Abstract
The effects of acid mine drainage (AMD) in a stream and groundwater near an abandoned copper mine were characterized by physicochemical properties, bacterial community structure using denaturing gel gradient electrophoresis (DGGE), and microbial activity/diversity using Ecoplate technique. Based on DGGE fingerprints, the eubacterial community structures grouped into the stream water (GRS1, GRS2 and GRS3) and groundwater samples (GW1 and GW2), apparently based on differences in water temperature and the concentrations of dissolved oxygen, nitrate and sulfate. The most highly AMD-contaminated sample (GRS1) had additional α-Proteobacteria whereas the groundwater samples included additional β-Proteobacteria, suggesting the development of populations resistant to AMD toxicity under aerobic and anaerobic conditions, respectively. Community level physiological activities on the 31 Ecoplate substrates suggested that the activities decreased with increasing concentrations of sulfate and heavy metals derived from AMD. The Shannon index showed that microbial diversity was greatest in GRS2, and lowest in GRS1, and was probably related to the level of AMD.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Mine pollution, in particular heavy metals derived from active or abandoned mines, is a serious problem that has negative effects on the environment and human health. In Korea, about 2,006 mines have been developed since 1930, of which 1,276 have been abandoned (MCIE 2006). Since 2006, the Korean Ministry of Commerce, Industry and Energy has undertaken pollution prevention and restoration in about 1,344 mines (739 active mines, 215 abandoned coal mines and 390 abandoned metal mines) that posed serious pollution problems (MCIE 2006).
Mine facilities, tailings and waste rocks left near abandoned and active mines are major contamination sources of acid mine drainage (AMD), which is extremely detrimental to stream water, groundwater and soil due to high acidity, as well as high sulfate and heavy metal concentrations (Okabayashi et al. 2005). AMD, which has been reported to affect water quality in waterways in many countries (Herlihy et al. 1987; Harries 1997; Banks et al. 1997), is thought to significantly reduce the diversity and abundance within affected biological communities (Kelly 1988).
Several studies have investigated the bacteria associated with AMD. For example, Acidithiobacillus ferrooxidans (Kelly and Wood 2000; Tan et al. 2007), a well-known chemolithotrophic bacterial species, can utilize ferrous iron or reduced sulfur compounds as an energy source in the presence of sulfide minerals. In addition, A. thiooxidans (Hallberg and Johnson 2001), A. caldus (Hallberg and Johnson 2001), Acidiphilium cryptum (Kelly and Wood 2000), Leptospirillum ferrooxidans (Tan et al. 2007) and some archaebacteria (Edwards et al. 2000) have been found in AMD-influenced environments. These are sulfur and/or iron-oxidizing prokaryotes that predominate under sulfur and iron-rich conditions. Based on Ecoplate data, Weber et al. (2008) reported that the interstitial bacterial community in planted wetland mesocosms was not significantly affected by exposure to AMD. Although acidophilic chemolithotrophs and heterotrophs have been commonly found at acid mine sites (Baker and Banfield 2003; Hallberg and Johnson 2001; Léveillé et al. 2001), little is known about their roles, or the shifts that occur in microbial community structure, diversity or activity due to the presence of mine pollutants. This is primarily because the previous studies have generally been culture-based.
In the present study, the influences of AMD in stream and groundwater near an abandoned copper mine were characterized by determining the physicochemical properties of the water at these sites as well as the microbial community structure and activity. Denaturing gel gradient electrophoresis (DGGE) was used in genetic and metabolic fingerprinting based on 16S rDNA gene analysis to examine the effect of AMD on eubacterial community structure. This molecular biological method has the advantage of enabling analysis of unculturable species, which will lead to a better understanding of their role in natural environments (Tan et al. 2007). In addition, community-level physiological profile (CLPP) assays using the Ecoplate technique were utilized to investigate the effect of AMD on the functional potential and diversity of microorganisms in streams and groundwater. CLPP assays, which assess a microbial community’s capacity to metabolize specific sole carbon sources, are a method for characterizing microbial ecology in environmental samples (Miguel et al. 2007).
