Abstract
The composition of bacteria in the gastrointestinal tract of piglets is easily affected by environmental changes, particularly during the weaning period. Compound strains of Lactobacillus reuteri and Lactobacillus salivarius were supplemented to piglets during pre- and post-weaning to determine their effects in improving the growth performance and ameliorating the diarrhea rate and stress caused by antioxidation in piglets. A larger number of L. reuteri and L. salivarius colonized the distal segment of the ileum and the total numbers of Lactobacillus spp. and Bifidobacteria were higher in the ileal mucous membrane and cecal lumen with probiotics supplementation. The numbers of antioxidants and immune molecules increased, levels of cortisol and endotoxin reduced, and growth hormone and insulin-like growth factor 1 improved in the plasma following compound bacteria (CL) supplementation. Spearman’s and KEGG analysis of the bacterial operational taxonomic unit and antioxidative and immune indices and metabolic genes indicated that the body growth modulation by CL supplementation could be attributed to optimization of the intestinal bacterial composition; functional strains of L. delbrueckii, L. salivarius, L. formicilis, L. reuteri, and L. mucosae were positively correlated with body antioxidation and immunity derived by CL supplementation. Strains of L. agilis and L. pontis were diverse and negatively correlated with body antioxidation and immunity. Collectively, these results suggest that supplementation with CL could reduce stress and improve the growth performance of piglets during weaning by optimizing the intestinal bacterial composition.
Key points
• The colonization of L. reuteri and L. salivarius in ileal mucous membrane optimize bacterial composition of GIT, mainly some functional strains of Lactobacillus, L. delbrueckii, L. salivarius, L. formicilis, L. reuteri, and L. mucosae.
• The optimized bacterial composition of piglets in both ileal mucous membrane and cecal content improves body growth hormone level, immunity, and antioxidation, which is helpful to defend the stress. These benefits induce to increased growth performance of animal model piglets during weaning.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The microbiota of the mammalian gastrointestinal tract (GIT) plays evolutionarily conserved roles in the metabolism, immunity, development, and behavior of the host, and the microbiota composition is significantly altered during body growth and by changes in the diet and environment (Wampach et al. 2017; Clemente et al. 2012; Mitreva et al. 2012). The societal pressure to decline the number of nonhuman primates such as dogs and monkeys employed in biomedical research led to an increase in the use of pigs. Feeding pigs and collecting samples from them are easier when compared to other farm animals (El-Kadi et al. 2018).
The GIT microbiota of piglets is easily altered during the weaning period when the feed is altered or when the piglets are isolated from sows or transferred between groups, all of which cause physiological and psychological stress (Li et al. 2018; Wang et al. 2018). Hence, pathogens can infect piglets more easily during weaning, leading to high morbidity and low growth performance (Bloomfield et al. 2019). In addition, low levels of digestive enzymes and hydrochloric acid in the gastric juice secretions of piglets lead to insufficient feed digestion (Holtan et al. 2019). Together, these factors impede the growth performance of piglets during the pre- and post-weaning and group transfer periods. Some genera or strains of bacteria are conserved across animals of different age groups and species as core members of the GIT (Nicholson et al. 2012; Wang et al. 2019; Hou et al. 2020). Lactobacillus is a core genus in growing pigs and some other mammals (Yang et al. 2018). Some strains of Lactobacillus compose the native bacteria of the mucous membrane of the GIT; the native bacteria are known to play important roles in maintaining the balance of the intestinal milieu, which influences the bacterial colonization at different mucosal sites in the GIT and the secretion of organic acids, digestive enzymes, and bioactive peptides (Yang et al. 2018; Vemuri et al. 2019). Additionally, some strains of lactobacilli, such as Lactobacillus reuteri, Lactobacillus fermentum, Lactobacillus delbrueckii, and Lactobacillus rhamnosus, can influence the composition of the gut microbiota, which in turn modulates hormonal secretion through the brain-gut axis to reduce body stress and anxiety (Indrio et al. 2014; Warda et al. 2019). Therefore, dietary supplementation of weaning piglets with Lactobacillus spp. could optimize the composition of the gut microbiota by increasing the number of lactic acid bacteria and promoting diversity, thereby improving digestive function and alleviating stress. Lactobacillus reuteri and Lactobacillus salivarius are two probiotic bacterial strains that reside in the mucous membrane of the mammalian alimentary tract and are known to exert health benefits, such as optimization of the GIT microbiota composition, inhibition of pathogens, enhancement of intestinal barrier function, improvement of host immunity, and improvement of host digestive function via secretion of organic acids, peptides, and digestive enzymes (Yang et al. 2018; Yang et al. 2020a). Lactobacilli have strain-specific characteristics with diverse effects and interactions in the host (Wang et al. 2017). Therefore, while screening for probiotic GIT bacteria for use as supplements in animals, it is important to note that homogeneous origin of the isolated strains is crucial for host colonization.
Two strains of swine origin Lactobacillus sp., L. reuteri KT260178, and L. salivarius MH 517354, were isolated from the distal segment of ileal content of healthy local Wei finishing pigs from the Anhui province of China and stored in the China Center for Type Culture Collection. In our previous study, we found that L. reuteri KT260178 is a candidate probiotic bacterium based on its effects on the growth performance of neonatal piglets upon colonization of the mucous membrane of the distal jejunum and ileum (Yang et al. 2020a). The two strains of L. reuteri KT260178 and L. salivarius MH 517354 strongly affected the secretion of organic acids and digestive enzymes and inhibited pathogenic bacteria in vitro. Considering physiological characteristics of L. reuteri KT260178 and L. salivarius MH 517354, these strains would be more useful to the weaning piglets in combination than as single supplementations to promote the organic acid secretion and optimize the bacterial composition. Many studies have confirmed that the beneficial effects of probiotics on health, immunity, pathogen defense, and other biological functions are much stronger when using multiple strains compared with using single-strain administration. This finding could be attributed to the symbiotic relationships and synergism among strains (Chapman et al. 2011; Yang et al. 2017; Yang et al. 2020b).
In the study, we combined two strains of swine origin, L. reuteri KT260178 and L. salivarius MH 517354, and administered these to piglets between the pre- and post-weaning period to evaluate their effects on the intestinal bacterial composition and digestive function and their effectiveness in ameliorating the stress, morbidity, and decreased growth performance, which set an example to human beings. The structure and anatomy of the digestive tract neonatal pigs are similar to those of infant; hence, they are commonly used as an animal model in biochemical research (Davis et al. 1996). Supplementation with compound homologous probiotic bacteria could be extended to human infants to improve health.
