Abstract
Objective
Radiographic bone age assessment (BAA) is used in the evaluation of pediatric endocrine and metabolic disorders. We previously developed an automated artificial intelligence (AI) deep learning algorithm to perform BAA using convolutional neural networks. We compared the BAA performance of a cohort of pediatric radiologists with and without AI assistance.
Materials and methods
Six board-certified, subspecialty trained pediatric radiologists interpreted 280 age- and gender-matched bone age radiographs ranging from 5 to 18 years. Three of those radiologists then performed BAA with AI assistance. Bone age accuracy and root mean squared error (RMSE) were used as measures of accuracy. Intraclass correlation coefficient evaluated inter-rater variation.
Results
AI BAA accuracy was 68.2% overall and 98.6% within 1 year, and the mean six-reader cohort accuracy was 63.6 and 97.4% within 1 year. AI RMSE was 0.601 years, while mean single-reader RMSE was 0.661 years. Pooled RMSE decreased from 0.661 to 0.508 years, all individually decreasing with AI assistance. ICC without AI was 0.9914 and with AI was 0.9951.
Conclusions
AI improves radiologist’s bone age assessment by increasing accuracy and decreasing variability and RMSE. The utilization of AI by radiologists improves performance compared to AI alone, a radiologist alone, or a pooled cohort of experts. This suggests that AI may optimally be utilized as an adjunct to radiologist interpretation of imaging studies to improve performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Machine learning has emerged as a powerful technique in computer science to teach computers to autonomously find patterns in data and now underlies many large-scale software products including Google Translate [1], Alexa speech recognition [2], and mastering the game of Go [3]. Intense research has focused on applying these techniques to medical applications with recent successes in detecting diabetic retinopathy [4] and detecting malignant melanomas with an accuracy rivaling that of board-certified dermatologists [5]. While there has been much discussion in the lay press about the role of machine learning in radiology [6,7,8], no direct assessment of the impact of a machine-learning algorithm on the performance of a cohort of radiologists has been performed.
Radiographic bone age assessment (BAA) is a central part of the clinical workup of pediatric endocrine and metabolic disorders, in which the patient’s chronologic age is compared with their level of skeletal maturity based on a standardized reference. BAA in clinical practice is typically performed using either the Greulich and Pyle [9] or Tanner–Whitehouse [10] (TW2) methods by comparing a radiograph of the hand and wrist to an age-based atlas or determining age based on scoring specific radiographic features. In both cases, BAA is time-consuming and contains significant interrater variability among radiologists. BAA is an ideal application for automated image evaluation, as there is a single image—the left hand and wrist—and relatively standardized findings.
We have previously developed a fully automated, deep learning algorithm to perform bone age assessment (BAA) using convolutional neural networks (CNN) that achieved mean accuracies of 92.3% within 1 year for the female and male cohorts compared with radiology report reference [11]. As AI based interpretation tools enter radiology clinical practice, important unanswered questions include how these tools compare with radiologist performance and how they are best integrated into radiology practice.
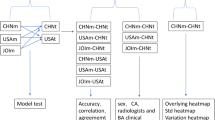
The purpose of this study is to compare the performance of a deep learning-based BAA algorithm to a cohort of pediatric radiologists and evaluate the impact of the implementation of a deep learning-based BAA algorithm on radiologist accuracy and variability when performing BAA on a set of standardized cases with and without access to AI interpretation (Fig. 1).
Methods
Patients
IRB approval was obtained for this retrospective, HIPAA-compliant study. We constructed a balanced cohort with ten representative cases for each class and gender, representing 280 cases ranging from 5 to 18 years from a cohort of 8325 radiographs previously used to train a deep learning CNN. The CNN was trained and validated (85:15) again without these 280 cases. These radiographs were then interpreted by the CNN, creating a predicted bone age and attention map for each.
Patient characteristics and indications
Self-reported demographic data for the 280 test cases are presented in Table 1. The distribution of chronologic ages roughly matches that of the bone age- and gender-matched test bone age cohort—ten patients per class and gender (Appendix Table 5). The predominant indications for BAA were evaluation of short stature (92/280 = 33%), monitoring of growth hormone therapy (52/280 = 19%), precocious puberty (57/280 = 20%), and research (38/280 = 19%). Please see appendix Fig. 6 for a detailed list of indications and appendix Table 5 for a graph of chronological age distribution.
