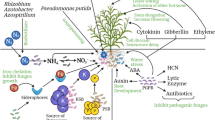
Abstract
Maize is an essential cereal crop and the third most essential food crop globally. The extensive dependence on pesticides and chemical fertilizers to control pests and increase crop yield, respectively, has generated an injurious impact on soil and animal health. Plant growth-promoting rhizobacteria (PGPR), which depict a broad array of bacteria inhabiting the root vicinity and root surface, have proven to be a better alternative. These organisms expressly or by implication foster the growth and development of plants by producing and secreting numerous regulatory compounds in the rhizosphere. Some rhizobacteria found to be in association with Zea mays rhizosphere include Bacillus sp., Azotobacter chroococcum, Burkholderia spp., Streptomyces spp., Pseudomonas spp., Paenibacillus spp., and Sphingobium spp. For this review, the mechanism of action of these rhizospheric bacteria was grouped into three, which are bioremediation, biofertilization, and biocontrol.
Graphical abstract

Key points
• Plant–microbe interaction is vital for ecosystem functioning.
• PGPR can produce volatile cues to deter ravaging insects from plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Zea mays is the most valuable grain crop globally, accounting for about 38.1% of all grains production, followed by wheat with a production of 29.1% and rice with a production of 20.8% (Costa et al., 2016). The United Nations report has it that maize accounts for about 50% of the calories and protein intake in Southern and Eastern Africa and 20% of the calories and protein intake in West Africa. It serves as feed and fodder for livestock and as food for humans. Steep corn liquor, one of its byproducts, is used to culture some microorganisms. Plastics, fabrics, and adhesives can be made from the starch of maize. Maize progressively serves as a primal matter required to produce ethanol, while its cobs are used as a source of fuel. Some challenges recorded in maize cultivation include pest and disease invasion, lack of tolerant cultivars, poor quality seeds, low soil fertility, lack of crop rotation, and intercropping. Classic examples of biotic stresses peculiar to maize cultivation are maize streak virus (MSV) and maize lethal necrosis (MLN) (Ray et al., 2013).
Over the years, different agricultural practices such as the use of pesticides and chemical fertilizers have been adopted to curb the challenges militating against the cultivation of cereal crops. Such practices have, in the long run, generated public health and environmental challenges. Though these challenges are not peculiar to maize cultivation, this review however centers on maize due to its comparative importance. More information is needed to improve maize production to meet up with its increasing demand because it is one of the most consumed food crops in the world. Understanding the molecular interaction between maize and some beneficial microbes geared towards improving crop yield or impeding disease conditions associated with maize can serve as a model for controlling such diseases in other crops. Considering the undesirable effects of pesticides and chemical fertilizers and the fact that they will perpetually increase as the human population increases, it is paramount to employ sustainable agricultural practices with minimal harmful environmental effects that will boost the production of maize.
One alternative is to opt for bioresources such as rhizobacteria. The word rhizobacteria describes plant root-associated bacteria that may be beneficial, neutral, or harmful to the host plant. These rhizobacteria occupy the rhizosphere, which is the soil with high proximity to a plant’s root, and are usually populated by a significant amount of rhizobacteria, which are economically important and utilize the organic exudates released from plant roots during chemical reactions (Odelade & Babalola, 2019).
Though the composition of the microbial community of the rhizospheric soil is similar to that of the bulk soil, the total abundance of microbes in the rhizospheric soil is higher (Alawiye & Babalola, 2019); it is populated with microorganisms and macroorganisms (including bacteria, viruses, fungi, protozoa, algae, microarthropods, and nematodes) and is characterized by a wide array of interactions, which can either be competitive, exploitative, neutral, commensal, or mutualistic (Jacoby et al., 2017). The low- and high-molecular-weight carbon compounds released from the root of plants into the surrounding soil to lubricate their root tips selectively influence the growth of microorganisms in this habitat by altering the chemistry of soil in this zone (Hartmann et al., 2009). The carbon derived from plants ameliorates the carbon limitation in the soil, resulting in increased bacterial activity and growth, with associated microbial community changes (Steinauer et al., 2016). Yang et al. (2017) compared the bacteria community in the rhizosphere of maize and bulk soil by sequencing the V3–V4 regions of the 16s rRNA gene on the Illumina system. They collected triplicate samples after maize had grown for 14 days, 35 days, and 63 days. Calculation of the Shannon diversity index using mothur yielded an average of 5.645 for rhizosphere soil and 5.501 for bulk soil. On average, the relative abundance of the genera Aeromicrobium was 0.307 for rhizospheric soil, while 0.19 was recorded for bulk soil. An average of 0.75 was recorded for the genera Burkholderia in the rhizospheric soil, while 0.43 was recorded for bulk soil. For Pseudomonas, an average of 1.68 was recorded for rhizospheric soil and 1.37 for bulk soil.
The term “plant growth-promoting rhizobacteria” (PGPR) describes a group of free-living bacteria present in the rhizosphere that provide some benefits to the plant. These bacteria show an adversarial or harmonious association between the soil, plants, and other microorganisms. PGPR can generally be categorized into two major groups: intracellular plant growth-promoting rhizobacteria (iPGPR) and extracellular plant growth-promoting rhizobacteria (ePGPR) (Viveros et al., 2010). The iPGPRs predominantly inhabit the specialized root cells’ nodular structures, while the ePGPRs primarily populate the rhizosphere (on the microenvironment near the root surface) or within the spaces between the root cortex cells (Viveros et al., 2010). The symbiotic bacteria that populate the rhizosphere of some maize species exhibit different favorable effects on the maize plant, which can include defending the health of the plant and promoting growth and productivity without leaving deleterious environmental effects (Akhtar et al., 2012; Raza et al., 2016). Although bacteria present in the maize rhizosphere bequeath immensely to ecological services such as biofertilization and biocontrol, there is insufficient information on their biotechnological prospects. Biotechnology is a rapidly emerging and significant field in technology due to its usefulness in food, health, and environmental sustainability (Björnberg et al., 2015). Hence, this review highlights the diversity of bacteria in the maize rhizosphere and their probable contributions to bioremediation, biofertilization, and biocontrol.
Underlying mechanism employed by PGPR in improving plant growth
PGPR residing in Zea mays rhizosphere vastly improves its growth by enhancing nutrient availability and accessibility (Tomer et al., 2016). It can either be through an established natural cascade of events that directly influence physiological processes and consequently promote growth or through indirect means, which ameliorate inimical environmental conditions that impede growth. Direct means include phosphates solubilization, nitrogen fixation, production of phytohormones (indole acetic acid [IAA], cytokinins, gibberellins, etc.), and production of 1-aminocyclopropane-1-carboxylate (ACC) deaminase. The indirect plant growth promotion mechanism includes the production of siderophore (which creates iron scarcity in the rhizosphere by chelating iron, thereby inhibiting proliferation of root pathogens), biofilm formation (exo-polysaccharides production), induced systemic resistance (ISR), antibiotics, and lytic enzymes production (Gupta et al., 2015). Some PGPR present in the rhizosphere of Zea mays and other crops have been reported to exhibit more than one mechanism of promoting plant growth. Enterobacter sp., for instance, produces IAA and solubilizes phosphates, while Pseudomonas sp. produces IAA and hydrogen cyanide and solubilizes phosphate. Furthermore, Agrobacterium sp. produces IAA, hydrogen cyanide, and siderophore and solubilizes phosphate (Chinakwe et al., 2019). In silico analysis of soil sample obtained from maize rhizosphere using antiSMASH v.3.0 and RAST revealed the presence of siderophore gene clusters and genes involved in the production of IAA, phosphate, and nitrogen metabolism (Babalola et al., 2019). Rhizoremediation of heavy metals that can impede plant growth is another benefit of PGPR; Enterobacter cloacae, for instance, can remove Pb and Cd present in the soil (Abedinzadeh et al., 2018). Figure 1 attempts to summarize some of the biological processes these rhizobacteria engage in to aid plant growth.
Rhizoremediation
There have been severe environmental consequences resulting from industrialization and global increase in population (Patnaik, 2018). Anthropogenic activities result in the emission of a broad array of chemicals that are hostile to human and ecosystem health. Some of these chemicals are pesticides, petroleum hydrocarbons (PHCs), polycyclic aromatic hydrocarbons (PAHs), halogenated hydrocarbons, solvents, and heavy metals (Abedinzadeh et al., 2018; Enagbonma & Babalola, 2019). The symbiosis, growth, and ultimate yield of crops can be remarkably retarded as a result of the plant’s uptake of metals, which are bountifully present in the soil (Ayangbenro & Babalola, 2017). These metals act as genotoxic substances by collapsing cell organelles and upsetting the membranes, thereby disrupting physiological processes such as photosynthesis or deactivating protein synthesis, respiration, and carbohydrate metabolism (Sharma & Talukdar, 1987).