Materials and methods
Animal studies
The experimental protocols in this study, including those related to animal husbandry and slaughter, were approved by the Institution of Animal Science and Welfare of Anhui Province (no. IASWAP2017056937). The experimental guidelines and treatment, housing, and husbandry conditions conformed to the Institutional Animal Care and Use Committee of China (National Research Council (US) 2011). A total of 45 sows (Landrace breed) with the same parity and body weight were randomly allocated to three groups (n = 10 each). The sows were artificially inseminated by six Duroc boars of similar body weight and age. The expected date of piglet birth of the selected sows was the same. Thirty sows that gave birth on the same day were selected. The number of neonatal piglets was adjusted to 12 for each sow (initial average birth weight 1.37 kg). When the neonatal piglets reached the age of 14 days, all 30 sows with 300 piglets (10 piglets with each sow, body weight nearly 3.0 kg, piglets with highest and lowest body weight were eliminated) were allocated to three groups: control, compound L. reuteri KT260178 and L. salivarius (CL), and aureomycin (antibiotic) supplementation groups. The experiment was carried out for 42 days. The feed duration was 28 days, from May 15 to June 12, 2017. Sows and piglets were individually housed in farrowing pens (2.2 × 1.8 m) with crates and slatted floors. Heating pads were additionally installed for the piglets. The farrowing room temperature was maintained at 20 °C. All 300 piglets were separated from their sows at 21 days, and groups were transferred to the nursery house at day 28 after birth. Piglets were administered iron supplements by intramuscular injection of iron dextran at days 3 and 10 after birth. All piglets had free access to water and creep feed without any probiotics, antibiotics, or other medicines added during the experiment. A standard immune procedure was implemented throughout the trial.
Diet for each group
Lactobacilli reuteri KT260178 and L. salivarius MH 517354 were isolated from the ileum of healthy local Wei pigs in the Anhui province of China by our research group. L. reuteri was stored in the China General Microbiological Culture Collection Center (CGMCC), Beijing, China; the strain number was 17718. L. salivarius was stored in the China Center for Type Culture Collection (CCTCC), Wuhan University, Wuhan, China, and the strain number was M2015660. The two strains of Lactobacilli sp. were mixed and grown in de Man, Rogosa, and Sharpe medium after inoculation at 1%:1%. The bacterial cells were harvested after approximately 18 h of fermentation. The second solid fermentation was used to increase the live number of bacterial cells, in which a mixture of corn and soybean meal powder was employed to incorporate the suspended fermented liquid containing the live bacterial cells; the ratio of mass to volume was 1:0.6. To obtain a solid direct microbial additive, the mixture was dried at a constant temperature of 45 °C; the numbers of live L. reuteri and L. salivarius were 6.0 × 108 and 3.0 × 108 colony-forming units per gram (CFU/g), respectively. Piglets in the control group were fed the basal diet. The compound probiotic L. salivarius and L. reuteri were supplemented in the basal diet to piglets between suckling and nursery period in the trial. The diets fed during the suckling and nursery stage were different. The composition and nutrient analysis results for the basal diet of suckling piglets and nursery pig are shown in supplementary material (Table S1) (NRC (National Research Council) 1998). In the CL group, 0.5 kg of dried feed additive CL was added to a 99-kg basic mass feed. A blender was used to mix the additives uniformly for 10 min to attain a homogeneous mixture. After inoculation in the CL group, live L. reuteri and L. salivarius reached 3.0 × 106 and 1.5 × 106 CFU/g, respectively. The diet with aureomycin supplementation was prepared in the same manner as for the CL groups, with 150 μg/kg aureomycin in the feed. All food fed to the piglets was prepared once every 7 days. The compositions of the basic diets are shown in the supplementary material. The sows were individually fed and had free access to water throughout the experimental period. The diets for lactating sows were identical and formulated according to the National Research Council requirements for gestating and lactating sows (Table S1) (NRC (National Research Council) 1998); this diet contained no probiotics, antibiotics, or other medicine. The way of feeding for the diet in all three groups was the same. Piglets were fed three times a day during the trial, 6:00 am, 11:30 am, and 5:00 pm. Piglets in the sucking period were allowed ad libitum to feed and water in the whole experimental duration.
Growth performance and sample collection
Piglets in every replicate from each treatment group were weighed both on the transfer days (experimental day 14, ED 14) and at the end of the experiment (ED 28). Average daily gain (ADG) was calculated according to the formula: ADG = body increase (g)/number of days (ED 14 or 28). The number of piglets with diarrhea in each group was recorded to calculate the diarrhea rate according to the following formula: diarrhea rate = sum of number in each group for 28 days/(20 × 11 × 28) × 100%. At the end of the experiment, one piglet from every replicate was selected, and 10 mg/kg ketamine hydrochloride was intramuscularly injected. Blood samples were collected from sacrificed piglets by bleeding of the carotid artery. Tissues of the distal ileum and cecum were removed under aseptic conditions, stored in sterile plastic tubes in boxes packed with ice, and immediately transported to our laboratory for microbiota quantification by the plate-counting method.
Plate-counting of microbiota assay
The samples of the distal ileum were cut open and washed with sterilized physiological saline (pH 7.0). Samples of ileal mucous membrane were scratched from the ileum using a slide. The ileal mucous membrane and cecal lumen contents (0.4 g each) were prepared and 10-fold dilutions were prepared with sterilized saline. The bacterial composition of the ileal mucous membrane and cecal lumen contents in all groups was determined using the plate method. Eosin methylene blue agar was used for total Escherichia coli (which mainly resides in cecal lumen or contents, abundant number or transfer of E. coli into other segment of intestine caused illness), a Salmonella-Shigella agar plate was used for Salmonella (pathogenic bacteria, resides in intestine and content), and MRS agar was used for total Lactobacillus (Yang et al. 2020b; Mountzouris et al. 2007). The assays were repeated three times.