Materials and methods
Study site description
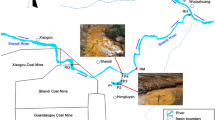
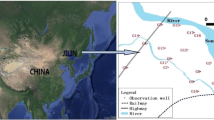
Guryong (G) Mine, an abandoned copper mine which produced 5,470 tons of the metal, is located in the north part of Kyongnam, South Korea. The site is approximately 50 km2 (60% forest, 35% agricultural and 3% residential). A main stream flows from south to north, with several tributaries of the main stream flowing east to west or west to east. The main stream flows into the Nakdong River, which in turn flows to the South Sea. The area contains several kinds of minerals including andesite, tuff, biotite granite, and sedimentary rock. The G Mine is located in the upper part of the main stream and is relatively large compared to the many other small abandoned mines and mineralized zones that occur in the area. A quarry for mining aggregates is located to the north side of the waterways.
The major environmental impacts of the G Mine are associated with the mine tailings heap, the mine waste rock pile, an unclosed pit head, and the mine waste rock square. The major pollution problems are from heavy metals in leachates from the mine tailings heap and mine waste rock pile, which contaminate stream waters, sediments and agricultural soils.
Sampling and analysis
To examine hydrogeochemical and microbial effects of mine tailings and waste rocks, samples were obtained from two groundwater wells installed in the mine tailings heap (GW1 and GW2), and sites on a stream affected by AMD (GRS1), a stream unaffected by AMD (GRS2), and a larger stream receiving the flow from the stream on which GRS1 was located (GRS3) (Fig. 1).
The groundwater and stream water samples were analyzed in the field for temperature, pH, redox potential (Eh), electroconductivity (EC), dissolved oxygen (DO) and alkalinity. Other physicochemical properties were measured in the laboratory (Table 1).
Bacterial community structure analysis
The bacterial community structure in the water samples was analyzed using the polymerase chain reaction (PCR) of 16S rDNA sequences followed by DGGE. Genomic DNA was extracted from approximately 0.5 g of each collected cells using the BIO101 FastDNA SPIN KIT for soil (Q-Biogene, USA). PCR was used to amplify a 177 base pair portion of the 16 s rDNA using bacterial primers 341FGC (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG-3′) and 518R (5′-ATT ACC GCG GCT GCT GG-3′). All reactions were carried out in 25 μl volumes containing 20 pmol of each primer, 10 mM of each deoxynucleoside triphosphate, 12.5 mg/ml of bovine serum albumin, 2.5 μl of 10′ PCR buffer (20 mM Tris-HCl, 100 mM KCl, pH 8.0), 2 μl of 25 mM MgCl2, 0.5 U of Taq DNA polymerase (Promega, USA), and 1 μl of the DNA extract. The amplification conditions were 93°C for 2 min, 35 cycles of 92°C for 1 min, 55°C for 1 min and 68°C for 45 s, followed by a final extension at 72°C for 2 min (Kim et al. 2006).
DGGE was performed with the D-Code™ System (Bio-Rad, USA) using a 16′ 16 cm 8% (w/v) polyacrylamide gel maintained in 7 L of 0.5′ TAE buffer (20 mM Tris, 10 mM acetate, 0.5 mM Na2EDTA, pH 7.8). Gradient gels were prepared with 40 and 60% denaturant (100% denaturant contains 7 M urea and 40% formamide, v/v). Gels were run at 60°C and 50 V for 11.5 h, stained with ethidium bromide, and banding was recorded with a Mupid-21 gel imager (Cosmo Bio, Japan) (Kim et al. 2006). DGGE images were used for microbial community structure comparisons using GelCompar II software (version 3.5; Applied Maths, Belgium) and the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) clustering using the Jaccard coefficient, based on band position.
DNA sequencing and phylogenic analysis
Bands were excised from the gel and mixed with 20 μl of sterile deionized water, and DNA was extracted by freeze-thaw treatment (freezing at −20°C for 10 min and thawing at 65°C for 3 min, repeated three times). The resulting DNA extract (1 μl) was used as the template for nested PCR amplification with the 341 forward primer (lacking the GC clamp) and the 518 reverse prime (the universal eubacterial primers), under the PCR conditions described above. The amplified products were purified with the QIAquick PCR purification kit (Qiagen, Germany), and 1 μl of the PCR product was cloned into the pGEM-T vector (Promega, USA), following the manufacturer’s protocol. Plasmid clones were amplified in E. coli according to standard procedures, extracted from broth cultures using the Wizard® Plus SV Miniprep DNA purification system (Promega, USA), and the DNT insert was excised with EcoRI. The cloned PCR fragments (100 μl) were sequenced using the T7 (5′-TAA TAC GAC TCA CTA CAG GG-3′) and SP6 (5′-ATT TAG GTG ACA CTA AGA AT-3′) primers and an ABI Prism model 373A automated DNA sequencer (Perkin Elmer, USA). The sequences obtained were compared with the GenBank database using the Basic Local Alignment Search Tool (BLAST) algorithm feature of the National Center for Biotechnology Information (NCBI) website, which identified the sub-class level of each sequence.