Remediation is the act of reversing, degrading, or outrightly removing pollutants from the environment. It can also involve the reversal of environmental damages the pollutants might have caused. Bioremediation is the breakdown of organic contaminants and hydrocarbon by organisms in the soil such as bacteria, fungi, and plants to enhance life (Olawale et al., 2020). The customary methods applied to remediate soils that are contaminated (such as routine excavation, incineration, transportation to dedicated landfills, stabilization, and ion exchange or coagulation filtration) are costly and disrupt the contaminated sites (Haritash & Kaushik, 2009). Although the use of plants alone in bioremediation (phytoremediation) has recorded a level of success, the idea of using plants in combination with plant-associated bacteria (rhizoremediation) offers more prospects for remediation and ecosystem sustainability (Kumar et al., 2017). The rhizoremediation technique involves the use of rhizospheric microbial communities for the biodegradation of pollutants.
Abedinzadeh et al. (2018) reported a high concentration of heavy metals-resistant bacteria possessing several plant growth-promoting (PGP) qualities in the endorhiza and rhizosphere of maize plants irrigated with municipal and industrial wastewater. They conducted an assay to determine the influence of these heavy metals (Cd, Pb, Cr, Ni, Copper [Cu], Cobalt [Co], and Zn) on the growth of selected bacteria isolated from the rhizosphere soil and root of maize plant irrigated with municipal and industrial wastewater. The isolates were cultured on mineral salt medium (MSM) streaked with different concentrations of heavy metals. E. cloacae could get rid of 88.95 % of Pb and Cd from the medium by adsorption to its cell wall, followed by active transmembrane transport, chelation or sequestration of metal ions, and accumulation within the cell. This strain further displayed a high capability to inhabit the root surface of maize under stress conditions of heavy metal and non-stress conditions. There was an observable increase in root hairs and length in maize plants inoculated with strains of E. cloacae under heavy metal stress compared to a plant that was not inoculated with E. cloacae. There was, however, no observable sign of disease and wilting in maize seedlings inoculated with the strain.
Arid and semiarid world regions are usually faced with the challenge of irrigation water. This leads to weak plant growth due to drought stress. Achromobacter xylosidans (rhizobacteria that produce 1-aminocyclopropane-1-carboxylate ACC, deaminase), when used with biochar (0.75%), significantly improves carotenoid content, total chlorophyll, stomata conductance, and photosynthetic rate of maize under drought stress (Danish et al., 2020).
The efficacy of the rhizoremediation process can be enhanced in numerous ways such as the expression and conservation of genetically modified plant microbial systems, altering the ratio of the contaminant to the actual amount of the contaminant that enters biological receptors, and the presence of root exudates. The selection of biosurfactant-producing bacteria found in the rhizosphere of the plants can also facilitate pollutant removal efficacy (Płociniczak et al., 2011). Kuiper et al. (2004) characterized biosurfactant-producing bacteria (inhabiting a region contaminated with PAHs) that facilitate the solubilization of PAHs, thereby enhancing biodegradation by microbes. This becomes imperative since some bioremediation microbes exhibit positive chemotaxis in the direction of the pollutants (Bisht et al., 2010). Consequently, the mutual action of chemotaxis and biosurfactant can enhance bacterial proliferation and spread of microbes in polluted regions in such a way that additional zones of the polluted site can be cleaned (Gerhardt et al., 2009).
Freshly contaminated soils usually tend to be much more responsive to rhizodegradation compared to field contaminated soil that has undergone a lengthened time of aging (Phillips et al., 2006). It was opined by Wenzel (2009) that poor rhizodegradation in a contaminated field and aged spiked soils was due to low bioavailability (the extent and rate of adsorption by microbes) of the contaminant. This has a significant influence on rhizodegradation applicability and the generation of statistical information from freshly or shortly aged spiked soil material. In areas where the main limitation is low bioavailability, other strategies such as the inoculation of degrader strains to enhance rhizodegradation will likely fail. Microbial treatments of contaminated soil that appear to be effective in laboratory experiments proved otherwise on field trials with aged contaminated soil (Child et al., 2007). This points out over again the significance of bioavailability and the sample size of the experiment. In view of the previously mentioned factors, improved rhizodegradation entails more tactical methods. The degradation proficiency of the isolated microbe may be enhanced by inducing a nutrition preference for the isolated strains. Narasimhan et al. (2003) identified a compound from root exudate (phenylpropanoids) that produced a nutrition prejudice in favor of improved degradation of polychlorinated biphenyls (PCB). Plant roots also play the role of tilling of the soil to improve aeration in soil and to slot in additives (nutrients) (Kuiper et al., 2004).
Bacteria present in the environment contaminated with heavy metals have higher resistance to the toxic effects of the heavy metals compared to bacterial isolates from a non-contaminated environment (Arivalagan et al., 2014). This is because microorganisms develop physiological, structural, and biochemical characteristics that aid the adsorption of the heavy metals from the rhizosphere (Fashola et al., 2016).
Biofertilizer
One common strategy to improve crop yield is to increase the fertility of the soil, and this can be achieved through the use of biofertilizers. Biofertilizers consist of living microbes that inhabit the interior part of root cells or the rhizosphere of the plant when applied to seeds, plant surfaces, or soil and support growth by enhancing the host plant accessibility to primary nutrients (Vessey, 2003). They are more cost effective and environmentally safe compared to chemical fertilizers (Reddy et al., 2020). This is in concordance with the findings of Amogou et al., 2019; they observed that the combined use of Serratia marcescens and Pseudomonas putida with 50% of the recommended dose of NPK was more economical in terms of crop yield. Rhizobacteria can heighten the nutrients available to plants by means of nitrogen fixation or solubilizing phosphorus. Iwuagwu et al. (2013) demonstrated that Azotobacter and Azospirillium could improve the growth of Zea mays L. They reported that plant height, root length, stem diameter, fresh weight, dry weight, and chlorophyll content increased by 49%, 16%, 33%, 18%, 46%, and 39%, respectively, when treated with the rhizobacteria.
In a similar study by Wahyudi et al. (2019), some groups of Actinomycetes (Streptomyces spp.) were isolated from the maize rhizosphere using humic acid–vitamin agar to determine their ability to promote plant growth in vitro. This test was conducted on 30 isolates using the following parameters: (i) IAA production, (ii) promotion of maize sprout growth, (iii) phosphate solubilization, and (iv) growth in N-free medium. It was observed that all the isolates produced IAA and grew on an N-free medium. In contrast, only 9 (30%) of the isolates promoted maize sprout growth, and 21 (70%) isolates solubilized phosphate in the Pikovskaya medium.
In a bid to promote maize productivity in an area characterized by accelerated climate change as a result of anarchic exploitation of forests, Amogou et al. (2019) conducted a field study in ferruginous soil in the north of Benin to determine the effects of Pseudomonas cichorii, Bacillus panthothenicus, Pseudomonas putida, Pseudomonas syringae, and Serratia marcescens on the growth and yield of maize. The study area is of low soil fertility and weakly acidic in nature. The 5 strains were isolated and characterized from the rhizosphere of maize from different ecological zones of northern and central Benin following the protocol described by Agbodjato et al. (2015) and stored at −20°C in Muller–Hinton broth with glycerol. Some seeds were planted without PGPR or mineral fertilizers to serve as the control. Others were planted with a particular strain of the test PGPR, while some were planted with PGPR and 50% of the recommended dose of NPK fertilizer. Seedlings treated with S. marcescens exhibited over 41.09% increase in height compared to control and a 3.14% increase in height compared to plant treated with 100% NPK. Treatments with 100% NPK, S. marcescens + 50% NPK, and P. putid + 50% NPK exhibited stem diameters of 2.14 cm, 2.06 cm, and 2.01 cm respectively. Observation of the effect of PGPR on the nutritional status of maize plants revealed that seedlings inoculated with S. marcescens had (1.239% ± 0.09%) of nitrogen, while seedlings treated with 100% NPK had (1.232% ± 0.05%). Furthermore, P. putida and P. cichorii + 50% NPK resulted in a 25% increase in phosphorus uptake, while treatment with P. cichorii resulted in a 12.35% and 10.85% increase in uptake of potassium (K) and magnesium (Mg), respectively. The result of fresh biomass indicated that treatment with S. marcescens + 50% NPK resulted in a 144.28% increase in the fresh weight of aboveground biomass and 213.34% of the fresh weight of root compared with controls.