Quantitative real-time PCR assay
L. reuteri KT260178 and L. salivarius MH 517354 were fermented in MRS medium respectively. Colonies were counted with the plate method under a microscope to obtain samples of 1 × 104, 105, and 106. Then, a tenfold dilution series (1 × 104, 105, and 106) of L. reuteri and L. salivarius was prepared. Total RNA in each dilution was extracted. The standard curve of serial dilution was generated based on mean cycle threshold values of quantitative real-time polymerase chain reaction. The method was carried out according to our previous study (Yang et al. 2020a). The primer sequences were as follows: L. reuteri KT260178—forward 5′ AACTCCCTGAAATGACAGTGAAG 3′, reverse 5′ TGACTGAACACTAACCCGAACCT 3′. L. salivarius MH 517354: forward 5′ GACTCACTGACATGACAGTGACG 3′, reverse 5′ ACGCTGAGCACTAACGCGACAC 3′. Samples (0.2 g) of ileal mucous membrane were prepared to extract total RNA to evaluate colonization by L. reuteri KT260178 and L. salivarius MH 517354 cells according to our previously reported method (Yang et al. 2020a). Reverse transcription was performed with a GoScript Reverse System (Invitrogen, Carlsbad, CA, USA). First-strand cDNA was synthesized by incubating a reaction mixture containing 11 μL RNA and 1 μL RNase-free dH2O at 70 °C for 3 min, followed by 0 °C for 5 min. A dNTP mixture (1 μL; 10 mmol/L), 4 μL GoScript 5× reaction buffer, 1 μL GoScript reverse transcriptase, 1.5 μL Mg2+ (25 mM), and 0.5 μL RNase inhibitor were combined in a total volume of 20 μL and incubated at a 37 °C in a water bath. Primers were designed according to the 16S ribosomal RNA (16S rRNA) of L. reuteri KT260178 and L. salivarius MH 517354 submitted to NCBI. Amplification was performed in a 20-μL mixture containing 10 μL of 2 × qPCR SYBR Premix Ex-Taq, 2 μL template cDNA, 0.5 μL each primer (10 μmol/L), and 7 μL PCR-grade water. The cycling protocol was as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s, and one cycle for melting curve analysis consisting of 95 °C for 60 s, 65 °C for 60 s, and 95 °C for 1 s. The amplification curve was generated based on the dilution of the standard curve of the L. reuteri KT260178 and L. salivarius MH 517354 cultures.
DNA extraction, PCR amplification, and sequencing
Samples (0.25 g) of the ileal mucous membrane and cecal lumen were prepared. Microbial DNA was extracted from these samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s protocol. Eight samples of each group were chosen, with two that remained as standby instead of the unqualified DNA. The final DNA concentration and purification were determined with a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Waltham, MA, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) with a thermocycler PCR system (GeneAmp 9700, Applied Biosystems, Foster City, CA, USA). PCR was conducted as follows: 3 min of denaturation at 95 °C, 27 cycles of 30 s at 95 °C, 30 s for annealing at 55 °C, and 45 s for elongation at 72 °C, and a final extension at 72 °C for 10 min. PCR was performed in triplicate 20-μL mixtures containing 4 μL of 5 ×FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu polymerase, and 10 ng of template DNA. The resulting PCR products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor™-ST (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, CA, USA) according to standard protocols. The Illumina sequencing raw data has been deposited into Sequence Read Archive database (SRP) of national center for biotechnology information (NCBI) of the USA, and the number is SUB6850760. The BioSample accession is SAMN13870560.
16S rRNA amplicon data processing and statistics
Diversity metrics were calculated using the core-diversity plugin within QIIME2. Feature level alpha diversity indices and operational taxonomic units (OTUs) were used to estimate the microbial diversity within an individual sample. Beta diversity distance measurements were performed with unweighted UniFrac to investigate the structural variation in the microbial communities across samples and then visualized via principal coordinate analysis (PCoA). Co-occurrence analysis was performed by calculating Spearman’s rank correlations between predominant Lactobacillus and the network plot. Additionally, the potential KEGG Ortholog (KO) functional profiles of microbial communities were predicted with PICRUSt.
Determining of plasma antioxidants, immune-related molecules, and hormones
Blood samples were collected in 5.0-mL sterile heparinized tubes and then centrifuged at 3000×g for 10 min to collect the plasma for immune assays. Plasma levels of antioxidant factors, including malondialdehyde (MDA), total antioxidant capacity (T-AOC), superoxide dismutase (SOD), and glutathione peroxidase (GPX), and immune-related molecules, interferon-α (INF-α) and interferon-β (IFN-β), were measured with enzyme-linked immunosorbent assay (ELISA). All ELISA kits of MDA, T-AOC, SOD, GPX, INF-α, and IFN-β were purchased from Nanjing Jiancheng Bioengineering Institute, Nanjing, China. Plasma hormonal levels of cortisol, endotoxin, growth hormone, and insulin-like growth factor 1 were evaluated by ELISA (kits were purchased from Nanjing Jiancheng Bioengineering Institute).
Statistical analysis
Statistical analyses of the data were performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean values with their standard errors. Differences between groups were compared by analysis of variance. Differences between means were assessed by Tukey’s honest significant difference test for post hoc multiple comparisons. A t test was used to identify taxonomic features of microbiological DNA sequence. A P value < 0.05 was considered statistically significant. Spearman’s correlation analysis was used to determine relationships between parameters. STAMP software was applied to detect differentially abundant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways among groups with false discovery rate correction. A P value (corrected) < 0.05 was considered to indicate statistical significance.
Results
Body growth
All piglets were weighed in both groups on ED 14 and ED 28. The average body weights of piglets supplemented with CL were 0.52 kg higher than those of controls on ED 14 and 0.93 kg higher than those of controls (P < 0.01) on ED 28, revealing no significant differences from those of the antibiotic group (P > 0.05; Fig. 1(A)). Additionally, the results of ADG were consistent with those for body weight, and CL-supplemented piglets showed a higher average daily gain of body weight compared to those in the control group both at ED 14 (P < 0.05; Fig. 1(B)) and ED 28 (P < 0.01; Fig. 1(B)). he diarrhea rate in the groups was CL group < antibiotic group < control group (P < 0.01; Fig. 1(C)) at both ED 14 and ED 28.
Growth performance of three groups. The different superscript capital letters in the same classification of column mean significant difference at 0.01 levels (P < 0.01); the different superscript lower case letters mean significant difference at 0.05 levels (P < 0.05). (A) Body weight of piglets in three groups. (B) ADG of piglets in three groups. (C) Diarrhea rate of piglets in three groups
Colonization of Lactobacillus sp.
The probiotic Lactobacillus sp. can colonize the mucous membrane of the ileum, as demonstrated by the qPCR assay. A larger number of L. reuteri and L. salivarius was detected in the ileal mucous membrane samples both on ED 14 and on 28 (Fig. 2a, b) in probiotics supplementary group than those of control and antibiotic groups (P < 0.01). The number of L. reuteri and L. salivarius in antibiotic group showed decreased tendency compared with control, without significance (P > 0.05). The total number of Lactobacillus sp. and Bifidobacterium in the ileal mucous membrane and cecal content was the highest in all groups (P < 0.01) (Table 1). In contrast, the number of Lactobacilli sp. and Bifidobacterium in the antibiotic-supplemented group was lower than that in other groups, and the other enumerated bacteria, such as Escherichia coli (conditionally pathogenic) and Salmonella (pathogenic), were significantly reduced compared to that in the control. Consistent with this result, Salmonella and E. coli counts were minimally reduced in the CL-supplemented group.