Image processing and training
Our architecture first normalizes input images to have black backgrounds and a uniform size (512 × 512 pixels), then uses a preliminary segmentation CNN based on LeNet-5 with a 32 × 32 imaging patch size and stride of 4 to automatically segment the hand and remove extraneous data such as background artifacts, collimation, and annotation markers. The segmented and normalized image then enters the vision pipeline and has contrast-limited adaptive histogram equalization (CLAHE), denoising, and sharpening applied to enhance bony details, and finally is passed to the classification CNN for skeletal age classification. The classification CNN is based on an ImageNet pre-trained GoogLeNet fine-tuned on our train dataset by applying data augmentation with geometric (rotation, resizing, and shearing) and photometric (contrast and brightness) transformations to avoid overfitting. After holding out 280 images for testing, 15% of images were randomly selected for validation, and the remaining 6838 were utilized to train the CNN. All training was performed with a mini-batch stochastic gradient descent with a mini-batch size of 96 using 0.001 base learning rate, gamma of 0.1, and momentum term of 0.9, and weight decay of 0.005. The best CNN models were selected based on the validation loss values.
For visualization of the network as well as to provide AI interpretive information for the radiologists, attention maps were generated using the occlusion method [12]. This method iteratively slides a small patch across an image, presents the occluded images to the network, and creates two-dimensional attention maps based on the change in classification probability.
Image interpretation
Six board-certified, subspecialty trained pediatric radiologists from three academic medical centers with a mean of 13.8 years clinical experience (S.J.W. 21 years, R.L. 18 years, R.S. 16 years, M.S.G. 14 years, J. N. 9 years, and H.I.G. 5 years) interpreted the 280 test radiographs first using the GP atlas method. Three radiologists were randomly chosen to be presented the automated BAA results (including attention maps) and asked to report the BAA using the GP atlas and the additional AI information to test the effect of AI on BAA.
Reference standard
Bone age is ultimately a consensus evaluation with reference to a representative atlas, making it difficult to define a true gold standard reference. As a result, reference bone ages were determined using two different methods: (1) an independent reviewer and (2) a normalized mean. The independent reviewer was a radiologist (initials [removed for peer review]) who was not part of the six-member cohort. This reviewer had access to AI attention maps, AI bone age, individual rater cohort scores, and the clinical reports. The reviewer also compared all of these results to Gilsanz and Ratib’s Digital Atlas of Skeletal Maturity [13], and selected the GP atlas timepoint closest to the GR BAA. The second method used the normalized mean by taking the mean of the six raters and selecting the closest GP time point.
Statistical analysis
Quantitative variables are presented as means with ranges. Accuracies were reported as the exact same result or accuracy within 1 year. Bone age accuracy and root mean squared error (RMSE) were used as measures of accuracy. 2 was used to test for exact accuracy statistical significance, and two-tailed t test was used for RMSE statistical significance. Intraclass correlation coefficient (ICC) based on two-way random average measures was chosen to evaluate inter-rater variation amongst the radiologists without and with AI as a measure of variability. Statistical differences were considered significant at p < 0.05.
Experimental environment
All experiments were run on a Devbox (NVIDIA Corp, Santa Clara, CA, USA) containing four TITAN X GPUs with 12GB of memory per GPU [22], and on Nvidia deep learning frameworks, including Nvidia-Caffe (0.16.1) and Nvidia DIGITS (5.1). Excel 360 and MedCalc version 17.9 were used for statistical analysis.
Results
AI and cohort accuracies when compared to the independent reviewer reference
AI RMSE was 0.548 years and mean single-reader RMSE was 0.704 years, ranging from 0.544–0.902. AI exact accuracy was 73.2 and 98.9% within 1 year. Mean single-reader accuracy was 62.6%, ranging from 50.7–74.6% (Table 2). For context, the original clinical reports had an exact accuracy of 68.6% and an RMSE of 0.633 years when compared to the independent reviewer reference.
Impact of pairing AI with radiologists
Radiologists who utilized AI had pooled RMSE decrease from 0.684 to 0.525 years, all individually decreasing—0.607 to 0.531 for rater 4, 0.902 to 0.551 for rater 5, 0.544 to 0.493 for rater 6 (Fig. 2). Combined AI and radiologist interpretation resulted in higher accuracy than AI alone or the six-reader cohort mean.