Rhizobacteria that hydrolyze starch occupy a critical part in plant nutrient uptake. In research to isolate and characterize bacteria that possess the ability to degrade starch from the rhizosphere of maize plants, Burkholderia cepacia, the predominant bacteria in maize of Italian agricultural fields, was found to ferment dextrose, galactose, mannose, sucrose, and xylose (Chatterjee et al., 2019). The research which was carried out in India employed the pour plate technique on NA to isolate bacteria from Zea mays L. soil samples. Burkholderia ambifaria isolated from maize plants’ rhizosphere was responsible for a notable increase in shoot growth and the growth of root and root hair (Brito et al., 2018). Strains of Burkholderia spp. can degrade starch into a soluble form by the production of amylase enzymes, which promotes the growth of maize plants (Chatterjee et al., 2019).
Nitrogen fixation
Nitrogen ranks as the fourth most vital element that constitutes the dry mass of the plant and also an imperative nutrient required for the growth of the plant (Aerts & Chapin, 2000; Munees & Mulugeta, 2014). Plants establish endosymbiotic associations with rhizobacteria and form root nodules where resident rhizobacteria fix atmospheric nitrogen. This symbiotic nitrogen fixation is extremely important because it improves crop production and supplies a great amount of fixed nitrogen to ecosystems (Shimoda et al., 2020). Nitrogen is a fundamental nucleotide, amino acid, and membrane lipid constituent. The most common form of this element is not readily accessible to plants because it is in gaseous form (N2) (Pujic & Normand, 2009). The amount of nitrogen in the atmosphere introduced again in biorhythm yearly through the means of biological nitrogen fixation (BNF) amounts to about 175 million. Rhizobacteria carry out BNF by converting atmospheric nitrogen (N2) to ammonia (NH3), which is the form plants can access. The process of nitrogen fixation by rhizobacteria is represented in Fig. 2.
Normally, the nitrogen-fixing microorganisms gain from the fixed nitrogen without excreting its compounds. However, after they die and decompose, nitrogen is made available to plants. The nitrogen accumulated in the dead organism is first converted to ammonium and then to nitrates. Consequently, fixing microorganisms also helps to accumulate nitrogen compounds over time (Vitousek et al., 2002).
Several studies have been done to identify PGPR that aids nitrogen fixation in maize plants. Fukami et al. (2017) and Pereg et al. (2016) highlighted the ability of Azospirillum to fix atmospheric N2 and substitute for the use of N fertilizers when associated with grain crops such as maize (Zea mays L.), rice (Oryza sativa L.), and wheat (Triticum aestivum L.). Richard et al. (2018) evaluated Azospirillum sp. isolated from the rhizosphere of maize plants for ammonia production and nitrogen fixation using the micro-Kjeldahl method. Their results revealed that Azospirillum possesses high nitrogenase activity indicative of the possibility of using this bacteria as a biofertilizer to improve soil fertility for improved and efficient maize cultivation.
Similarly, Bacillus pumilus, Klebsiella sp., Klebsiella pneumoniae, and Acinetobacter sp. were evaluated for N2 fixing abilities in a greenhouse experiment with reduced fertilizer N input (a third of recommended fertilizer N rate); the N2 fixation abilities of PGPR in association with maize were determined by 15N isotope dilution technique at two harvests. The results showed that dry biomass of top, root, and ear; total N content; and bacterial colonization in non-rhizosphere, rhizosphere, and endosphere of maize roots were influenced by PGPR inoculation, with the plants inoculated with B. pumilus giving the highest N2 fixing ability of 30.5%. Beyond fixing N2, these isolates were also reported to be engaged in N remobilization and delayed plant senescence in maize which results in greater grain production (Kuan et al., 2016).
Relatedly, Shimoda et al. (2020) testified that successful symbiotic nitrogen fixation (SNF) could substitute for nitrogen fertilizers in agricultural lands. A similar study done in nitrogen-depleted fields of Oaxaca, Mexico, proved that the mucilage associated with the aerial roots of Sierra Mixe maize can aid a complex diazotrophic microbiome that can encode active nitrogenase, and the fixed nitrogen (29 to 82% of the plant nitrogen) can efficiently travel from the nitrogen-fixing microbiota to host plants (Deynze et al. 2018). Furthermore, biologically fixed nitrogen consumes about 25 to 30% less energy than chemical fertilizers (Iwuagwu et al., 2013). Ali et al. 2012 also reported that the application of plant growth-promoting rhizobacteria in place of chemical fertilizers in maize increased plant height and biological yield. Following field research in southern Benin, Adjanohourn et al. (2011) opined that there are PGPRs that are crop species specific and suitable for species-dependent enhancement. They observed that Azospirillium lipoferum, Pseudomonas putida, and Pseudomonas fluorescens are the most suitable maize crop-specific PGPRs that can be applied to advance field maize crop yield. This is also in concordance with the findings of Shaharoona et al. (2006) and Biari et al. (2008). They observed that Pseudomonas aeruginosa and P. fluorescens improve shoot fresh biomass by 23.40% and 59.57%, respectively, under field soil conditions.
Phosphate solubilization
After nitrogen, the second mineral element, whose deficiency significantly retards plant growth, is Phosphorus (Nisha et al., 2014). This element constitutes approximately 0.2% of plant dry weight and is a cardinal building block of phospholipids, phytin, and nucleic acids. It is also very important in respiration, transfer and storage of energy, photosynthesis, seed formation, and division and elongation of the cell (Sagervanshi et al., 2012). Phosphorus is absorbed by plants in soluble forms that are mono and dibasic (H2PO4-, HPO4) (Razaq et al., 2017). Regrettably, about 95–99% of soil phosphorus are present as insoluble organic phosphates (phosphomonesters, phosphotriesters, and inositol phosphate) or insoluble inorganic phosphates (apatite) that the plants are unable to assimilate (Khan et al. 2008; Perez-Montano et al., 2014). Precipitation with cations Ca2+ and Mg2+ in basic soils and with Fe3+ and Al3+ in acidic soils immobilizes phosphorus (El-Komy 2005). Consequently, on applying this soluble inorganic phosphate to cultivated soil, a large percentage is speedily immobilized and turns out to be inaccessible by the plant (Vikram & Hamzehzarghani, 2008). However, some PGPR can solubilize the insoluble soil phosphate to a form plants can utilize. This group of PGPR is known as “phosphate solubilizing bacteria, PSB.” Examples of PSB associated with Zea mays are Paenibacillus sp., Pseudomonas sp., Bacillus subtilis, and Sphingobium sp. These strains can solubilize AlPO4, Ca3(PO4)2, and FePO4 to release soluble phosphorus that can be assimilated by plants (Li et al., 2017; Olanrewaju & Babalola, 2019). Mosimann et al. (2017) reported that treatment of maize seedling with Pseudomonas sp. yielded an increase in the biomass of maize plants grown on acid soil with the low phosphorus content.
PSB can also excrete extracellular enzymes to mineralize the organic phosphate that is insoluble. Examples of such enzymes are phosphatases (which catalyze phosphoric esters hydrolysis), C-P lyases, and phytases (Panhwar et al., 2012). Worthy of note is the fact that mineralization and solubilization can co-occur within the same PSB (Tao et al., 2008). Yazdani et al. (2009) stated that the use of PSB could reduce the application of phosphorus to 50% without having an effect on the seed yield of maize. Therefore, the availability and absorption rate of phosphorus in the rhizosphere is increased by the plants’ inoculation with PSB. The procedure for phosphate solubilization is illustrated in Fig. 3.
Extracellular polysaccharides production
The production of extracellular (exopolysaccharides [EPS]), intracellular, and structural polysaccharides is one of the ways Zea mays rhizobacteria promote its growth. These polysaccharides form hydrated gels around the cells, constituting the boundary between the rhizobacteria and their direct environment. The exopolysaccharides aggregate soil and change its porosity, thereby controlling the soil water transported to the roots, soil aeration, and root growth (Redmile-Gordon et al., 2020). EPS envelop and safeguard the roots against infection by pathogenic microorganisms. In the condition of salt stress, the EPS contribute to reducing the salinity of the rhizosphere by chelating cations available in the root zone (Khana, 2015).