Overall bacterial community structure
The bacterial composition in the ileal mucous membrane and cecal content of piglets in the antibiotic group were significantly influenced by aureomycin supplementation. We compared the CL and control groups on ED 14 and ED 28 to determine the effects of supplementation with the two Lactobacillus spp. and the influence of antioxidation on the composition of the microbiota. The composition of microbiota of the CL and control groups was analyzed by sequencing of the bacterial 16S rRNA V3 and V4 regions. High-throughput pyrosequencing of the samples (n = 6) produced a total of 1,365,826 raw reads. After removing low-quality sequences, 468,232 clean tags were identified as a total of 484 OTUs in six samples. This sequencing depth nearly reflected the total microbial species richness, and most OTUs were present at low abundance (Fig. 3a). In total, 365 and 387 OTUs were obtained from the ileal mucous membrane and 392 and 413 OTUs from the cecal content in the control and CL groups, respectively. The microbiota in the ileal mucous membrane of piglets was mostly comprised by three phyla, i.e., Bacteroidetes, Firmicutes, and Proteobacteria, which collectively accounted for 98.92% and 98.57% of the bacterial abundance on ED 14 and ED 28 (Fig. 3b). The colonic microbiota of piglets on ED 14 and ED 28 was also chiefly constituted by Bacteroidetes, Firmicutes, and Proteobacteria, which accounted for more than 90% of the total microbiological abundance. Actinobacteria, Fusobacteria, Verrucomicrobia, and Saccharibacteria constituted the remaining 10% abundance of the microbiota (Fig. 3b).
The OTU of bacterial community of three groups in phylum level. The different superscript capital letters in the same color of column in the same experimental duration mean significant difference at 0.01 levels (P < 0.01), the different superscript lower case letters mean significant difference at 0.05 levels (P < 0.05). a The bacterial community of three groups in phylum level. b Differences in bacterial community of three groups in phylum level
Differences in diversities of bacteria in ileum and cecum
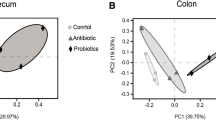
To determine the phylogenetic variations caused by CL supplementation and age of piglets, we measured unweighted UniFrac distances. Analysis of similarities of the UniFrac-based principal coordinate analysis revealed significant differences in the bacterial community structure among different groups (illustrated in the PCoA plot in Fig. 4). In the PCoA of bacterial OTUs, the samples of cecal content in the CL group both in ED 14 and in ED 28 clustered together more than those in the control group. The situation in the ileal mucous membrane was different than that in the cecum. The samples of bacterial OTUs in the two groups clustered adjacently. However, the cluster of samples in the CL groups at two time points ED 14 and ED 28 was closer than that in the control group. The longer distance among spots of samples indicated that their bacterial communities were more different and unstable.
To further elucidate the dynamics and differences between the gut bacterial communities of the CL supplementation and control groups, the OTUs at the phylum and genus levels were characterized (Fig. 3). At the phylum level, CL supplementation increased Bacteroidetes population in the ileal mucous membrane on ED 14 (CL group 35.14% and control group 30.05%) (Fig. 3a) (P < 0.01). Similarly, the relative abundances of Proteobacteria (CL group 10.45% and control group 7.46%) and Firmicutes in the CL group were higher than those in the control group on ED 14 (CL group 53.32% and control group 61.08%) (Fig. 3b). The same results were observed for the cecal content. The relative abundances of Bacteroidetes and Proteobacteria in the CL group were significantly decreased compared to those in the control, and the abundance of Firmicutes was improved (P < 0.01). On ED 28, the relative abundances of Bacteroidetes (22.06%) and Proteobacteria (3.57%) in the ileal mucous membrane were reduced compared with those on ED 14 in both groups (19.06% and 2.76%, respectively). The relative abundances of Bacteroidetes and Proteobacteria in CL were significantly higher than those in the control. In contrast, the abundances of Firmicutes in the two groups on ED 28 were increased compared to those on ED 14. The results of microbiological composition of the cecal content were correlated with those of the ileal mucous membrane. The main phyla were Firmicutes, Bacteroidetes, and Proteobacteria, accounting for 67.52%, 21.82%, and 4.78% of the relative abundances in the CL group and 71.41%, 19.32%, and 3.28% in the control group on ED 14, respectively. The results on ED 28 showed that Firmicutes, Bacteroidetes, and Proteobacteria comprised 72.59%, 16.29%, and 3.82%, respectively, of the CL group microbiota and 77.93%, 12.29%, and 2.63%, respectively, of the control group microbiota (Fig. 5a). CL-supplemented piglets both on ED 14 and on ED 28 showed higher microbial diversity than control group piglets. Actinobacteria, Fusobacteria, Verrucomicrobia, and Saccharibacteria were more abundant in the CL group, showing high microbial proportions (Fig. 5b).
The different superscript capital letters in the same color of column in the same experimental duration mean significant difference at 0.01 levels (P < 0.01); the different superscript lower case letters mean significant difference at 0.05 levels (P < 0.05). a The bacterial community of three groups in genus level. b Differences in bacterial community of three groups in genus level
We further compared the microbial community at the genus level. The main genera were Lactobacillus, Escherichia-Shigella, Peptoclostridium, Acinetobacter, Ruminococcaceae UCG-014, Streptococcus, Clostridium sensu stricto 1, Bacteroides, Roseburia, and Helicobacter in the ileum and cecum, accounting for 99% of the relative abundance (Fig. 5a). Piglets in the CL group showed higher relative abundances for the genera Lactobacillus, Acinetobacter, Ruminococcaceae UCG-014, Bacteroides, and Helicobacter. However, lower relative abundances of Escherichia-Shigella, Streptococcus, Peptoclostridium, Clostridium sensu stricto 1, Fusobacterium, Roseburia, and Veillonella were observed. Lactobacillus and Escherichia-Shigella were opposite in their ratio. The proportions of Lactobacillus in the ileal mucous membrane at both experimental time points were 20.14% and 16.14%, showing higher values than in the control (12.70% and 11.23%; P < 0.01) (Fig. 5b). The proportion of Escherichia-Shigella decreased by 2.78% and 4.61% compared to in the control (P < 0.01). The relative abundances of Escherichia-Shigella in the cecal content on ED 14 and ED 28 in the CL group were 0.12% and 2.74% and in the control group were 0.31% and 12.63%, respectively (Fig. 5b). The relative abundance of Lactobacillus in the cecal content of piglets with CL supplementation was significantly increased at both time points.