Effect persistence with an alternative measure of ground truth
Similar improvements in accuracy and RMSE persisted when normalized cohort mean rating was used as the reference (Fig. 3). Radiologists paired with AI had increases in accuracy and RMSE (Table 3).
Interrater variation
Intraclass coefficients ICC(2,k) were calculated amongst the three radiologists exposed to AI. ICC without AI was 0.9914 (95% CI 0.9894 to 0.9930) and with AI was 0.9951 (95% CI 0.9940 to 0.9960). For comparison, ICC among the three radiologists who were not exposed to AI was 0.9908 (95% CI 0.9888 to 0.9925), similar to the other three radiologists without AI, but worse than the AI-assisted radiologists. Bland–Altman plots were generated and revealed decreased spread of ratings and decreased limits of agreement when paired with AI (Fig. 4).
AI performance variation based on patient ethnicity
Performance of the AI algorithm was evaluated based on the self-reported ethnicity/race of the patients. AI RMSE of the combined cohort was 0.548 years, with 0.551 years in Caucasian children and 0.542 years in non-Caucasian children (Table 4; p = 0.891).
Discussion
Machine learning-derived approaches have great potential for application in medicine, allowing rapid and scalable systems to perform complex analysis of medical data [14]. While most work has focused on applications of computer vision to natural images, these techniques can also be applied to medical images such as detection of malignant skin lesions [5] or diabetic retinopathy screening [15]. Recent work has demonstrated systems to detect tuberculosis on chest radiographs [16], stage and predict prognosis of COPD [17], and identify anatomic structures on abdominal CT [18]. These techniques have also been applied outside imaging by using a machine learning model to perform automated stratification of indeterminate breast lesions into surgical and observation groups, avoiding surgery in 30% of cases [19].
Fully automated BAA for use in the clinical setting has been a goal in computer vision and radiology research dating back to at least 1989 [20]. While most prior approaches have utilized hand-crafted features extracted from regions of interest [21], our approach utilizes transfer learning with a pre-trained CNN to automatically extract key features from all bones present in the hand and wrist, without the limitations imposed by hand-crafted features.
One of the challenges in BAA study design is the inherent variability in radiologist clinical interpretation of bone age radiographs, which makes selection of an appropriate reference standard difficult. For our study, we chose two different reference standards: (1) an independent radiologist reviewer and (2) a normalized mean cohort value from the six pediatric radiologists. We believe that a cohort-based reference standard is the most valid reference that best reflects the range of BAA in clinical practice. The six pediatric radiologists spanned three large academic medical centers and enabled a robust assessment of BAA intrinsic variation (measured as RMSE). Our six-radiologist mean cohort RMSE for BAA without AI was 0.661 years, comparable to previously published RMSE values—ranging from a mean RMSE of 0.96 years in a British cohort [22] and 0.59 years in the ATLAS dataset [23] to 0.51 ± 0.44 years [24] in a recent analysis in a Korean cohort. Thus we believe that our baseline radiologist BAA performance can be considered consistent with standard clinical radiologist performance.
An important result of our work is that AI BAA performance is at a level comparable to pediatric radiologists, similar to recently reported work by Larson et al. [8]. AI achieved an RMSE of 0.601 years, which was not significantly different from the cohort mean RMSE and is comparable with the previously reported values of RMSE intrinsic to BAA. In addition, AI had comparable BAA accuracy compared to the pediatric radiologist reader cohort, with no significant difference in exact (68.2 and 63.6%) or within 1 year (98.6 and 97.4%) accuracies, respectively. The slight but not significantly increased accuracy achieved by AI compared to the reader cohort could reflect a small degree of overfitting given that four of the six raters also provided interpretations for the initial training dataset.