Other means rhizobacteria aid biofertilization are by phytostimulation, indole acetic acid production, and ethylene regulation.
Biocontrol
Biocontrol agents (BCAs) aid the control of phytopathogenic microbes inhabiting the soil via secretion of metabolites that are antagonistic (lytic enzymes, siderophores, volatile compounds, hydrogen cyanide, and antibiotics), induction of systemic resistance, and by competition for nutrients and space (Tariq et al., 2014). Ectocarpus fasciculatus, Streptomyces hygroscopicus, Pseudomonas fluorescens, P. putida, P. aeruginosa, and Azospirillum lipoferum inhibit the growth of the mycelial of Aspergillus ochraceus and Fusarium verticillioides that are pathogens of maize (Noumavo et al. 2015). P. aeruginosa and P. fluorescens are extremely antagonistic against F. verticillioides (responsible for 52.24% of the inhibition of mycelial growth) and A. ochraceus (responsible for 58.33% of the inhibition of mycelial growth) (Noumavo et al., 2016). Furthermore, Akhtar et al. (2018) reported that the pathogen Fusarium oxysporum could be controlled by inoculation of maize seeds (grown in soil infested with the pathogen and fertilized with NPK) with Serratia sp.
The most common fungal pathogen of maize is Fusarium verticillioides (Fv) (Sacc.) Nirenb. It causes a disease condition in maize known as stalk, ear, and root rot (SERR) and is accountable for enormous global fiscal losses (Hernandez-Rodriguez et al., 2008). Farming practices such as monocultivation of maize have led to a high prevalence of this disease along with crop fatalities as a result of Fv in the Sinaloa state of Mexico (Figueroa-Lopez et al., 2016). Besides the effect of this infection on grain yield, it can also be responsible for poor grain quality (Figueroa-Lopez et al., 2016).
Beneficial microbes can also influence plant–insect interaction. Females of Ostrinia nubilalis (Hubner) rely on volatile cues to locate and oviposit on maize plants. Larvae of this insect have been reported to cause significant damage to maize ears and result in a drastic economic loss of fresh sweet corn. Disi et al. (2018) opined that treatment of maize with PGPR could alter the production of volatile organic compounds that serve as a signal to the insect. They investigated the production of the volatile cue by inoculating maize seedlings with the following PGPR strains that were isolated from the maize rhizosphere: Bacillus velezensis, B. pumilus, Fictibacillus solisalsi, and Bacillus mojavensis. They observed that O. nubilalis females differentiated between PGPR-treated and untreated maize plants and laid significantly fewer eggs on PGPR-treated plants.
Comprehensive insight into the microbial ecology and diversity present in the rhizosphere of maize would aid the improvement of the health of field crops and develop efficient biological control strategies to lessen our dependency on chemical pesticides (Filion et al., 2004). The practice of using biological antagonists to control pathogens has proven to be a more sustainable agronomic practice. This has been adopted recently on a commercial scale, while numerous experimental approaches are being developed to optimize the efficacy of the process (Souza et al., 2015).
Antibiotic production
The production of antibiotics is a very efficient and well-studied biological control mechanism of PGPR against phytopathogens. The mode of action is usually by preventing the proliferation of plant pathogens (Shilev, 2013). Examples of identified antibiotics are oomycin A, amphisin, pyoluteorin, 2,4-diacetylphloroglucinol (DAPG), pyrrolnitrin, tropolone, cyclic lipopeptides, phenazine, and tensin produced by Pseudomonads (Loper & Gross, 2007). Other examples are kanosamine, oligomycin A, xanthobaccin, and zwittermicin A taken from Bacillus spp., Stenotrophomonas spp., and Streptomyces spp. to inhibit proliferation of phytopathogens of fungal origin (Compant et al., 2005). Furthermore, Bacillus subtilis A1, Bacillus velezensis A3, and Bacillus subtilis A29 have been reported to synthesize antifungal and antimicrobial compounds, such as bacillaene, bacilysin, bacillibactin, difficidin, fengycin, subtilosin A, surfactin, and macrolactin (Babalola et al., 2019). The structure, mode of action, and specificity of most of the antibiotics have been well studied with some of the biocontrol strains commercially available. However, a major challenge with depending on antibiotic-producing bacteria as BCAs is that a good number of phytopathogens can become resistant to specific antibiotics with frequent use. To surmount this challenge, a few researchers recommend the utilization of biocontrol strains that synthesize antibiotics and hydrogen cyanide (HCN). Some biocontrol agents secrete enzymes such as proteases, cellulases, chitinases, lipases, and β-1,3-glucanases which can lyse part of some pathogenic fungi cell walls (Hayat et al., 2010). These enzymes can control the proliferation of an array of disease-causing fungi, including Fusarium oxysporum, Phytophthora spp., Botrytis cinerea, Pythium ultimum, Sclerotium rolfsii, and Rhizoctonia solani (Nadeem et al., 2013).
Iron chelation (siderophores production)
Some bacterial strains produce siderophores as a means of biocontrol to prevent fungal pathogens from gaining access to available iron. Iron is vital to living organisms; it is involved in different vital biological activities. Most enzymes employed in oxygen transfer (hemoglobin) or electron transfer (mitochondrial respiration) engage iron as the cofactor (Thomine & Languar, 2011). Although it has been observed that iron is present in large amounts in ground rock, this enormous amount of iron, however, is present as ferric ions (Fe3+). Consequently, living organisms (bacteria, plants, etc.) can assimilate only a very little quantity (Ammari & Mengel, 2006). Rhizobacteria have come up with various iron uptake strategies including the production of siderophores to adapt to their environment and overall survival. The molecular weight of siderophores is low (400 to 1500 Da), with a unique affinity for Fe3+ (Ka between 1023 and 1052) and able to bind the complex Fe siderophores with membrane receptors to support the absorption of iron by plants and microorganisms (Hider & Kong, 2010). Siderophores are also useful in the preparation of fertilizer to facilitate the growth of plants by regulating iron intake. Sah et al. (2017) planted maize seeds and treated them with Pseudomonas aeruginosa to observe how these rhizobacteria can influence iron acquisition in maize. The number of seeds, root, and shoot and cob length of maize plants were analyzed after 70 days of planting. The results showed that the plants treated with Pseudomonas aeruginosa were taller and sturdier than the control. The shoot length of the control was 103 cm, while the root length was 7 cm. When there is iron deficiency, Pseudomonas produces siderophore, which helps to chelate and transport Fe into the roots of the plant and aids plant growth (Sharma & John, 2003).
Production of lytic enzymes
The cell walls of fungi can be degraded by a number of PGPR by the secretion of hydrolytic enzymes, which include β-glucanases, dehydrogenases, chitinases, lipases, proteases, phosphatases, pectinolyases, hydrolases, cellulases, and exo- and endo-polygalacturonases (Jadhav & Sayyed, 2016). In an in vitro experiment conducted by Sharma et al. (2009), several strains of Pseudomonas displayed antifungal activity against three fungi zoospores. The researchers ascertained that the antifungal activity was a result of the production of rhamnolipid, which lysed the plasma membrane of the zoospores fungi. This lytic property of PGPR helps to eliminate the pathogen, thereby protecting the plant against biotic stress (Noumavo et al., 2016).
Pseudomonas spp. produce a wide range of lytic enzymes, siderophores, cyanide, and antibiotics (Weller et al., 2007) that displays inhibitory effects on pathogens, including Fv. A good number of secondary metabolites that display antifungal effects on different plant pathogens are produced by the Bacillus genus (Raaijmakers & Mazzola, 2012). Bacillus spp. employ different mechanisms to inhibit this fungal pathogen (Shafi et al., 2017). Examples of such are nutrient antagonism (Kamilova et al. 2005), antifungal lipopeptides production (Hazarika et al., 2019), and lytic enzyme production such as chitinases that can prevent fungal hyphal extension by cell wall degradation (Kishore et al. 2005).
In a study by Olanrewaju and Babalola (2019), Bacillus subtilis, Streptomyces heliomycini, Pseudomonas sp., Streptomyces griseoflavus, and Streptomyces globisporus were shown to exhibit antagonistic effect against Fusarium graminearum, a phytopathogen.