Antioxidant and immune-related molecule levels
To investigate the protective roles of CL on oxidative molecules caused by stress, MDA, SOD, T-AOC, and GPX in the plasma were measured on ED 14 and ED 28 (Table 2). With antibiotic and CL supplementation, the concentration of MDA in the plasma was decreased significantly compared to that in the control at both time points (P < 0.01). The MDA content in the CL group was the lowest, showing values 4.16 and 6.13 μmol/L lower than that in the control on ED 14 and ED 28, respectively. SOD, CAT, and GPX activities in CL-supplemented piglets were significantly higher than those in the control (P < 0.05); there were no differences in SOD and CAT activities between the antibiotic and control groups (P > 0.05). The plasma levels of immune-related molecules are shown in Table 3. IFN-α levels in the CL group were higher than those in the aureomycin supplementation group (P < 0.01) and higher than those in control piglets (P < 0.01). Similar results were obtained for IFN-β, with the highest level detected in CL-supplemented piglets (P < 0.01), showing the highest value among the three groups.
Hormonal levels
The plasma hormonal levels of cortisol, endotoxin, growth hormone, and insulin-like growth factor 1 are shown in Table 3. At ED 14 and ED 28, the plasma level of cortisol in piglets with CL supplementation was significantly lower than that in the two groups without Lactobacilli sp. supplementation (P < 0.05). There were no differences between the control and antibiotic groups (P > 0.05). The level of plasma endotoxin in the CL group was very significantly reduced compared to in the antibiotic group (P < 0.01), which was lower than that in the control group (P < 0.05) at both time points. The levels of growth hormone and insulin-like growth factor 1 in the CL-supplemented group were significantly increased compared to those in the control and antibiotic groups at both time points, which were 1.13 and 59.92 ng/mL higher than in the control group, respectively, on ED 28 (P < 0.01). There were no differences between the control and antibiotic groups (P > 0.05).
Comparison of metabolic pathway abundance
To determine the relationship between bacteria colonizing the ileal mucous membrane and antioxidant and immune-related molecule levels, Spearman’s correlation analysis was performed. The antioxidant and bacterial abundance in ileal mucous membrane samples on ED 28 is shown in Fig. 6. The strains of L. delbrueckii, L. salivarius, L. formicilis, L. reuteri, and L. mucosae were positively correlated with plasma antioxidation and SOD, GPX, and CAT levels and negatively correlated with the MDA concentration. However, the strains of L. agilis and L. pontis were diverse and negatively correlated with the levels of SOD, GPX, and CAT. The strains of L. delbrueckii, L. salivarius, and L. formicilis were positively correlated with IFN-β levels. However, L. agilis was negatively correlated with INF-α and IFN-β levels. Additionally, L. pontis was negatively correlated with INF-α levels. L. salivarius was positively correlated with both the antioxidative and immune indices.
To further study which metabolic genes were altered after treatment with probiotics, KEGG pathways among the samples between the CL and control groups on ED 28 were analyzed. In terms of metabolic pathways, genes regulating the metabolism of cofactors and vitamins (P < 0.01), other amino acids (P < 0.01), lipid (P < 0.01), terpenoids and polyketides (P < 0.05), transport, and catabolism (P < 0.01) were more abundant in the CL-supplemented group than in the control group (Fig. 7a, b). However, the genes related to membrane transport (P < 0.01), replication, and repair (P < 0.01) were less abundant in the CL group than those in the control group.
KEGG pathways among samples between CL and control groups on ED 28. STAMP software was applied to detect the differentially abundant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways among groups with false discovery rate correction. *mean significant difference at 0.05 levels (P < 0.05), **mean significant difference at 0.01 levels (P < 0.01). a KEGG pathways among samples between CL and control groups on ED 28. b Differences in gene expression of three groups
Discussion
Effect of CL administration on body growth of piglets
The supplemental doses of probiotics on piglets were 106–107 CFU per gram of feed, which were confirmed by some researches (Mountzouris et al. 2010; Lei et al. 2013; Yang et al. 2020a, 2020b). Based on our previous study and documented references, we carried out this experiment on piglets in the dose of 4.5 × 106 CFU/g in combined use of L. salivarius and L. reuteri during the pre- and post-weaning and group transfer periods to nursery. The results of growth performance (data of final body weight and average daily gain) indicated that piglets with CL supplemented are better than those of control. There were no significant differences between the antibiotic group and the CL group. Probiotic Lactobacillus in multi-strain combining strains capable of reducing antigen-load, improving the intestinal barrier, and eliciting a regulated immune response could potentially have stronger overall effects than single-strain on gastrointestinal barrier function and homeostasis and restore the ecological balance, during antibiotic administration and other unfriendly conditions (Branning et al. 2009). In use of improved and stress and growth performance of body, different studies have confirmed positive effects on health when multi-strain probiotics are used, due to the symbiosis among strains (Ouwehand et al. 2018).
It was reported that residence time of Lactobacillus sp. in the intestine was about 3 days, then the number of bacteria reduced in no supplementation, and reached normal levels in 3–4 days (Lee et al. 2004), which indicated that the rate of Lactobacillus sp. inclusive in feed is 3 days after continuously feeding for 7 days for colonizing probiotic bacteria in intestine. The inclusive rate of probiotics was also determined by the physiological factors of animal in different growing periods. In the special period, such as stress and early growth period for the shortage secretion of digestive enzymes, the continuously supplemented with certain strains of Lactobacillus sp. is essential to help the body to improve digestion, overcome the stress, and establish optimized bacterial composition which improved the body health and growth (Yang et al. 2018; Yang et al. 2020a).
The composition of bacteria in the gastrointestinal tract of piglets is easily affected by environmental changes, particularly during the weaning period. Both physiological and psychological stresses can cause oxidative damage in the body. The free radical chain reaction is the most widely accepted theory of inflammation (Farmer et al. 1942). Free radicals, other reactive oxygen species, and toxic products produced by oxidation can attack biological molecules, causing serious damage, including cellular damage. The MDA level is a marker of oxidative stress (Engin et al. 2010). Enzymes such as SOD, GPX, and CAT contribute to antioxidant defense and therefore can serve as biomarkers for evaluating the efficacy of nutritional interventions (Ogawa et al. 2011).
A usable strain of isolated Lactobacillus must be able to survive in and effectively colonize the GIT, particularly, the intestinal mucous (Yang et al. 2020a; Rao et al. 2016). Our qPCR results indicated that both L. reuteri and L. salivarius colonized the mucous membrane of the distal segment of the ileum, and the finding is supported by previous plate assays (Sattler et al. 2014; De Martinis et al. 2007). Once L. reuteri and L. salivarius colonize the ileal mucous, they can utilize nutrition in the intestine for propagation. The probiotic induced the secretion of metabolic enzymes, small peptides, and organic acids; optimized the composition of the gut microbiota; and promoted various beneficial interactions within the host piglets. These effects collectively improved nutrient absorption and alleviated stress through the brain-gut axis (Audet 2019; Ejtahed and Hasani-Ranjbar 2019). The growth performance of the piglets supplemented with CL improved, while diarrhea rate decreased, partly because of the probiotic effects of L. reuteri and L. salivarius. This result agrees with the findings of our previous study (Yang et al. 2020a).