Analysis of microbial community using ecoplate
The Ecoplate (Biolog Ecoplate™, USA) technique was used to analyze the functional diversity of microbial communities of individual samples. An Ecoplate consists of 96 wells comprising three sets of 31 substrates and control wells not containing substrate. The tetrazolium dye in the wells turns violet in response to microbial respiration of the substrate as a sole carbon source, enabling comparative evaluation of substrate utilization among microbial communities. Ecoplate wells were inoculated with groundwater (150 μl) and the plates were incubated at 20°C. The optical density (OD) at 595 nm of solution in each well was measured every 24 h during incubation using a microtiter plate reader (Multiskan Ascent, Thermo Labsystem, Finland), and the results were analyzed for comparative substrate utilization patterns according to sample and time. Incubation was continued for 11 days, at which time the OD had stabilized.
Statistical analysis
The average well color development (AWCD) with time was calculated from the ODs of each Ecoplate well for all samples in relation to the control, according to the equation of Garland and Mills (1991).
where C is the OD595 nm value of the substrate well, R is the OD595 nm value of control well, and n is the number of substrates (31). Using these values, the potential utilization of various carbon sources by microbial communities could be assessed.
Principal component analysis (PCA) (SPSS 12.0K for Windows) was conducted to statistically compare carbon source utilization by the microbial communities in each sample, using the value obtained from Eq. 2.
where C id is the OD595 nm value of each well on day i, R id is the OD595 nm value of the control on day i, and AWCD id is the value of AWCD on day i. The range of PCA data calculated using Eq. 2 was 0–4, with values less than 1 meaning that C–R is less than AWCD. The similarity in carbon source utilization could be obtained using PCA values. These represent sample differences, which are shown on the graph divided by two axes (principal component 1, PC1; principal component 2, PC2). Samples on the same section of the PCA graph are similar in terms of carbon source utilization (Garland and Mills 1991).
In addition, the Shannon index (representing species diversity) was calculated across time and samples using the following equation:
where H is the Shannon index, and P i is the OD595 nm value for each substrate divided by the total OD595 nm value of all substrates, expressed as P i = C/∑(C−R).
Results
Physicochemical properties of waters polluted with AMD
Table 1 shows the physicochemical data for all water samples (stream water and groundwater) influenced by AMD except the background water sample. The temperature of the stream water samples varied from 23–28°C, and was higher than the groundwater samples (17–19°C) as the former were collected in summer. The GW1 and GW2 samples were obtained from the mine tailings heap region and were low in pH, although this may have varied according to the depth of the groundwater or the amount of sulfide minerals present. GRS1 had a pH of 3.51 and its EC was relatively low compared to GW1 and GW2, since precipitation of many chemical components, or mixing with pure stream water, caused a reduction in the solute concentration. Yellow boy (a Fe-precipitate) was observed in the AMD stream (GRS1,) and white boy (an Al-precipitate resulting from increased pH) was found in the mixing area (GRS3).
While the AMD-unaffected stream water (GRS2) was classified as (Ca + Na)-HCO3 in a Piper Diagram (data not shown), all other samples were identified as type CaSO4, derived perhaps from increased sulfate from the oxidation of pyrite in mine tailings. GRS3 was positioned between GRS1 and GRS2 in the diagram, which was probably due to the expected dilution effect. There was no difference between GW1 and GW2. DO levels were higher in the stream water samples than groundwater samples, and were proportional to nitrate levels.