Dressing seed with biocontrol agents is a suitable technique to restrain phytopathogens in the rhizosphere and spermosphere (Pereira et al., 2007). Bacterial inoculants have been proven to promote plant growth by antagonizing soil-borne phytopathogens such as Fv. In Argentina, root rot caused by Fv has been controlled by Bacillus subtilis and Pseudomonas cepacia (Cavaglieri et al., 2005). Burkholderia spp. encourage the growth of the plant and repress disease conditions triggered by Fv in maize plant (Hernandez-Rodriguez et al., 2008), while species such as Enterobacter hormaechei and Bacillus amyloliquefaciens lessen the fumonisin accumulation and Fv infection in maize kernels (Pereira et al., 2010). For efficiency in biocontrol, there is the need to develop control microbes that are normal flora of the soils where the plant of interest is grown. Augmentation of an ecosystem with a huge quantity of “exotic” microorganisms can upset a native ecosystem and lead to negative ecological effects on the overall microbiota of the rhizosphere (David et al., 2017). Additionally, once microbiological control agents are released, they may not only suppress phytopathogens. Still, they may also have a deleterious effect on other rhizobacteria that hitherto are of importance to the host plant. Figueroa-Lopez et al. (2016) demonstrated that some bacterial strains inhibit plant pathogens that affect rhizospheric and endophytic bacteria populations.
Production of hydrogen cyanide and volatile compounds
Volatile compounds such as hydrogen cyanide (HCN) are also generated by PGPR to antagonize plant pathogens. Noumavo et al. (2016) reported the secretion of HCN by rhizospheric organisms. P. corrugata displayed antagonistic action against Fusarium oxysporum and Alternaria alternata pathogens of maize and some other plants (Trivedi and Pandey 2008). The antagonism was a result of the volatile compounds produced, even though P. corrugata also can produce hydrolytic enzymes under in vitro culture conditions. Bacillus megaterium antagonized two test plant pathogens, A. alternata and F. oxysporum, by producing volatile compounds (Trivedi and Pandey 2008). Olanrewaju and Babalola (2019) also reported that Streptomyces globisporus present in Zea mays rhizosphere also produces hydrogen cyanide.
Induction of systemic resistance
Plants’ defense mechanism can be triggered by PGPR in a phenotypical manner, which is akin to plants’ usual defense response when exposed to a pathogenic organism (Pieterse et al., 2009). This phenomenon, which is known as induced systemic resistance (ISR), makes the plant resilient and readily prepared against an imminent attack by any pathogen (Van Loon, 2007). Rhizobacteria induction of systemic resistance is seen as a favorable biocontrol mechanism against plant diseases (Razaq et al., 2017). The ISR can be triggered by several microorganisms, including gram-positive bacteria such as B. pumilus or gram-negative bacteria such as Pseudomonas fluorescens, P. aeruginosa, and P. putida and enterobacteria such as Serratia (Serratia plymuthica, Serratia marcesens) or Pantoea agglomerans (Beneduzi et al., 2012). The ISR does not only protect the plants against several pathogens, including viral, bacterial, and fungal pathogens, but also protects the plant against some diseases caused by nematodes and insects (Choudhary et al. 2007). Several bacterial metabolites, including lipopolysaccharides (LPS), cyclic lipopeptides, siderophores, homoserine lactones, 2,4-diacetylphloroglucinol, and some volatile compounds such as 2,3-butanediol and acetoin, can trigger ISR (Doornbos et al. 2012).
Competition for nutrients, space, and iron
It may be intricate to expressly validate how the competition for nutrients, space, and iron can control pathogenic attacks. However, the indirect pieces of evidence reveal that the perpetual competition between PGPR and pathogens might lessen the prevalence and level of damage caused as a result of the plant disease condition. The swift and overwhelming colonization of plant roots by PGPR (which populates the plant pathogens’ infection sites) and consumption of the bulk of the nutrients available retard the spread of pathogens. It is, therefore, evident that competition for nutrients is an important means of biological control because high rhizobacteria biomass mounts pressure on the available nutrients. The presence of flagella for mobility, lipopolysaccharide (LPS), chemotaxis, synthesis of macromolecules and vitamins, and usage of root exudates comparatively place PGPR at an advantage on plant root colonization (Lugtenberg & Kamilova, 2009). Table 1 enumerates PGPR associated with Zea mays and how they aid plant growth.
Molecular mechanisms of the interaction between PGPR and maize plant
The scientific study of the set of metabolites and the small molecule substrates, intermediates, and products of cell metabolism involved in the interaction between maize and its associated PGPR can aid our understanding of how these rhizobacteria support plant growth or serve as phytopathogen biocontrollers. Detailed information on the interactions between plant, toxicogenic microbes, and beneficial microbes could serve as a reference point in the formulation of new strategies to improve crop protection, quality, and yields (Adeniji et al., 2019). Most fungal and bacterial plant-borne pathogens produce bioactive metabolites that undermine the availability of healthy harvested crops for human and animal consumption. Examples of such metabolites are albicidin, alternariol, citrinin, coronatine, fumonisin, RS-toxin, and toxoflavin produced by Xanthomonas sp., Alternaria sp., Penicillium sp., Pseudomonas sp., Fusarium sp., Rhizoctonia sp., and Burkholderia sp., respectively. These phytopathogens are known to be responsible for cereal mildews, and fusariosis or smuts/spots, and consequently have a global impact on the human population, especially those who rely on cereals, such as maize, as a staple food source (Franco et al., 2019).
Benzoxazinoids (BXs), a tryptophan-derived heteroaromatic metabolite, influence the metabolic regulation and differentiation of maize roots and a vast group of secondary root metabolites (Adeniji et al., 2020; Meihls et al., 2013). The effects of BX-regulated root metabolites with BX-dependent rhizosphere microbiota were studied by Cotton et al. (2019), using an untargeted ultra-high performance liquid chromatography with quadrupole time-of-flight mass spectrometry metabolomics analysis, to evaluate the influence of the BXs metabolites on the maize root metabolome. They compared the metabolome of wild-type (WT) BX-regulated maize (Zea mays cv. W22) root and a BX-deficient W22 mutant (the mutation brought about by inserting transposons at three different steps [BX1, BX2, and BX6] of the BX biosynthesis pathway). It was observed from the results of the WT and BX root profiling that the BX1 and BX2 mutations significantly impacted the root metabolome.
Metabolomics creates an opportunity to identify phytopathogen-specific metabolic biomarkers or plant disease/defense biomarkers and then use them to monitor disease progression or the infectivity of phytopathogens. Additionally, phytopathogen control can be achieved by isolating or synthesizing functional biomolecules associated with the biocontrol organisms, which can be used to arouse antiphytopathogenic mechanisms in plants. For example, specific biomarkers and or biochemical processes identified during maize–Fusarium graminearum–Bacillus amyloliquefaciens interaction would most likely contribute to a better understanding of the metabolic regulation of all the interacting living systems, providing valuable insights potentially useful in plant breeding, metabolic bioengineering, robust secondary metabolite-producing beneficial microbes, and cultivation/production of biofungicides which can be used in maize cultivation.
Plant growth-promoting rhizobacteria can mediate drought tolerance by modulating metabolic signaling and stress-responsive genes. The molecular pathways engaged in PGPR-mediated drought stress tolerance in maize plant was studied by SkZ et al. (2018). They observed the differential gene response between Pseudomonas putida strain FBKV2 and maize interaction under drought stress using Illumina sequencing. This was done by generating RNA Seq libraries from leaf tissue of maize seedlings treated with P. putida strain FBKV2 and subjected to drought stress. They also collected the same from leaf tissue of maize seedlings not treated with the strain and subjected to drought stress to serve as the control. On mapping the libraries with the maize genome database to identify the differentially expressed genes (DEGs), the expression studies revealed downregulation of ethylene biosynthesis (ET), superoxide dismutase, abscisic acid (ABA) and auxin signaling, catalase, and peroxidase in seedlings treated with FBKV2. On the contrary, genes conferred by P. putida strain FBKV2, which could act as key elements in drought tolerance, were observed to be upregulated. Examples included genes involved in β-alanine and choline biosynthesis, heat shock proteins, and late embryogenesis abundant (LEA) proteins. Also noteworthy was the expression of genes encoding benzoxazinoid (BX) biosynthesis which acts as the chemoattractant P. putida strain FBKV2. This was further confirmed by green fluorescent protein-labeled P. putida strain root colonization studies.
