Abstract
Streptomyces avermitilis is well known as the producer of anthelmintic agent avermectins, which are widely used in agriculture, veterinary medicine, and human medicine. aveI encodes a TetR-family regulator, which is the homolog of AtrA. It was reported that deletion of aveI caused enhanced avermectin production. In this study, we investigated the regulatory function of the AveI in S. avermitilis. By binding to the 15-nt palindromic sequence in the promoter regions, AveI directly regulates at least 35 genes. AveI represses avermectin production by directly regulating the transcription of the cluster-situated regulator gene aveR and structural genes aveA1, aveA3, and aveD. AveI represses oligomycin production by repressing the CSR gene olmRII and structural genes olmC. AveI activates melanin biosynthesis by activating the expression of melC1C2 operon. AveI activates morphological differentiation by activating the expression of ssgR and ssgD genes, repressing the expression of wblI gene. Besides, AveI regulates many genes involved in primary metabolism, including substrates transport, the metabolism of amino acids, lipids, and carbohydrates. Therefore, AveI functions as a global regulator in S. avermitilis, controls not only secondary metabolism and morphological differentiation, but also primary metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Streptomyces is the known producer of more than 70% antibiotics used in medicine, veterinary practice, and agriculture. Genome sequencing has revealed that some Streptomyces strains harbor tens of putative gene clusters responsible for secondary metabolites production (Choudoir et al. 2018; Craney et al. 2013). Individual antibiotic is usually biosynthesized by a large gene cluster, including cluster-situated regulator (CSR) encoding gene or genes. Various pleiotropic regulators sense developmental state, nutrient availability, diverse stresses, and then transmit the signals to the CSR genes to regulate antibiotic biosynthesis (Liu et al. 2013a; van Wezel and McDowall 2011). Elucidation of the antibiotic regulatory networks will provide strategies for increasing antibiotic productivity and “awakening” cryptic antibiotic production.
S. avermitilis is well known as the producer of 16-membered macrolide avermectins. Avermectins and its derivative, ivermectin, have potent anthelmintic activities and are widely used in human and veterinary medicine, and agriculture (Ikeda and Omura 1997). In addition to avermectins, S. avermitilis also produces 26-membered macrolide oligomycin and polyene macrolide filipin (Ikeda et al. 2014; Xu et al. 2010). aveR is the only CSR gene of avermectin biosynthetic gene cluster, activating transcription of ave structural genes (Guo et al. 2010; Kitani et al. 2009). Similar to other CSR genes, the expression of aveR is also under complex regulatory networks. Several regulators have been shown to directly regulate the expression of aveR. The response regulator PhoP of two-component PhoR–PhoP system, which controls phosphate metabolism, represses aveR transcription by binding to a PHO box located downstream of aveR transcriptional start site (Yang et al. 2015). A pseudo γ-butyrolactone receptor homolog AvaR2 (Zhu et al. 2016) and a butenolide-type autoregulator receptor homolog AvaR1 (Zhu et al. 2017) both repress avermectin production by binding to the promoter region of aveR. GlnR, the master regulator of nitrogen metabolism, directly stimulates avermectin production through aveR (He et al. 2016). Redox-sensing regulator Rex directly represses avermectin production by binding to the ROP in the aveR promoter region (Liu et al. 2017). SAV742, an AraC-family regulator, inhibits avermectin production through direct control of ave structural genes (aveA1-aveD, aveA4-orf1, and aveF), rather than the CSR gene (Sun et al. 2016). Some regulators have been demonstrated to control avermectin production indirectly, including SAV3818 (Duong et al. 2009), SAV7471 (Liu et al. 2013b), SAV576 and SAV577 (Guo et al. 2013, 2014), and AveT (Liu et al. 2015).
AveI (SAV4110) is a homolog of AtrA (actinorhodin-associated transcriptional regulator) from S. coelicolor A3(2) (Chen et al. 2008; Uguru et al. 2005). Deletion of aveI in S. avermitilis led to increased avermectin B1a production, which could be complemented by either aveI or its homologous gene atrA-c from S. coelicolor (Chen et al. 2008). Comparative transcriptomic analyses between aveI deletion mutant and WT showed that AveI affected expression of avermectin, oligomycin, and filipin biosynthetic gene clusters. The genes involved in precursor biosynthesis for antibiotics were upregulated, while genes involved in protein synthesis and fatty acid metabolism were downregulated in aveI mutant (Chen et al. 2009). These results suggested that AveI may function as a global regulator controlling not only secondary metabolism, but also primary metabolism. However, so far, the direct gene targets of AveI remain to be identified.