Another goal of our study was to assess the impact of AI on pediatric radiologist BAA performance. To do this, we asked pediatric radiologists to interpret bone age radiographs before and after access to AI input. Our results also show that access to AI improves the accuracy and decreases the variability of subspecialty-trained pediatric radiologists BAA. Among the three radiologists who were paired with AI, the mean RMSE decreased from 0.661 to 0.508 years. Mean accuracy increased from 63.8 to 74.5% when compared to AI accuracy of 68.2%. All individual radiologist+AI RMSEs statistically decreased below that of AI or the radiologists alone, while accuracy statistically increased for two out of the three (Table 3). Importantly, the improvement that AI provides for pediatric radiologist BAA accuracy and variability is observed when compared with two different reference standards (both an independent reviewer and normalized cohort mean). Our study design (six independent evaluations of 280 standardized cases) accounts for the fact that a true reference BAA standard in clinical practice should incorporate both accuracy as well as intrinsic variation among different radiologists. These results build on recent data by Kim et al. [25] demonstrating that AI can help trainees improve their accuracy and interpretation speed when paired with a neural network-based bone age classifier.
Much attention has been focused on the potential of AI to replace humans in performing complex visual tasks, including radiologic interpretation [6, 7]. Our results indicate that performance is optimized when AI is deployed in conjunction with radiologist interpretation. Further studies are needed to elucidate the ways in which AI and radiologist image interpretation synergizes, but it is likely that AI is more helpful in cases that are not easily mapped to a specific timepoint. In this way, perhaps AI can be used in other areas of radiology as a time-saving tool to allow radiologists to spend more time on challenging cases.
In addition, our dataset included an ethnically diverse patient population, allowing us to compare AI performance across different ethnic groups. Our results show that AI demonstrates similar BAA performance across different ethnicities, providing good evidence of its generalizability.
Our system was directly embedded into both PACS and our computerized dictation system to aid interpretation and reduce burdens to use (Fig. 5). The system consists of a webapp that allows the radiologist to view the AI BAA prediction, easily scroll through the reference Greulich and Pyle images, and make the final determination while generating a structured report with Brush foundation standard deviations. The system saves time by avoiding table lookups and transcription errors while also keeping the radiologist focused on the images rather than distracting their attention to the atlas or the reporting system.
Strengths of our system include a diverse population and multiple experienced readers to provide a robust ground truth. Limitations of our system include a single site as the source of the training dataset and the intrinsic use of BAA in patients with suspected disease. As our experimental design specifically tried to determine the impact of AI’s interpretation on radiologist accuracy and agreement, our study design required immediate interpretation with and without AI, precluding time measurements to compare interpretation acceleration. Additionally, our retrospective design precludes evaluating the impact of higher accuracy on subsequent patient care. Further investigations should utilize multi-site training data and normal healthy patients, while preserving the ability to measure time-savings and the impact of improved BAA on subsequent clinical care as well as assessing whether improvement over time is consistent.
Conclusions
AI performs similarly to practicing pediatric radiologists for BAA. The utilization of AI by radiologists improves performance compared to AI alone, a radiologist alone, or a pooled cohort of experts. This suggests that AI may optimally be utilized as an adjunct to radiologist interpretation of imaging studies, suggesting a model for how AI may best be utilized in radiology.
References
Johnson M, Schuster M, Le QV, et al. Google’s multilingual neural machine translation system: enabling zero-shot translation. arXiv [csCL]. 2016. http://arxiv.org/abs/1611.04558.
Maas R, Rastrow A, Goehner K, Tiwari G, Joseph S, Hoffmeister B. Domain-specific utterance end-point detection for speech recognition. In: Interspeech 2017. 2017. https://doi.org/10.21437/interspeech.2017-1673.
Silver D, Schrittwieser J, Simonyan K, et al. Mastering the game of go without human knowledge. Nature. 2017;550(7676):354–9. https://doi.org/10.1038/nature24270.
Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–10. https://doi.org/10.1001/jama.2016.17216.
Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–8. https://doi.org/10.1038/nature21056.
Lewis-Kraus G. The Great A.I. Awakening. The New York Times. https://www.nytimes.com/2016/12/14/magazine/the-great-ai-awakening.html. Published December 14, 2016. Accessed 23 Oct 2017.
Mukherjee S. A.I. Versus M.D. The New Yorker. https://www.newyorker.com/magazine/2017/04/03/ai-versus-md. Published March 27, 2017. Accessed 23 Oct 2017.
Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology. 2017;170236. https://doi.org/10.1148/radiol.2017170236.
Greulich WW, Pyle SI. Radiographic atlas of skeletal development of the hand and wrist. Am J Med Sci. 1959;238(3):393. https://doi.org/10.1097/00000441-195909000-00030.