The small and large intestine play key roles in digestion and nutrient absorption. The composition of the microbiome of the ileum and cecum contributes to various metabolic reactions, including the synthesis of amino acids, lipids, carbohydrates, vitamin B, and short-chain fatty acids (Liu et al. 2014; Munoz-Tamayo et al. 2011). An abundant and diverse intestinal microbiota improves health by alleviating the oxidation caused by stress (Peixoto et al. 2018; Bonfili et al. 2018).
Effect of CL administration on GIT bacterial composition of piglets
Aureomycin was chosen as the positive control to compare the growth performance with the compound L. reuteri and L. salivarius supplementation, which was to determine the efficiency of compound use of those two strains, while the bacterial composition conducted by the plate method assay indicated that the number of tested bacteria reduced significantly compared with the control. Considering the adverse effect of aureomycin supplemented on composition of bacteria, bacterial composition between the probiotic supplementation and the control was compared. The OTUs of the bacterial community of the ileal mucous membrane and cecal content indicated that the microbiological composition of the CL group was diverse. The PCoA results of the unweighted UniFrac distance metric on bacterial community showed that the OTUs from piglets supplemented with CL clustered together at both ED 14 and ED 28. However, OTUs from the control showed dispersion at both time points. The main influence on bacterial diversity is exerted by CL supplementation and age (Guevarra et al. 2019; Hu et al. 2016). These results indicate that piglets in the CL group were significantly affected by age or CL feeding during the experimental period. Additionally, the structure of the microbial community in the ileum and cecum varied widely, indicating their different functions in the body. Lactobacillus and Escherichia-Shigella had the greatest influence, consistent with the results of the plate method to evaluate the ileal mucous membrane and cecal contents. Escherichia coli as a colibiota constitutes normal GIT flora, which mainly reside in the cecal lumen or contents and whose abundant number in the cecum or transfer into other segment of the intestine can cause illness (Touchon et al. 2009). Salmonella is a pathogenic bacteria that reside in the intestine and cecal contents. In this study, the number of Escherichia coli and Salmonella was significantly reduced in the ileal mucous membrane and cecal lumen contents with compounds L. reuteri, and L. salivarius supplemented, and the number of total Lactobacillus and Bifidobacterium significantly increased indicated by the plate method assay. The results, coupled with the sequencing assay, suggested that the optimized bacterial flora was achieved via Lactobacillus supplementation.
Antioxidative and immune functions were found to be related to strains of L. delbrueckii, L. salivarius, L. formicilis, L. reuteri, and L. mucosae. In our results, greater colonization by L. salivarius and L. reuteri colonization in the ileal mucous membrane contributed to lower abundances of conditional and pathogenic bacteria owing to the optimized bacterial composition (Mackos et al. 2013; Galley et al. 2017; Cervantes-Barragan et al. 2017; Belkaid and Hand 2014). The results suggested that more probiotic Lactobacillus resided in the mucosal membrane contributing to body antioxidation and immunity, which reduce the damage caused by stress.
Relationship between CL administration and immunity of piglets
The supplemented effects of L. reuteri and L. salivarius were cumulative to the body. It is true that in both experimental points ED 14 and ED 28, the number of Lactobacillus colonized was significantly improved. The probiotic effects of bacteria contributed to the body must be more manifested in longer feeding duration. In order to confirm and highlight the probiotic effect of L. reuteri and L. salivarius, the experimental point ED 28 was chosen to carry out Spearman’s correlation analysis. In the KEGG pathway analysis of samples on ED 28, the genes encoding molecules involved in regulating the metabolism of piglets supplemented with CL were significantly enriched (Lopez et al. 2012; Zhao et al. 2019). The results of the study suggested that genes regulating the metabolism of cofactors and vitamins, other amino acids, lipid, terpenoids and polyketides, transport, and catabolism were more enhanced with CL supplemented. Both L. reuteri and L. salivarius secrete related metabolic bioactive peptides and lactate and hydrogen peroxide can colonize in distal segments of the ileum. The development of microstructures in the small intestine, such as intestinal villi and crypt of piglets, was enhanced with CL supplementation at the concentration of 106–107 CFU/g. The intestinal bacterial structure is a crucial producer of vitamins that play a key role in host health (Wang et al. 2019), implying the importance of increased gut bacterial vitamin B metabolism (Hu et al. 2016). All of these factors were helpful for body nutrition absorption. Molecules related to lipid and amino acid metabolism, which influence the secretion of hormones involved in growth, such as cortisol, endotoxin, growth hormone, and insulin-like growth factor 1, were increased, thereby improving growth performance and reducing stress (Jamilian et al. 2018; Falcinelli et al. 2017). The colonization of L. reuteri and L. salivarius in the ileal mucous membrane reduced the infection of pathogen and optimized the commensal bacterial structure, which alleviated stress and enhancement of immunity and antioxidation. These benefits could contribute to a lower diarrhea rate in CL supplementary piglets in weaning and group transferred periods.
In conclusion, the study revealed that probiotic L. salivarius and L. reuteri ameliorated stress and improved growth performance of weaning piglets via gut microbiota optimization. This study also suggested that functional strains of L. delbrueckii, L. salivarius, L. formicilis, L. reuteri, and L. mucosae were positively correlated with body antioxidation and immunity with CL supplementation; the strains of L. agilis and L. pontis were diverse and negatively correlated. However, the related metabolic signals and molecular pathways affected with CL supplementation need investigation in the future. The benefits of probiotic L. salivarius and L. reuteri were elaborated and can be used as an alternative to antibiotic drug in the production of weaning piglets. These discoveries can inform strategies for the rearing of healthier weaning pigs and for human consumption and can have important economic implications.