Conclusion
One readily available key for the sustainable promotion of maize production and global food security is the exploitation of rhizobacteria. Soil microbial community offers an unlimited potential to resolve myriads of challenges militating against crop production. The major requirements for improved maize production are nutrient availability (N and P) and disease control. Fortunately, P solubilizing and N2 fixing bacteria inhabit the natural population of Zea mays rhizobacteria. These attributes are pertinent growth-promoting traits for Zea mays growing in areas facing incessant erosion and soil degradation. The control of phytopathogens can be achieved effectively by the use of biocontrol agents (BCAs) producing hydrolytic enzymes, antibiotics, siderophores; induction of systemic resistance or by the production of hydrogen cyanide; and competition for space, iron, and nutrients. Hydrolytic enzymes, for instance, can selectively lyse the cell wall of phytopathogens without harming the plant tissues. This makes BCAs more eco-friendly, sustainable, and safer than chemical agents. All of these potentials can be harnessed in an economical and eco-friendly manner to get desired results without negatively disrupting the ecosystem. Consequently, there is a need for significant research efforts to boost the practical use of rhizobacteria in combating agricultural challenges.
References
Abedinzadeh M, Etesami H, Alikhani HA (2018) Characterization of rhizosphere and endophytic bacteria from roots of maize (Zea mays L.) plant irrigated with wastewater biotechnological potential in agriculture. Biotechnol Rep 20:1–12
Adeniji AA, Loots DT, Babalola OO (2019) Bacillus velezensis: phylogeny, useful applications, and avenues for exploitation. Appl Microbiol Biotechnol 103:3669–3682
Adeniji AA, Babalola OO, Loots DT (2020) Metabolomic applications for understanding complex tripartite plant-microbes interactions: strategies and perspectives. Biotechnol Rep 25:1–9
Adjanohourn A, Allagbe M, Noumavo PA, Gotoechan-Hodonou H, Sikirou R, Dossa KK, GleleKakai R, Kotchoni SO, Baba-Moussa L (2011) Effects of plant growth promoting rhizobacteria on field grown maize. J Anim Plant Sci 11(3):1457–1465
Aerts R, Chapin FS (2000) The mineral nutrition of wild plants revisited: a re-evaluation of processes and patterns. Adv Ecol Res 30:1–67
Agbodjato NA, Noumavo PA, Baba-Moussa F, Salami HA, Sina H, Sezan A, Bankole H, Adjanohoun A, Baba-Moussa L (2015) Characterization of potential plant growth promoting rhizobacteria isolated from maize (Zea mays L.) in Central and Northern Benin (West Africa). Appl Environ Soil Sci 2015:1–9
Akhtar N, Qureshi MA, Iqbal A, Ahmad MJ, Khan KH (2012) Influence of Azotobacter and IAA on symbiotic performance of rhizobium and yield parameters of Lentil. J Agric Res 50:361–372
Akhtar N, Naveed M, khalid M, Ahmad N, Rizwan M, Siddique S (2018) Effect of bacterial consortia on growth and yield of maize grown in Fusarium infested soil. Soil Environ 37(1):35–44
Alawiye TT, Babalola OO (2019) Bacteria diversity and community structure in typical plant rhizosphere. Divers 11:179–190
Ali A, Akhtar N, Khan BA, Khan MS, Rasul A, Zaman SU, Khalid N, Waseem K, Mahmood T, Ali L (2012) Acacia nilotica: A plant of multipurpose medicinal uses. J Med Plants Res 6(9):1492–1496
Ammari T, Mengel K (2006) Total soluble iron in soil solutions of chemically different soils. Geoderma 136:876–885
Amogou O, Dagnenonbakin G, Agbodjato NA, Noumavo PA, Solako KV, Adoko MY, kakai RG, Adjanohoun A, Baba-Moussa L (2019) Applying rhizobacteria on maize cultivation in Northern Benin: effects on growth and yield. Agric Sci 10:763–782
Arivalagan P, Singaraj D, Haridass V, Kalianan T (2014) Removal of cadmium from aqueous solution by batch studies using Bacillus cereus. Ecol Eng 71:728–735
Ayangbenro AS, Babalola OO (2017) A new strategy for heavy metal polluted environments: a review of microbial biosorbents. Int J Environ Res Public Health 14:94–110
Babalola OO, Ayangbenro AS, Olanrewaju OS (2019) Draft genome sequences of three rhizospheric plant growth-promoting bacteria. Microbiol Resours Announc 8. https://doi.org/10.1128/MRA.00455-19
Beneduzi A, Ambrosini A, Passaglia MD (2012) Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet Mol Biol 35(4):1044–1051
Biari A, Gholami A, Rahmani HA (2008) Growth promotion and enhanced nutrient uptake of maize (Zea mays L.) by application of plant growth promoting rhizobacteria under green house and two different field soil condition. J Biol Sci 8:1015–1020
Bisht S, Pandey P, Sood A, Sharma S, Bisht NS (2010) Biodegradation of naphthalene and anthracene by chemotactically active rhizobacteria of Populus deltoides. Braz J Microbiol 41:922–930
Björnberg KE, Jonas E, Marstorp H, Tidaker P (2015) The role of biotechnology in sustainable agriculture: views and perceptions among key actors in the Swedish food supply chain. Sustainability 7:7512–7529
Brito TS, Buss LA, Carvalho JPF, Eberling T, Martinez A, Guimaraes VF, Chaves EID (2018) Growth promotion of Burkholderia ambifaria associated to nitrogen fertilization in the initial development of corn. J Agric Sci 10(6):123–135
Cavaglieri L, Orlando J, Rodriquez MI, Chulze S, Etcheverry MG (2005) Biocontrol of Bacillus subtilis against Fusarium verticillioides in-vitro and at the maize root level. Microbiol Res 156:748–754
Chatterjee P, Roy P, Mandal P, Chatterjee S (2019) Isolation, phenotypic and molecular characterization of Burkholderia sp. (strain, PCS1) from maize fields exhibiting starch hydrolysis ability. Biotech Commun 12(1):66–72
Child R, Miller CD, Liang Y, Sims RC, Anderson AJ (2007) Pyrene mineralization by Mycobacterium sp. strain in a barley rhizosphere. J Environ Qual 36:1260–1265
Chinakwe E, Mberede C, Ngumah C (2019) Plant growth promoting rhizobacteria from local and hybrid maize (Zea mays) varieties. AFST 20:387–392
Choudhary DK, Prakash A, John BN (2007) Induced systemic resistance (ISR) in plants: mechanism of action. Indian J Microbiol 47(4): 289–297
Cohen AC, Travaglia CN, Bottini R, Piccoli PN (2009) Participation of abscisic acid and gibberellins produced by endophytic Azospirillum in the alleviation of drought effects in maize. Botanique 87:455–462
Compant S, Duffy B, Nowak J, Clement C, Barka EA (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action and future prospects. Appl Environ Microbiol 71(9):4951–4959
Costa CT, Teodoro I, Silva S, Cunha FN, Teixeira B, Soares FA, Morais WA, Silva NF, Gomes FH, Cabral LB (2016) Agronomic performance, production components and agricultural productivity of maize (Zea mays L.) cultivars. Afr J Agric Res 11(43):4375–4383
Cotton TA, Pétriacq P, Cameron DD, Meselmani MA, Schwarzenbacher R, Rolfe SA, Ton J (2019) Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J 1
Danish S, Satfar-ul-Hyle M, Mushin F, Hussain M (2020) ACC-deaminase producing plant growth promoting rhizobacteria and biochar mitigate adverse effects of drought stress on maize growth. PLoS One 15(4):1–14
David P, Thebault E, Anneville O, Duyck P, Chapuis E, Loeville N (2017) Impacts of invasive species on food webs. Rev Empir Data 56:1–60
Deynze AV, Zamora P, Delaux P, Heitmann C, Jayaraman D, Rajasekar S, Graham D, Maeda J, Gibson D, Schwartz KD, Berry AM, Bhatnagar S, Jospin G, Darling A, Jeannotte R, Lopez J, Weimer BC, Eisen JA, Shapiro H, Ane J, Bennett AB (2018) Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLOS Biology 16(8):1–21
Disi JO, Zebelo S, Kloepper JW, Fadamiro H (2018) Seed inoculation with beneficial rhizobacteria affects European corn borer (Lepidoptera: Pyralidae) oviposition on maize plants. Entomol Sci 21:48–58
Doornbros RF, Loon LC, Bakker P (2012) Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. Agron Sustain Dev 32:227–243