The TetR-family regulator AtrA is highly conserved in Streptomycetes. AtrA was first characterized as a transcription activator in S. coelicolor A3(2), stimulating the transcription of actII-ORF4, the pathway-specific activator gene of actinorhodin biosynthetic gene cluster (Uguru et al. 2005). AtrA-c also directly activates the expression of nagE2 and ssgR, which encode the major permease for N-acetylglucosamine and the activator for cell division-related gene ssgA (Kim et al. 2015; Nothaft et al. 2010). AtrA-gr of S. griseus binds to an inverted repeat between two AdpA-binding sites upstream promoter of strR, the transcriptional activator gene for streptomycin production. AtrA-gr only has a conditionally positive effect on streptomycin biosynthesis, probably through stimulating the AdpA-dependent transcriptional activation of strR (Hirano et al. 2008). AtrA-p in S. pristinaespiralis positively regulates pristinamycin production by directly stimulating the transcription of two CSR genes of pristinamycin gene cluster, spbR, and papR5 (Wang et al. 2015). AtrA-r in S. roseosporus positively controls daptomycin production by binding directly to the promoter region of dptE (Mao et al. 2015). S. globisporus AtrA-gl stimulates lidamycin production by binding to the promoter regions of lidamycin CSR genes, sgcR1 and sgcR2. The DNA-binding activity is inhibited by interaction with heptaene, a biosynthetic intermediate of lidamycin (Li et al. 2015).
Though AtrA has a common role in regulating antibiotic production in Streptomycetes, the mechanism how AveI (AtrA homolog) modulates avermectin production is still unknown. Besides, unlike the positive control of antibiotic production by AtrA in most cases, which is quite unusual for a TetR-family regulator (usually functioning as a repressor), AveI has a negative regulatory role in avermectin production. In this study, we provide evidences that AveI serves as a global regulator in S. avermitilis. AveI negatively regulates avermectin and oligomycin production by directly binding to the promoter regions of CSR and structural genes, positively regulates melanogenesis and morphological differentiation, and controls a variety of genes involved in primary metabolism.
Materials and methods
Strains and growth conditions
The S. avermitilis strains used in the present study included ATCC31267 (wild-type strain), DaveI (an aveI deletion mutant of WT), CaveI (a complementation strain of aveI deletion by aveI), and CAU69 (an avermectin high-producer). S. avermitilis strains were grown at 28 °C on YMS agar for sporulation, in modified liquid YEME medium (25% sucrose) for growth of mycelia for protoplast preparation, and on RM14 medium for regeneration of protoplasts (Macneil and Klapko 1987). Seed medium and fermentation medium I were used for avermectin production and fermentation medium II was used for growth analysis as described previously (Jiang et al. 2011). E. coli JM109 and Rosetta (DE3) were used for plasmid construction and protein expression, respectively. E. coli strains were cultured at 37 °C in LB medium.
Gene deletion and complementation
To produce an aveI null mutant, two DNA fragments flanking aveI gene were amplified by PCR using ATCC31267 genomic DNA as template. A 521-bp fragment upstream (position − 480 to + 41 from start codon) and a 563-bp fragment downstream (position − 42 to + 521 from stop codon) of aveI were amplified with primers aveI-up-1/aveI-up-2 and aveI-dw-1/aveI-dw-2, respectively (Table S1). The amplified fragments were recovered, digested by EcoRI/XbaI and XbaI/HindIII, respectively, and were cloned into pKC1139 to generate the aveI deletion vector pKCDaveI. The resulting plasmid was introduced into S. avermitilis ATCC31267 protoplasts. Double-crossover mutants were selected as described previously (Yang et al. 2015). The mutants were confirmed by PCR using primers listed in Table S1. The gene-deleted strain was termed as DaveI. To complement DaveI, a DNA fragment (1606 bp) carrying aveI gene with its promoter was amplified by PCR using primers aveI-C-1 and aveI-C-2 (Table S1). The fragment was recovered, digested by EcoRI/BamHI, and then ligated to the integrative vector pSET152 to produce vector pSET-aveI, which was introduced into DaveI to obtain complemented strain.
Fermentation and HPLC analysis of avermectin and oligomycin production
Fermentation conditions and HPLC analysis of avermectin and oligomycin production were performed as described previously (Luo et al. 2014).