Ehrenberg ASC. J R Stat Soc Ser C Appl Stat. 1977;26(1):80. https://doi.org/10.2307/2346874.
Lee H, Tajmir S, Lee J, et al. Fully automated deep learning system for bone age assessment. J Digit Imaging. 2017. https://doi.org/10.1007/s10278-017-9955-8.
Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T, editors. Computer vision – ECCV 2014. Lecture Notes in Computer Science. Springer International Publishing; 2014. p. 818-833. https://doi.org/10.1007/978-3-319-10590-1_53.
Gilsanz V, Ratib O. Hand bone age: a digital atlas of skeletal maturity. Berlin Heidelberg: Springer; 2011. https://doi.org/10.1007/978-3-642-23762-1.
Abuzaghleh O, Barkana BD, Faezipour M. Noninvasive real-time automated skin lesion analysis system for melanoma early detection and prevention. IEEE J Transl Eng Health Med. 2015;3:2900310. https://doi.org/10.1109/JTEHM.2015.2419612.
van Grinsven MJJP, van Ginneken B, Hoyng CB, Theelen T, Sanchez CI. Fast convolutional neural network training using selective data sampling: application to hemorrhage detection in color fundus images. IEEE Trans Med Imaging. 2016;35(5):1273–84. https://doi.org/10.1109/TMI.2016.2526689.
Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017:162326. https://doi.org/10.1148/radiol.2017162326.
González G, Ash SY, Vegas Sanchez-Ferrero G, et al. Disease staging and prognosis in smokers using deep learning in chest computed tomography. Am J Respir Crit Care Med. 2017. https://doi.org/10.1164/rccm.201705-0860OC.
Lee H, Troschel FM, Tajmir S, et al. Pixel-level deep segmentation: artificial intelligence quantifies muscle on computed tomography for body morphometric analysis. J Digit Imaging. 2017. https://doi.org/10.1007/s10278-017-9988-z.
Bahl M, Barzilay R, Yedidia AB, Locascio NJ, Yu L, Lehman CD. High-risk breast lesions: a machine learning model to predict pathologic upgrade and reduce unnecessary surgical excision. Radiology. 2017:170549. https://doi.org/10.1148/radiol.2017170549.
Michael DJ, Nelson AC. HANDX: a model-based system for automatic segmentation of bones from digital hand radiographs. IEEE Trans Med Imaging. 1989;8(1):64–9. https://doi.org/10.1109/42.20363.
Thodberg HH, Kreiborg S, Juul A, Pedersen KD. The BoneXpert method for automated determination of skeletal maturity. IEEE Trans Med Imaging. 2009;28(1):52–66. https://doi.org/10.1109/TMI.2008.926067.
King DG, Steventon DM, O’Sullivan MP, et al. Reproducibility of bone ages when performed by radiology registrars: an audit of Tanner and Whitehouse II versus Greulich and Pyle methods. Br J Radiol. 1994;67(801):848–51. https://doi.org/10.1259/0007-1285-67-801-848.
Cao F, Huang HK, Pietka E, Gilsanz V. Digital hand atlas and web-based bone age assessment: system design and implementation. Comput Med Imaging Graph. 2000;24(5):297–307. http://www.ncbi.nlm.nih.gov/pubmed/10940607
Kim SY, Oh YJ, Shin JY, Rhie YJ, Lee KH. Comparison of the Greulich-Pyle and Tanner Whitehouse (TW3) methods in bone age assessment. J Korean Soc Pediatr Endocrinol. 2008;13(1):50–5. https://www.koreamed.org/SearchBasic.php?RID=0113JKSPE/2008.13.1.50&DT=1
Kim JR, Shim WH, Yoon HM, et al. Computerized bone age estimation using deep learning-based program: evaluation of the accuracy and efficiency. AJR Am J Roentgenol. 2017:1-7. https://doi.org/10.2214/AJR.17.18224.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None
Additional information
This work has been accepted for presentation at RSNA 2017 and awarded an RSNA Trainee Research Prize.
Rights and permissions
About this article
Cite this article
Tajmir, S.H., Lee, H., Shailam, R. et al. Artificial intelligence-assisted interpretation of bone age radiographs improves accuracy and decreases variability. Skeletal Radiol 48, 275–283 (2019). https://doi.org/10.1007/s00256-018-3033-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00256-018-3033-2