References
Audet MC (2019) Stress-induced disturbances along the gut microbiota-immune-brain axis and implications for mental health: does sex matter? Front Neuroendocrinol 11:100772
Belkaid Y, Hand TW (2014) Role of the microbiota in immunity and inflammation. Cell 157:121–141
Bloomfield MA, McCutcheon RA, Kempton M, Freeman TP, Howes O (2019) The effects of psychosocial stress on dopaminergic function and the acute stress response. Elife. 8:e46797
Bonfili L, Cecarini V, Cuccioloni AM, Berardi S, Scarpona S, Rossi G, Eleuteri AMS (2018) LAB51 probiotic formulation activates SIRT1 pathway promoting antioxidant and neuroprotective effects in an AD mouse model. Mol Neurobiol 55:7987–8000
Branning C, Hakansson A, Ahrne S, Jeppsson B, Molin G, Nyman M (2009) Blueberry husks and multi-strain probiotics affect colonic fermentation in rats. Br J Nutr 101:859–870
Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, Cella M, Gordon JI, Hsieh CS, Colonna M (2017) Lactobacillus reuteri induces gut intraepithelial CD4(+) CD8αα(+) T cells. Science 357:806–810
Chapman C, Gibson G, Rowland I (2011) Health benefits of probiotics: are mixtures more effective than single strains? Eur J Nutr 50:1–27
Clemente JC, Ursell LK, Parfrey LW, Knight R (2012) The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270
Davis TA, Burrin DG, Fiorotto ML, Nguyen HV (1996) Protein synthesis in skeletal muscle and jejunum is more responsive to feeding in 7- than in 26-day-old pigs. Am J Physiol Endocrinol Metab 270:E802–E809
De Martinis EC, Duvall RE, Hitchins AD (2007) Real-time PCR detection of 16S rRNA genes speeds most-probable-number enumeration of foodborne listeria monocytogenes. J Food Prot 70:1650–1655
Ejtahed HS, Hasani-Ranjbar S (2019) Neuromodulatory effect of microbiome on gut-brain axis; new target for obesity drugs. J Diabetes Metab Disord 18:263–265
El-Kadi SW, Boutry C, Suryawan A, Gazzaneo MC, Orellana RA, Srivastava N, Nguyen HV, Kimball SR, Fiorotto ML, Davis TA (2018) Intermittent bolus feeding promotes greater lean growth than continuous feeding in a neonatal piglet model. Am J Clin Nutr 108:830–841
Engin KN, Yemisci B, Yigit U, Agachan A, Coskun C (2010) Variability of serum oxidative stress biomarkers relative to biochemical data and clinical parameters of glaucoma patients. Mol Vis 16:1260–1271
Falcinelli S, Rodiles A, Hatef A, Picchietti S, Cossignani L, Merrifield DL, Unniappan S, Carnevali O (2017) Dietary lipid content reorganizes gut microbiota and probiotic L. rhamnosus attenuates obesity and enhances catabolic hormonal milieu in zebrafish. Sci Rep 7:5512
Farmer EH, Bloomfiel GF, Sundralingam A, Sutton DA (1942) The course and mechanism of autoxidation reactions in olefinic and polyolefinic substances, including rubber. Trans Faraday Soc 38:348–356
Galley JD, Mackos AR, Varaljay VA, Bailey MT (2017) Stressor exposure has prolonged effects on colonic microbial community structure in Citrobacter rodentium-challenged mice. Sci Rep 7:45012
Guevarra RB, Lee JH, Lee SH, Seok MJ, Kim DW, Kang BN, Johnson TJ, Isaacson RE, Kim HB (2019) Piglet gut microbial shifts early in life: causes and effects. J Anim Sci Biotechnol 10:1
Holtan SG, Shabaneh A, Betts BC, Rashidi A, MacMillan ML, Ustun C, Amin K, Vaughn BP, Howard J, Khoruts A, Arora M, DeFor TE, Johnson D, Blazar BR, Weisdorf DJ, Wang J (2019) Stress responses, M2 macrophages, and a distinct microbial signature in fatal intestinal acute graft-versus-host disease. JCI Insight 5:129762
Hou Q, Zhao F, Liu W, Lv R, Khine WWT, Han J, Sun Z, Lee YK, Zhang H (2020) Probiotic-directed modulation of gut microbiota is basal microbiome dependent. Gut Microbes:1–20. https://doi.org/10.1080/19490976.2020.1736974
Hu J, Nie Y, Chen J, Zhang Y, Wang Z, Fan Q, Yan X (2016) Gradual changes of gut microbiota in weaned miniature piglets. Front Microbiol 7:1727
Indrio F, Di Mauro A, Riezzo G, Civardi E, Intini C, Corvaglia L, Ballardini E, Bisceglia M, Cinquetti M, Brazzoduro E, Del Vecchio A, Tafuri S, Francavilla R (2014) Prophylactic use of a probiotic in the prevention of colic, regurgitation, and functional constipation: a randomized clinical trial. JAMA Pediatr 168:228–233
Jamilian M, Mansury S, Bahmani F, Heidar Z, Amirani E, Asemi Z (2018) The effects of probiotic and selenium co-supplementation on parameters of mental health, hormonal profiles, and biomarkers of inflammation and oxidative stress in women with polycystic ovary syndrome. J Ovarian Res 11:80
Lee YK, Ho PS, Low CS, Arvilommi H, Salminen S (2004) Permanent colonization by Lactobacillus casei is hindered by the low rate of cell division in mouse gut. Appl Environ Microbiol 70:670–674
Lei K, Li YL, Yu DY, Rajput IR, Li WF (2013) Influence of dietary inclusion of Bacillus licheniformis on laying performance, egg quality, antioxidant enzyme activities, and intestinal barrier function of laying hens. Poult Sci 92:2389–2395
Li Y, Guo Y, Wen Z, Jiang X, Ma X, Han X (2018) Weaning stress perturbs gut microbiome and its metabolic profile in piglets. Sci Rep 8:18068
Liu H, Zhang J, Zhang S, Yang F, Thacker PA, Zhang G, Qiao S, Ma X (2014) Oral administration of Lactobacillus fermentum I5007 favors intestinal development and alters the intestinal microbiota in formula-fed piglets. J Agric Food Chem 62:860–866
Lopez P, Gonzalez-Rodriguez I, Sanchez B, Ruas-Madiedo P, Suarez A, Margolles A, Gueimonde M (2012) Interaction of Bifidobacterium bifidum LMG13195 with HT29 cells influences regulatory-T cell associated chemokine receptor expression. Appl Environ Microbiol 78:2850–2857
Mackos AR, Eubank TD, Parry NM, Bailey MT (2013) Probiotic Lactobacillus reuteri attenuates the stressor-enhanced severity of Citrobacter rodentium infection. Infect Immun 81:3253–3263
Mitreva M, The Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy humanmicrobiome. Shows the enormous heterogeneity in phylogenetic composition of healthy human microbiota and relative stability of metabolic pathways. Nature 486:207–214
Mountzouris KC, Tsirtsikos P, Kalamara E, Nitsch S, Schatzmayr G, Fegeros K (2007) Evaluation of the efficacy of a probiotic containing Lactobacillus, Bifidobacterium, Enterococcus, and Pediococcus strains in promoting broiler performance and modulating cecal microflora composition and metabolic activities. Poultry Sci 86:309–317