RNA extraction and qRT-PCR analysis
Mycelia of S. avermitilis from fermentation medium I or YMS agar were collected, frozen in liquid nitrogen, and ground to a fine powder. RNA was extracted using TRIzol reagent (Tiangen, Beijing, China) following the manufacturer’s instructions, and was treated with DNase I (TaKaRa, Shiga, Japan) to remove chromosomal DNA contamination. RNA samples (2 μg each) were reverse transcribed by M-MLV (RNase H−; TaKaRa), and qRT-PCR analysis was performed using FastStart Universal SYBR Green Master (ROX) by an ABI 7900HT Sequence Detection System with primer pairs listed in Table S1. PCR included a 10 min preincubation at 95 °C, followed by 40 cycles of denaturation at 95 °C for 10 s, and annealing and extension at 60 °C for 30 s. 16S rRNA was used as the internal control. The relative expression level was calculated using the comparative Ct method. Gene expression was determined in triplicate.
Overexpression and purification of His6-AveI
The 859 bp coding region of aveI was amplified using primers aveI-E-1 and aveI-E-2, digested by EcoRI/BamHI, and then cloned into pET28a (+) to produce pET-aveI. After confirmation by DNA sequencing, pET-aveI was introduced into E. coli Rosetta (DE3) for overexpression of AveI with a His6 tag at N terminus. E. coli Rosetta (DE3) harboring pET-aveI was grown at 37 °C in LB with 50 μg ml−1 kanamycin to an OD600 of 0.4–0.6, induced by 0.1 mM IPTG, and incubated for 5 h at 28 °C. The cells were harvested, washed, re-suspended in lysis buffer (20 mM Tris base, 500 mM NaCl, 5 mM imidazole, 5% glycerol, pH 7.9), and disrupted by sonication on ice. After centrifugation, His6-AveI was purified from the supernatant using Ni2+-NTA resin (Qiagen, Germany) according to the manufacturer’s protocol.
Electrophoretic mobility gel shift assays (EMSAs)
EMSAs were carried out using a DIG Gel Shift Kit (2nd Generation, Roche) according to the manufacturer’s instructions. DNA probes used for EMSA were amplified by PCR with the primers listed in Table S1 and labeled with Digoxigenin-11-ddUTP using recombinant terminal transferase. The binding mixture (20 μl) contained 0.3 nM DIG-labeled DNA probe, varying quantities of His6-AveI, and 1 μg of poly[d(I–C)]. After incubation at 25 °C for 30 min, protein-bound and free probes were separated by electrophoresis on 5.0% native polyacrylamide gels with 0.5 × TBE buffer as running buffer. Then, DNA probes were electroblotted onto a positively charged nylon membrane, and the signals were detected by chemiluminescence and recorded on X-ray film.
DNase I footprinting assays
For DNase I footprinting assays, DNA fragments were amplified by PCR with FAM-labeled primers (Table S1) and purified from the agarose gel. 400 ng FAM-labeled probes and varying quantities of His6-AveI were incubated at 25 °C for 30 min in a 25 μl volume. 0.017 U DNase I was added to the mixture. After incubation at 37 °C for 40 s, the digestion was stopped with 10 μl of 0.2 M EDTA (pH 8.0). DNA samples were extracted and subjected to capillary electrophoresis. Electrophoregrams were analyzed using GeneMarker soſtware v2.2.0.
Results
AveI represses avermectin and oligomycin production, and stimulates melanogenesis and morphological differentiation
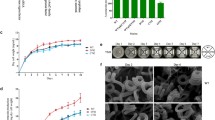
Chen et al. (2008) have reported that deletion of aveI enhanced avermectin production in S. avermitilis. To fully assess the function of AveI in secondary metabolism, morphological differentiation, and primary metabolism, we constructed the aveI deletion mutant (DaveI) in S. avermitilis wild-type strain by homologous recombination. Deletion of aveI did not affect growth in fermentation media (Fig. S1). Compared to the WT strain, avermectin and oligomycin productions of DaveI were ~ 3.1-fold and 1.47-fold of those in WT, and the productions were restored to the WT levels when an aveI gene was introduced into DaveI (Fig. 1a). When the strains were grown on RM14 medium, which favor melanin production, DaveI grew slower and produced less melanin than WT, while melanin production was restored to WT levels in the complemented strain of DaveI (Fig. 1b). When grown on YMS media, DaveI mutant displayed delays of aerial hyphae formation and sporulation in comparison with WT and complemented strain (Fig. 1b). These findings indicated that AveI had a negative regulatory role in both avermectin and oligomycin production, but a positive regulatory role in melanogenesis and morphological differentiation.