GW1 had the lowest pH value (2.70), and more dissolved components were found in the groundwater samples than in stream water samples. In particular, levels of Ca, Mg and SO4 were much higher in groundwater, and heavy metals (Mn, Fe, Zn, Pb, Ni) were relatively higher. The higher concentration of Fe seemed to be due to dissolution of pyrites or secondary Fe-oxidizing minerals. With the exception of GW1, Cu concentrations in samples were generally much higher than in the AMD-unaffected stream water (GRS2). The low Cu concentration in GW1 might be due to the Cu being heterogeneously distributed in the tailings layer. The concentrations of Li were high (474 and 537 μg/L, respectively) in GW1 and GW2, but low (24–48 μg/L) in stream waters. It may be that as Li ions produced by the dissolution of silicate minerals flowed into the groundwater and then into the stream, they were removed by precipitation reactions at elevated oxygen levels. Pb was relatively abundant in groundwater and was removed by co-precipitation, which occurred at the same time as precipitation of metallic oxides of AMD.
Overall, the level of AMD in stream water (GRS1 > GRS3 > GRS2) was proportional to the levels of acidity, Eh and EC, and also to the levels of almost all measured ions and metals (Ca, Mg, Na, K, Si, Fe, Al, Mn, F, Cl, NO3, SO4, Zn, Pb, Ni, Ba, Cu, Sr, and Li) with the exception of HCO3. In particular, AMD-contaminated groundwater (GW1 and GW2) showed much higher values for EC, Ca, Mg, SO4, Pb, Sr and Li than the stream water, indicating that AMD had more influence in the groundwater.
Bacterial community structure and phylogenetic similarity analysis
DGGE fingerprints were compared and a similarity tree was obtained (Fig. 2). The closest relationship in band profiles was found between GRS2 and GRS3 (45.46% similarity), and both were similar in pH and had similarly low concentrations of several metals. The second most similar samples were GRS1 and GRS2 (41.67% similarity). GRS3 and GW2 (the mixed main stream and the aquifer under the tailings heap, respectively) were least similar (28.57% similarity) (Table 2). In the phylogenetic analysis (Table 3), the sample which was most highly contaminated with AMD (GRS1) included three major bacterial groups: γ-Proteobacteria (56%), α-Proteobacteria (33%) and unidentified bacteria (11%). Similarly, most bacteria in GRS2 and GRS3 were γ-Proteobacteria (86% and 100%, respectively). For the groundwater samples (GW1 and GW2), γ-Proteobacteria predominated, followed by β-Proteobacteria. Connotatively, the stream water samples and groundwater samples were grouped, respectively, on the basis of phylogenetic comparison (Fig. 2b). In particular, the sample most highly contaminated with AMD (GRS1) had additional α-Proteobacteria, and the groundwater samples (GW1 and GW2) included additional β-Proteobacteria.
Microbial community level physiological characteristics
Heterotrophic microbial activity in samples (based on OD measurements of substrate use in Ecoplates, and AWCD values) was in the order GRS2 > GRS3 > GRS1 > GW1 > GW2 (Fig. 3). This result was expected as it corresponded positively with the DO level and negatively with AMD concentration (Table 1). The Shannon index indicated three groups: Group 1 (GRS2), Group 2 (GRS3, GW1, GW2), and Group 3 (GRS1) (Fig. 4). Given that the Shannon index estimates microbial diversity, it is evident that diversity was affected by the AMD contamination level, with Group 1 corresponding to uncontaminated water, Group 2 with moderately contaminated water, and Group 3 with heavily contaminated water. PCA analysis, representing differential utilization of individual substrates, showed that GRS1 was different from all the other samples (Fig. 5). This result suggests that substrate utilization patterns were heavily influenced by the toxicity of AMD, since GRS1 was the most highly contaminated water. In addition, there were no significant differences in data for days 8 and 11 (data not shown).
Discussion
This study investigated shifts in microbial community structure, activity and diversity due to the impact of AMD in a mine area, using DGGE and Ecoplate methods. Stream water was sampled from three sites with different levels of AMD contamination, as well as groundwater from two sites. Comparison among stream water samples and groundwater samples was difficult because the respective environmental conditions were very different.
As expected, the sample most affected by AMD (GRS1) had high concentrations of Fe and sulfate derived from pyrite (FeS2) through oxidation and microbially mediated reactions (Table 1). For the stream water samples, the order of concentration levels of all the other measured components (ions and heavy metals, with the exception of K) was GRS1 > GRS3 > GRS2, and depended on the level of AMD contamination (Table 1). As metal release from tailings may lead to AMD (Salomons 1995; El Khalil et al. 2008), the level of AMD acidity will be proportional to the level of ions and metals. Also, this order of AMD concentrations might influence the Eh (redox potential) values because they have the same order. The Eh values looked like to be dependant on redox potential components such as DO and SO4, showing the same order as well.