El-Komy HMA (2005) Coimmobilization of Azospirillum lipoferum and Bacillus megaterium for successful phosphorus and nitrogen nutrition of wheat plants. Food Technol Biotechnol 43(1):19–27
Enagbonma BJ, Babalola OO (2019) Environmental sustainability: a review of termite mound soil material and its bacteria. Sustainability 11:1–10
Fashola MO, Ngole-Jeme VM, Babalola OO (2016) Heavy metal pollution from gold mines: environmental effects and bacterial strategies for resistance. Int J Environ 13:1047–1067
Figueroa-Lopez AM, Cordero-Ramirez JD, Martinez-Alvarez JC, Lopez-Meyer M, Lizarraga-Sanchez GJ, Felix-Gastelum R, Castro-Martinez C, Maldonado-Mendoza E (2016) Rhizospheric bacteria of maize with potential for biocontrol of Fusarium verticillioide. SpringerPlus 5:330–342
Filion M, Hamelin RC, Bernier L, St-Arnaud M (2004) Molecular profiling of rhizosphere microbial communities associated with healthy and diseased black spruce (Picea mariana) seedlings grown in a nursery. Appl Environ Microbiol 7(6):3541–3551
Franco LT, Petta T, Rottinghaus GE, Bordin K, Gomes GA, Oliveira CAF (2019) Co-occurrence of mycotoxins in maize food and maize-based feed from small-scale farms in Brazil: a pilot study. Mycotoxin Res 35:65–73
Fukami J, Ollero FJ, Megías M, Hungria M (2017) Phytohormones and induction of plant stress tolerance and defense genes by seed and foliar inoculation with Azospirillum brasilense cells and metabolites promote maize growth. AMB Express 7:153–160
Gerhardt KE, Huang XD, Glick BR, Greenberg BM (2009) Phytoremediation and rhizoremediation of organic soil contaminants: potential and challenges. Plant Sci 176:20–30
Gupta G, Parihar SS, Ahirwar NK, Snehi SK, Singh V (2015) Plant growth promoting rhizobacteria (PGPR): current and future prospects for development of sustainable agriculture. JMBT 7:96–102
Haritash AK, Kaushik CP (2009) Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater 169:1–15
Hartmann A, Schmid M, Van Tuinen D, Berg G (2009) Plant-driven selection of microbes. Plant Soil 321:235–257
Hayat R, Ali S, Amara U, Khalid R, Ahmed I (2010) Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol 60:579–598
Hazarika DJ, Goswami G, Gautom T, Parveen A, Das P, Barooah M, Boro RC (2019) Lipopeptide mediated biocontrol activity of endophytic Bacillus subtilis against fungal phytopathogens. BMC Microbiol 19:71–84
Hernandez-Rodriguez A, Heydrich-Perez M, Acebo-Guerrero Y, Velazquez- del Valle, MG, Hernandez-Lauzardo AN (2008) Antagonistic activity of Cuban native rhizobacteria against Fusarium verticillioides (Sacc.) Nirenb. In maize (Zea mays L.) Appl. Soil Ecol. 39(2):180–186
Hider RC, Kong X (2010) Chemistry and biology of siderophores. Nat Prod Rep 27(5):637–657
Hussain A, Hasnain S (2009) Cytokinin production by some bacteria: its impact on cell division in cucumber cotyledons. Afr J Microbiol Res 3:704–712
Iwuagwu M, Chukwuka KS, Uka UN, Amandianeze MC (2013) Effects of biofertilizers on the growth of Zea mays L. AJMBES 15(2):235–240
Jacoby R, Peikert M, Succurro A, Koprivova A, Kopriva S (2017) The role of soil microorganisms in plant mineral nutrition-current knowledge and future directions. Front Plant Sci 8:1617
Jadhav HP, Sayyed RZ (2016) Hydrolytic enzymes of rhizospheric microbes in crop protection. MOS Cell Sci Rep 3(5):135–136
Kamilova F, Validov S, Azarova T, Mulders I, Lugtenberg B (2005) Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ Microbiol 7:1809–1817
Khana V (2015) ACC-deaminase and EPS production by salt tolerant rhizobacteria augment growth in chickpea under salinity stress. IJBSM 6:558–565
Khan ZR, Amudavi DM, Midega CAO, Wanyama JM, Pickett JA (2008) Farmers' perceptions of a 'push-pull' technology for control of cereal stemborers and Striga weed in western Kenya. Crop Protection 27(6):976–987
Kishore GK, Pande S, Podile AR (2005) Chitin-supplemented foliar application of Serretia marcescens gps 5 improves control of late leaf spot disease of groundnut by activating defence-related enzymes. J Phytopathol 153(3):169–173
Kuan KB, Othman R, Abdul Rahim K, Shamsuddin ZH (2016) Plant growth-promoting rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilisation of maize under greenhouse conditions. PLoS One 11(3):1–19
Kuiper I, Lagendijk EL, Bloemberg GV (2004) A beneficial plant microbe interaction. Mol Plant-Microbe Interact 17:6–15
Kumar SS, Kadier A, Malyan SK, Ahmad A, Bishnoi NR (2017) Phytoremediation and rhizoremediation: uptake, mobilization and sequestration of heavy metals by plants. In: Singh DP, Singh HB, Prabha R (eds) Plant microbes interactions in agro-ecological perspectives. Springer Nature, Singapore, pp 367–394
Li Y, Liu X, Hao T, Chen S (2017) Colonization and maize growth promotion induced by phosphate solubilizing bacterial isolates. Int J Mol Sci 18:12–53
Loper JE, Gross H (2007) Genomic analysis of antifungal metabolite production by Pseudomonas fluorescens Pf-5. Eur J Plant Pathol 119:265–278
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Meihls LN, Handrick V, Glauser G, Barbier H, Kaur H, Haribal MM, Lipka AE, Gershenzon J, Buckler ES, Erb M (2013) Natural variation in maize aphid resistance is associated with 2, 4-dihydroxy-7-methoxy-1, 4-benzoxazin-3- one glucoside methyltransferase activity. Plant Cell 25:2341–2355
Mosimann C, Oberhansli T, Ziegler D, Nassal D, Kandeler E, Boller T, Mader P, Thonar C (2017) Tracing of two Pseudomonas strains in the root and rhizoplane of maize, as related to their plant growth-promoting effect in contrasting soils. Front Microbiol 7:2150–2164
Munees A, Mulugeta K (2014) Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J King Saud Univ Sci 26:1–20
Nadeem E, Olin SS, Hill LC, Hoagwood KE, Horwitz SM (2013) Understanding the components of quality improvement collaboratives: a systematic literature review. Milbank Q 91(2):354–394
Narasimhan K, Basheer C, Bajic VB, Swarup S (2003) Enhancement of plant microbe interactions using a rhizosphere metabolomics driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol 132:146–153
Nisha K, Devi P, Vasandha S, Sunitha K (2014) Role of phosphorus solubilizing microorganisms to eradicate phosphorus deficiency in plants: a review. Int J Sci Res 4(7):1–5
Noumavo PA, Agbodjato NA, Gachomo EW, Salami HA, Baba-Moussa F, Adjanohoun A, Kotchoni SO, Baba-Moussa L (2015) Metabolic and biofungicical properties of maize rhizobacteria for growth promotion and plant disease resistance. Afr J Biotechnol 14:811–819
Noumavo PA, Agbodjato NA, Baba-Moussa F, Adjanohoun A, Baba-Moussa LL (2016) Plant growth promoting rhizobacteria: beneficial effects for healthy and sustainable agriculture. Afr J Biotechnol 15(27):1452–1463
Odelade KA, Babalola OO (2019) Bacteria, fungi and archaea domains in rhizospheric soil and their effects in enhancing agricultural productivity. IJERPH 16:1–19
Olanrewaju OS, Babalola OO (2019) Bacterial consortium for improved maize (Zea mays L.) production. Microorganisms 7(11):519
Olawale O, Obayomi KS, Dahunsi SO, Folarin O (2020) Bioremediation of artificially contaminated soil with petroleum using animal waste: cow and poultry dung. Cogent Eng 7:1–15
Panhwar QA, Othman R, Rahman ZA, Moen S, Ismail MR (2012) Isolation and characterization of phosphate-solubilizing bacteria from aerobic rice. AJB 11(11):2711–2719
Patnaik R (2018) Impact of industrialization on environment and sustainable solutions–reflections from a south Indian region. Earth Env Sci 120:1–9
Pereg L, Luz E, Bashan Y (2016) Assessment of affinity and specificity of Azospirillum for plants. Plant Soil 399:389–414
Pereira C, Camougrand N, Manon S, Sousa MJ, Corte-Real M (2007) ADP/ATP carrier is required for mitochondrial outer membrane permeabilization and cytochrome C release in yeast apoptosis. Mol Microbiol 66(3):571–581
