1. We thank David Izquierdo and Ninon Burgos for their assistance using their algorithms. We also thank Sylvia Schachoff and Claudia Meisinger for their technical assistance with the PET/MRI scanner.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The research leading to these results has received funding from the Seventh Framework Programme [FP7/2007-2013] under grant agreement n° 602621- Trimage, n° 294582- MUMI, and from the DFG grant no. FO 886/1–1.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
(DOCX 181 kb)
Supplementary Fig. 2
(DOCX 434 kb)
Supplementary Fig. 3
(DOCX 100 kb)
Supplementary Fig. 4
(DOCX 110 kb)
Supplementary Fig. 5
(DOCX 345 kb)
Supplementary Fig. 6
(DOCX 238 kb)
Supplementary Table 1
(DOCX 16 kb)
Supplementary Table 2
(DOCX 16 kb)
Supplementary Table 3
(DOCX 18 kb)
Supplementary Table 4
(DOCX 16 kb)
Supplementary Table 5
(DOCX 16 kb)
Rights and permissions
About this article
Cite this article
Cabello, J., Lukas, M., Rota Kops, E. et al. Comparison between MRI-based attenuation correction methods for brain PET in dementia patients. Eur J Nucl Med Mol Imaging 43, 2190–2200 (2016). https://doi.org/10.1007/s00259-016-3394-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-016-3394-5