Although GW1 and GW2 were sampled from groundwater at the similar sites (the tailings heap), they differed in some physicochemical properties (pH and Eh) and ion concentrations (Al, Mn, HCO3, Zn, and Cu), probably due to disparate tailings layers. This may explain why their DGGE profiles had relatively low similarity (39.13%) (Table 2). The AMD-contaminated groundwater (GW1 and GW2) had much higher values than the stream water samples for EC, Ca, Mg, SO4, Pb, Sr, Ni and Li, probably due to leaching from the tailings heap and soil particles. The high EC value seemed to be related to high salt concentrations in the groundwater, and the high heavy metal concentrations were likely to have been associated with the high sulfate concentrations. The lower concentrations of nitrate in groundwater compared to stream waters suggest that nitrate-reducing bacteria were utilizing nitrate under anaerobic conditions (low levels of DO in Table 2), usually induced by aerobic bacteria in groundwater environments. The pH was unexpectedly high in GW2 despite the concentration of sulfate being higher than in GW1. This suggests that there may have been some neutralization mechanisms affecting H+ ion reduction occurring in the groundwater due to complex environmental conditions, which our analyses did not detect. This may be only one reason why the Eh value was higher in GW1 (496 mV) than in GW2 (191 mV).
Concentrations of inorganic macrocomponents and heavy metals were greatest at the most AMD-polluted stream site (GRS1) (Table 1), which also showed the least number of DGGE bands (n = 14), suggesting a reduction in the diversity and abundance of biota (Kelly 1988). In contrast, GRS3 was less polluted than GRS1 and had the most DGGE bands (n = 27) of all sites including the AMD-unaffected site, GRS2. This result suggests the emergence of new populations of bacteria that grew well under the acidic conditions, whereas the indigenous bacteria were not significantly inhibited by the diluted pollutants. The presence of the 3 dominant α-Proteobacteria in the stream most highly contaminated with AMD (GRS1) suggests these bacteria were adapted to the acidic environment (Table 3), and they may be related to species of Acidiphilium spp., which are capable of ferric iron (Fe3+) reduction (Tan et al. 2007). Many γ-Proteobacteria species were found in all the samples, indicating that these bacteria are able to live under either natural or AMD conditions. The γ-Proteobacteria found in previous studies of acid mine sites included Acidithiobacillus spp., which are acidophilic chemolithotrophs (Nicomrat et al. 2006; Tan et al. 2007). GW2 was not comparable to other samples in terms of the dominant phylogenetic groups because only two clones were successfully sequenced (Table 3). The groundwater samples (GW1 and GW2) had additional β-Proteobacteria, suggesting that these were populations resistant to AMD toxicity under anaerobic conditions. Overall, two groups were evident in the similarity analyses of the eubacterial community structure, and were associated with stream and groundwaters, respectively (Fig. 2). This result may be due to the different environmental conditions at each site, which varied in 10 or more environmental parameters including water temperature, DO level, nitrate reduction activity, and sulfate concentration (Table 1).
The three bacterial groups found in this study, and commonly reported in comparable studies, were the α-Proteobacteria (Morales et al. 2005; Okabayashi et al. 2005; He et al. 2007; Tan et al. 2007; Zhang et al. 2007), the β-Proteobacteria (Morales et al. 2005; Okabayashi et al. 2005; Bruneel et al. 2006; Nicomrat et al. 2006; He et al. 2007; Tan et al. 2007; Zhang et al. 2007), and the γ-Proteobacteria (Okabayashi et al. 2005; Bruneel et al. 2006; Nicomrat et al. 2006; He et al. 2007; Tan et al. 2007; Zhang et al. 2007). It was not find some of the bacteria reported in similar studies, including the δ-Proteobacteria (Morales et al. 2005; Bruneel et al. 2006; Tan et al. 2007; Zhang et al. 2007), Actinobacteria (Morales et al. 2005; Okabayashi et al. 2005; Bruneel et al. 2006; Zhang et al. 2007), Nitrospira (Okabayashi et al. 2005; He et al. 2007; Tan et al. 2007), Verrucomicrobia (Okabayashi et al. 2005), Chlorobi (Okabayashi et al. 2005), and Gemmatimonadetes (Zhang et al. 2007). This may be due to the lack of 16S rDNA clones or/and the environmental conditions at the study sites.