Pereira H, Leadley P, Proenca V, Alkemade R, Walpole M (2010) Scenarios for global biodiversity in the 21st century. Science 330:1496–1501
Perez-Montano F, Alias-villegas C, Bellogin RA, Cerro P, Espuny MR, JimenezGuerrero I, Lopez FJ (2014) Plant growth promotion of cereal and leguminous agricultural important plants: from microorganism capacities to crop production. Microbiol Res 169(5):325–336
Phillips LA, Greer CW, Germida JJ (2006) Culture based and culture independent assessment of the impact of mixed and single plant treatments on rhizosphere microbial communities in hydrocarbon contaminated flare pit soil. Soil Biol Biochem 38:2823–2833
Pieterse CM, Leaon-Reyes A, Van der Ent S, Van Wees SC (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5:308–316
Płociniczak MP, Płaza GA, Seget ZP, Cameotra SS (2011) Environmental applications of biosurfactants: recent advances. Int J Mol Sci 12:633–654
Pujic P, Normand P (2009) The root symbiosis between Frankia bacterium actinorhizal plants. Biofutur 298:26–29
Raaijmakers J, Mazzola M (2012) Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu Rev Phytopathol 50:403–424
Ray D, Mueller ND, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS One 8(6):e66428
Raza W, Ling N, Yang L, Huang Q, Shen Q (2016) Response of tomato wilt pathogen Ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain Bacillus amyloliquefaciens SQR-9. Sci Rep 6:24856
Razaq M, Zhang P, Shen H (2017) Influence of nitrogen and phosphorus on the growth and root morphology of Acer mono. PLoS One 12(2):1–13
Reddy GC, Goyal RK, Puranik S, Waghmar V, Vikram KV, Sruthy KS (2020) Biofertilizers towards sustainable agricultural development. In: Varma A, Tripathi S, Prasad R (eds) Plant microbe symbiosis. Springer, Cham, pp 115–128pp
Redmile-Gordon M, Gregory AS, White RP, Watts CW (2020) Soil organic carbon, extracellular polymeric substances (EPS) and soil structural stability as effected by previous and current land-use. Geoderma 363:1–10
Richard PO, Adekanmbi AO, Ogunjobi AA (2018) Screening of bacteria isolated from the rhizosphere of maize plant (Zea mays L.) for ammonia production and nitrogen fixation. Afr J Microbiol Res 12(34):829–834
Sagervanshi A, kumara P, Nagee A (2012) Isolation and characterization of phosphate solubilizing bacteria from agricultural soil. Int J Life Sci Pharm Res 2:256–266
Sah S, Sing N, Sing R (2017) Iron acquisition in maize (Zea mays L.) using Pseudomonas siderophore. Biotechnol 7(2):121–128
Shafi J, Tian H, Mingshan J (2017) Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol Equip 31(3):446–459
Shaharoona AA, Muhammad AB, Azeem KA (2006) Performance of Pseudomonas spp. containing ACC-deaminase for improving growth and yield of maize (Zea mays L.) in the presence of nitrogenous fertilizer. Soil Biol Biochem 38:2971–2975
Sharma A, John BN (2003) Growth promoting influence of siderophore-producing Pseudomonas strains GRP3A and PRS9 in maize (Zea mays L.) under iron limiting conditions. Microbiol Res 158(3):243–248
Sharma A, Talukdar G (1987) Effects of metals on chromosomes of higher organisms. Environ Mutagen 9:191–226
Sharma M, Manoharlal R, Shukla S, Puri N, Prasad T, Ambudkar SV, Prasad R (2009) Curcumin modulates efflux mediated by yeast ABC multidrug transporters and is synergistic with antifungals. Antimicrob Agents Chemother 53(8):3256–3265
Shilev S (2013) Soil rhizobacteria regulating the uptake of nutrients and undesirable elements by plants. In: Arora NK (ed) Plant microbe symbiosis: fundamentals and advances. Springer, New Delhi, pp 147–150
Shimoda Y, Nishigaya Y, Yamaya-Ito H, Inagaki N, Umehara Y, Hirakawa H, Sato S, Yamazaki T, Hayashi M (2020) The rhizobial autotransporter determines the symbiotic nitrogen fixation activity of Lotus japonicus in a host-specific manner. PNAS 117(3):1806–1815
SkZ A, Vardharajula S, Vurukonda SKP (2018) Transcriptomic profiling of maize (Zea mays L.) seedlings in response to Pseudomonas putida strain FBKV2 inoculation under drought stress. Ann Microbiol 68:331–349
Souza RD, Ambrosini A, Passaglia LMP (2015) Plant growth-promoting bacteria as inoculants in agricultural soils. Genet Mol Biol 38:401–419
Steinauer K, Chatzinotas A, Eisenhauer N (2016) Root exudate cocktails: the link between plant diversity and soil microorganisms. Ecol Evol 6(20):7387–7396
Tao GC, Tian SJ, Cai MY (2008) Phosphate-solubilizing and mineralizing abilities of bacteria isolated from soils. Pedosphere 18:515–523
Tariq A, Mussarat S, Adnan M, AbdElsalam NM, Ullah R, khan AL (2014) Ethnoveterinary study of medicinal plants in a tribal society of Sulaiman range. Sci World J. 2014:1-11
Thomine S, Languar V (2011) Iron transport and signaling in plants. Annu Rev Plant Biol 54:99–131
Tomer S, Suyal DC, Goel R (2016) Biofertilizers: a timely approach for sustainable agriculture. In: Choudhary DK, Varma A, Tuteja N (eds) Plant-microbe interaction: an approach to sustainable agriculture. Pantnagar: Springer Nature, Singapore Pte Ltd, pp 375–395
Trivedi P, Pandey A (2008) Recovery of plant growth-promoting rhizobacteria from sodium alginate beads after 3 years following storage at 4 C. J Indl Microbiol Biotechnol 35(3):205–209
Van Loon LC (2007) Plant responses to plant growth-promoting rhizobacteria. Eur J Plant Pathol 119:243–254
Vessey JK (2003) Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255:571–586
Vikram A, Hamzehzarghani H (2008) Effect of phosphate solubilizing bacteria on nodulation and growth parameters of green gram (Vigna radiate L. Wilczek). Res J Microbiol 3(2):62–72
Vitousek PM, Cassman K, Cleveland C (2002) Towards an ecological understanding of biological nitrogen fixation. Biogeochemistry 87:1–45
Viveros OM, Jorquera MA, Crowley DE Gajardo G, Mora ML (2010) Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J Soil Sci Plant Nutr 10(3):293–319
Wahyudi AT, Priyanto JA, Fijrina HN, Mariastuti HD, Nawangsih AA (2019) Streptomyces spp. from rhizosphere of maize with potential as plant growth promoter. Biodiversitas 20(9):2547–2553
Weller DM, Landa BB, Mavrodi OV (2007) Role of 2,4-diacetylphloroglucinol-producing fluorescent. Pseudomonas spp In the defense of plant root. Plant Biol 9:4–20
Wenzel WW (2009) Rhizosphere processes and management in plant assisted bioremediation (phytoremediation) of soils. Plant Soil 321:385–408
Yang Y, Wang N, Guo X, Zhang Y, Ye B (2017) Comparative analysis of bacterial community structure in the rhizosphere of maize by high-throughput pyrosequencing. PLoS One 12(5):1–11
Yazdani M, Bahmanyar M, Pirdashti H (2009) Effect of phosphate solubilization microorganisms (PSM) and plant growth promoting rhizobacteria (PGPR) on yield and yield components of corn (Zea mays L.). WASET 37:90–92
Acknowledgements
OOB would like to thank the National Research Foundation, South Africa, for the grants (UID123634 and UID132595) that have supported research in her laboratory.
Funding
This study was funded by South Africa’s National Research Foundation grants (UID123634 and UID132595).
Author information
Authors and Affiliations
Contributions
EEI conceived and drafted the manuscript. OOB critically revised the various manuscript drafts and helped in the structuring and editing of the manuscript. Both authors approved its final version for publication.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any study with human participants or animals performed by any authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Imade, E.E., Babalola, O.O. Biotechnological utilization: the role of Zea mays rhizospheric bacteria in ecosystem sustainability. Appl Microbiol Biotechnol 105, 4487–4500 (2021). https://doi.org/10.1007/s00253-021-11351-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11351-6