Mountzouris KC, Tsitrsikos P, Palamidi I, Arvaniti A, Mohnl M, Schatzmayr G, Fegeros K (2010) Effects of probiotic inclusion levels in broiler nutrition on growth performance, nutrient digestibility, plasma immunoglobulins, and cecal microflora composition. Poult Sci 89:58–67
Munoz-Tamayo R, Laroche B, Walter E, Doré J, Duncan SH, Flint HJ, Leclerc M (2011) Kinetic modeling of lactate utilization and butyrate production by key human colonic bacterial species. FEMS Microbiol Ecol 76:615–624
National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals Guide for the care and use of laboratory animals, Eighth edn. The National Academies Press, Washington DC https://www.nap.edu/catalog/12910/guide-for-the-care-and-use-of-laboratory-animals-eighth. (2011)
Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S (2012) Host-gut microbiota metabolic interactions. Science 336:1262–1267
NRC (National Research Council) Nutrient requirement of swine, 10th edn. National Academy Press, USA http://www.nap.edu/catalog/2114.html. (1998)
Ogawa F, Shimizu K, Muroi E, Hara T, Sato S (2011) Increasing levels of serum antioxidant status, total antioxidant power, in systemic sclerosis. Clin Rheumatol 30:921–925
Ouwehand AC, Invernici MM, Furlaneto FAC, Messora MR (2018) Effectiveness of multistrain versus single-strain probiotics: current status and recommendations for the future. J Clin Gastroenterol 52(Suppl):1
Peixoto MJ, Domingues A, Batista S, Gonçalves JFM, Gomes AM, Cunha S, Valente LMP, Costas B, Ozório ROA (2018) Physiopathological responses of sole (Solea senegalensis) subjected to bacterial infection and handling stress after probiotic treatment with autochthonous bacteria. Fish Shellfish Immunol 83:348–358
Rao SC, Athalye-Jape GK, Deshpande GC, Simmer KN, Patole SK (2016) Probiotic supplementation and late-onset Sepsis in preterm infants: a meta-analysis. Pediatrics 137:e20153684
Sattler VA, Mohnl M, Klose V (2014) Development of a strainspecific real-time PCR assay for enumeration of a probiotic Lactobacillus reuteri in chicken feed and intestine. PLoS One 9:e90208
Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EP, Denamur E (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344
Vemuri R, Gundamaraju R, Shinde T, Perera AP, Basheer W, Southam B, Gondalia SV, Karpe AV, Beale DJ, Tristram S, Ahuja KDK, Ball M, Martoni CJ, Eri R (2019) Lactobacillus acidophilus DDS-1 modulates intestinal-specific microbiota, short-chain fatty acid and immunological profiles in aging mice. Nutrients 11:E1297
Wampach L, Heintz-Buschart A, Hogan A, Muller EEL, Narayanasamy S, Laczny CC, Hugerth LW, Bindl L, Bottu J, Andersson AF, de Beaufort C, Wilmes P (2017) Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front Microbiol 8:738
Wang H, Ni X, Liu L, Zeng D, Lai J, Qing X, Li G, Pan K, Jing B (2017) Controlling of growth performance, lipid deposits and fatty acid composition of chicken meat through a probiotic, Lactobacillus johnsonii during subclinical Clostridium perfringens infection. Lipids Health Dis 16:38
Wang Y, Xie Q, Sun S, Huang B, Zhang Y, Xu Y, Zhang S, Xiang H (2018) Probiotics-fermented Massa Medicata Fermentata ameliorates weaning stress in piglets related to improving intestinal homeostasis. Appl Microbiol Biotechnol 102:10713–10727
Wang X, Tsai T, Deng F, Wei X, Chai J, Knapp J, Apple J, Maxwell CV, Lee JA, Li Y, Zhao J (2019) Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 7:109
Warda AK, Rea K, Fitzgerald P, Hueston C, Gonzalez-Tortuero E, Dinan TG, Hill C (2019) Heat-killed lactobacilli alter both microbiota composition and behaviour. Behav Brain Res 362:213–233
Yang JJ, Qian K, Wu D, Zhang W, Wu YJ, Xu YY (2017) Effects of different proportions of two bacillus strains on the growth performance, small intestinal morphology, caecal microbiota and plasma biochemical profile of Chinese Huainan Partridge Shank chickens. J Integr Agric 16:1383–1392
Yang JJ, Qian K, Wang CL, Wu YJ (2018) Roles of probiotic lactobacilli inclusion in helping piglets establish healthy intestinal inter-environment for pathogen defense. Probiotics Antimicrob Proteins 10:243–250
Yang JJ, Wang CL, Liu LQ, Zhang MH (2020a) Lactobacillus reuteri KT260178 supplementation reduced morbidity of piglets through its targeted colonization, improvement of cecal microbiota profile, and immune functions. Probiotics Antimicrob Proteins 12:194–203
Yang JJ, Zhan K, Zhang MH (2020b) Effects of the use of a combination of two bacillus species on performance, egg quality, small intestinal mucosal morphology, and cecal microbiota profile in aging laying hens. Probiotics Antimicrob Proteins 12:204–213
Zhao S, Liu W, Wang J, Shi J, Sun Y, Wang W, Ning G, Liu R, Hong J (2019) Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J Mol Endocrinol 58:1–14
Funding
This study was financially supported by the fund of Anhui Academy of Agricultural Sciences Key Laboratory Project (No. 2019YL021), Anhui Science and Technology Key Project (No. 17030701008), Anhui Swine Industry Technology System Project (No. AHCYTX-05-09), National Key Research and Development Program of China (2016YFD0500509), and Science and Technology Program of Anhui Province (No. 1704A07020066).
Author information
Authors and Affiliations
Contributions
JY designed the study, fed the piglets and recorded the growth data, wrote the paper, and established the qRT-PCR assay. MZ and XP measured the levels of plasma antioxidant, immunity and hormonal indexes, and mRNA level. JW analyzed the data of plasma antioxidant, and immunity and hormonal indexes. CW and KH were involved in technical direction. We would like to thank Editage (www.editage.cn) for the English language editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
The experimental protocols in this study, including those related to animal husbandry and slaughter, were approved by the Institution of Animal Science and Welfare of Anhui Province (no. IASWAP2017056937). The experimental guidelines and treatment, housing, and husbandry conditions conformed to the Institutional Animal Care and Use Committee of China.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 76 kb)
Rights and permissions
About this article
Cite this article
Yang, J., Wang, C., Huang, K. et al. Compound Lactobacillus sp. administration ameliorates stress and body growth through gut microbiota optimization on weaning piglets. Appl Microbiol Biotechnol 104, 6749–6765 (2020). https://doi.org/10.1007/s00253-020-10727-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10727-4