GRS2 showed the greatest substrate usage by AWCD analysis, followed by GRS3 and GRS1. This order appeared to reflect an inverse relationship to AMD influence in stream water (Fig. 3), and suggested that microbial activity at GRS2 was least influenced by AMD. An explanation for all stream water samples being higher than groundwater samples is that the former maintained more aerobic conditions and supported more aerobic heterotrophic microbial activity in the aerobic Ecoplates (Table 1). In this study, it was found that AMD lowered the pH and levels of HCO3 − (an index of hydrocarbons/carbon source), and nitrate (nitrogen source), but raised the levels of all the other components (ions and heavy metals), which restricted microbial growth. In particular, the high concentrations of copper (Cu) in GRS1 (517 μg/L) and GW2 (951 μg/L) may have caused microbial inactivation due to acute toxicity (normal range 59.8–189 μg/L) (El Khalil et al. 2008). In conclusion, substrate utilization activities were highly influenced by pH and heavy metals, which appeared to be proportional to AMD levels.
Based on microbial diversity in substrate utilization, the Shannon index revealed three groups apparently associated with AMD. Although AWCD indicated GW1 and GW2 had more extreme conditions for microbial activity (Fig. 3) than other sites, the microbial diversities in GW1 and GW2 samples were higher than in GRS1 due to the high concentrations of electron acceptors and more tolerant circumstances with soil particles than water only. It can be assumed that various iron-, sulfate-, and manganese-reducing bacteria were present as well as other acid-resistant microorganisms. This result parallels the analysis of the PLFA patterns, which reflect metabolically more diverse microbial communities in contaminated aquifers or sediments than in uncontaminated ones (Fang and Barcelona 1998; Ben-David et al. 2004). As the number of DGGE bands, which implies richness of the microbial community, did not clearly reflect the diversity, it is suggested that evenness might have more influence on the diversity.
The PCA analysis showed very different patterns from the AWCD analysis, as GRS1 was separated from all the other samples, and the groundwater samples (GW1 and GW2) grouped with the stream water samples GRS2 and GRS3, suggesting that AMD had most influence on the utilization of individual substrates (Fig. 5). This is supported by the relative levels of bacteria according to phylogenetic groups, since only GRS1 included 3 members of the α-Proteobacteria (33% of all the dominant species) (Table 3). The results presented in Fig. 2 also support the PCA findings. On the other hand, there was no significant difference between the PCA values for days 8 and 11, indicating that the period of microbial growth did not affect the patterns of individual substrate utilization.
Conclusions
The effects of AMD, which is extremely detrimental to stream water, groundwater and soil, on the microbial activity and diversity in stream and groundwater near an abandoned copper mine were investigated. DGGE fingerprints of eubacterial community showed that water temperature, DO and sulfate concentrations and nitrate reduction activity among various parameters were key factors to characterize eubacterial community structure. The community level physiological activities were very sensitive to sulfate and heavy metal levels derived form AMD. In conclusion, this study indicates that AMD increased quantities of various dissolved components, changed bacterial community structure, and decreased microbial activity and diversity, but stimulated the occurrence of specific bacterial populations.
References
Baker BJ, Banfield JF (2003) Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44:139–152
Banks D, Younger PL, Ameson R, Iverson ER, Banks SB (1997) Mine-water chemistry: the good, the bad and the ugly. Environ Geol 32:157–174
Ben-David EA, Holden PJ, Stone DJ M, Harch BD, Foster LJ (2004) The use of phospholipid fatty acid analysis to measure impact of acid rock drainage on microbial communities in sediments. Microb Ecol 48:300–315
Bruneel O, Duran R, Casiot D, Elbaz-Poulichet F, Personné JC (2006) Diversity of microorganisms in Fe-As-rich acid mine drainage waters of Carnoulès, France. Appl Environ Microbiol 72:551–556
Edwards KJ, Bond PL, Gihring TM, Banfield JF (2000) An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 279:1796–1799
El Khalil H, El Hamiani O, Bitton G, Ouazzani N, Boularbah A (2008) Heavy metal contamination from mining sites in South Morocco: monitoring metal content and toxicity of soil runoff and groundwater. Environ Monit Assess 136:147–160
Fang J, Barcelona MJ (1998) Biogeochemical evidence for microbial community change in a jet fuel hydrocarbons-contaminated aquifer. Org Geochem 29:899–907
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level, sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359
Hallberg KB, Johnson DB (2001) Biodiversity of acidophilic microorganisms. Adv Appl Microbiol 49:37–84
Harries J (1997) Acid mine drainage in Australia. Office of the Supervising Scientist Report No. 125. Australia: Darwin
He Z, Xiao S, Xie X, Zhong H, Hu Y, Li Q, Gao F, Li G, Liu J, Qiu G (2007) Molecular diversity of microbial community in acid mine drainages of Yunfu sulfide mine. Exremophiles 11:305–314
Herlihy AT, Mills AL, Hornberger GM, Bruckner AE (1987) Sulfate reduction in freshwater sediments receiving acid mine drainage. Appl Environ Microbiol 49:179–186
Kelly M (1988) Mining and the freshwater environment. Elsevier, Cambridge, pp 43–63
Kelly DP, Wood AP (2000) Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus. Int J Syst Evol Microbiol 50:511–516
Kim J, Kang SH, Min KA, Cho KS, Lee IS (2006) Rhizosphere microbial activity during phytoremediation of diesel-contaminated soil. J Environ Sci Health A Tox Hazard Subst Environ Eng 41:2503–2516
Léveillé SA, Leduc LG, Ferroni GD, Telang AJ, Voordouw G (2001) Monitoring of bacteria in acid mine environments by reverse sample genome probing. Can J Microbiol 47:431–442
MCIE (Ministry of Commence, Industry and Energy) (2006) Mine Pollution Prevention Plan (2007–2011). Release copy of MCIE, Korea. http//www.mocie.go.kr. Accessed 3 March 2008
Miguel CS, Dulinskia M, Tate RLIII (2007) Direct comparison of individual substrate utilization from a CLPP study: a new analysis for metabolic diversity data. Soil Biol Biochem 39:1870–1877
Morales TA, Dopson M, Athar R, Herbert RB Jr (2005) Analysis of bacterial diversity in acidic pond water and compost after treatment of artificial acid mine drainage for metal removal. Biotechnol Bioeng 90:543–551
Nicomrat D, Dick WA, Tuovinen OH (2006) Assessment of the microbial community in a constructed wetland that receives acid coal mine drainage. Microb Ecol 51:83–89
Okabayashi A, Wakai S, Kanao T, Sugio T, Kamimura K (2005) Diversity of 16S ribosomal DNA-defined bacterial population in acid rock drainage from Japanese pyrite mine. J Biosci Bioeng 100:644–652
Salomons W (1995) Environmental impact of metals derived from mining activities: processes, predictions, prevention. J Geochem Explor 52:5–23
Tan GL, Shu WS, Hallberg KB, Li F, Lan CY, Huang LN (2007) Cultivation-dependent and cultivation-independent characterization of the microbial community in acid mine drainage associated with acidic Pb/Zn mine tailings at Lechang, Guangdong, China. FEMS Microbiol Ecol 59:118–126
Weber KP, Gehder M, Legge RL (2008) Assessment of changes in the microbial community of constructed wetland mesocosms in response to acid mine drainage exposure. Water Res 42:180–188
Zhang HB, Yang MX, Shi W, Zheng Y, Sha T, Zhao ZW (2007) Bacterial diversity in mine tailings compared by cultivation and cultivation-independent methods and their resistance to lead and cadmium. Microb Ecol 54:705–712
Acknowledgments
This research was supported by a grant (project: Application of groundwater ecology to characterization of groundwater systems and contaminant behavior) from the Korea Research Council of Public Science & Technology. S. Y. Koo and J. Y. Kim were financially supported by the Korea Science and Engineering Foundation (KOSEF) through the Advanced Environmental Biotechnology Research Center at Pohang University of Science and Technology (R11-2003-006-06001-0). E. H. Lee was financially supported by the KOSEF NRL Program grant funded by the Korea government (MEST) (R0A-2008-000-20044-0).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, J., Koo, SY., Kim, JY. et al. Influence of acid mine drainage on microbial communities in stream and groundwater samples at Guryong Mine, South Korea. Environ Geol 58, 1567–1574 (2009). https://doi.org/10.1007/s00254-008-1663-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00254-008-1663-8