Effects of aveI deletion on avermectin, oligomycin, and melanin production, and morphology. a Avermectin and oligomycin production in WT, DaveI, and CaveI. Values are mean ± SD of three replicate flasks cultured in FM-I. P values were determined by Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001. b Phenotype of aveI deletion mutant. The indicated strains were grown on YMS and RM14 media for 2, 4, or 6 days. DaveI, aveI deletion mutant. CaveI, complemented strain of DaveI
AveI regulates avermectin production by directly repressing aveR and ave structural genes
Avermectin was overproduced by deletion of aveI gene. To test whether AveI affects avermectin production through regulating ave genes, we carried out qRT-PCR analysis using RNAs isolated from DaveI and WT cultured in FM-1 for 2 days (exponential phase) and 6 days (stationary phase). The transcriptional levels of avermectin biosynthetic CSR gene aveR and structural genes aveA1, aveA3, aveA4, aveD, and aveF were increased in DaveI at both time points, particularly on 6 day (Fig. 2a). The augmented expression of ave genes is consistent with the avermectin overproduction in DaveI. The findings suggest that AveI represses the expression of ave genes. As TetR-family regulators are usually autoregulated, it was tested whether AveI regulates its own expression, we also examined the expression of aveI using the same RNAs. The transcriptional level of aveI was increased in DaveI at both 2 days and 6 days (Fig. 2a), indicating that aveI is negatively autoregulated.
AveI represses avermectin and oligomycin production, and activates melanogenesis. a qRT-PCR analysis of ave, olm, and melC genes in DaveI and WT. RNAs were prepared from cells cultured in FM-I for 2 and 6 days. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. b Binding of His6-AveI to the promoter regions of the above genes by EMSA. Concentrations of His6-AveI for probes: 0.15 and 0.3 μM. Competition assays were carried out using 500-fold excess of unlabeled specific (S) and non-specific (N) DNAs. Arrow: free probe
An N-terminal His6-tagged AveI protein was overexpressed in E. coli Rosetta (DE3) and purified for EMSAs (Fig. S2). The promoter regions of aveI, actII-ORF4, aveR, aveA3, aveA4, and aveF and the intergenic region of aveA1-aveD were DIG labeled for EMSAs. His6-AveI bound to the promoter region of aveI, and the shifted bands could be abolished by addition of 500-fold excess of unlabeled specific competitor DNA but not by unlabeled non-specific DNA, indicating that the binding was specific (Fig. 2b). AveI is negatively autoregulated by binding to its promoter region. His6-AveI could bind specifically to the promoter region of actII-ORF4 in S. coelicolor, implying that AveI and AtrA have similar binding site. His6-AveI bound specifically to the promoter regions of aveR and aveA3, and the intergenic region of aveA1-aveD, but not to the promoter regions of other ave genes (Fig. 2b). Therefore, AveI represses avermectin production by directly regulating the transcription of the CSR gene aveR and structural genes aveA1, aveA3, and aveD. The improved expression of aveA4 and aveF was probably caused by the enhanced expression of aveR.
AveI directly represses olm genes and activates expression of melC1C2 gene
EMSAs were also used to determine whether AveI directly regulates oligomycin biosynthesis and melanogenesis. His6-AveI bound specifically to the promoter regions of oligomycin biosynthetic CSR gene olmRII and structural gene olmC (Fig. 2b), but not to the promoter regions of CSR gene olmRI and other structural genes. The transcriptional levels of olmRII and olmC were increased in DaveI, in accordance with the increased oligomycin production in DaveI, indicating the negative role of AveI in regulating oligomycin production (Fig. 2a). S. avermitilis contains two melanin biosynthetic operons: melC1C2 and melC1-2C2-2. His6-AveI bound specifically to the promoter region of melC1C2 operon, but not to that of melC1-2C2-2 (Fig. 2b). The expression of melC1 was greatly reduced in DaveI (Fig. 2a), which was consistent with the decreased melanin production in DaveI. Thus, AveI positively controls melanogenesis through directly activating the expression of melC1C2.
AveI directly activates the expression of ssgR and ssgD genes and represses wblI gene
DaveI displayed delayed morphogenesis compared with WT. To identify the direct targets of AveI involved in morphological differentiation, we performed EMSAs using His6-AveI and the promoter regions of bldC, ftsZ, ssgD, ssgR, ssgY, wblB, and wblI. His6-AveI bound specifically to the promoter regions of ssgD, ssgR, and wblI, but not to the other promoters (Fig. 3a). qRT-PCR analysis showed that the transcriptional levels of ssgD and ssgR were decreased, while the transcriptional level of wblI was increased in DaveI (Fig. 3b), indicating that AveI affects morphological differentiation through directly activating ssgR and ssgD genes and repressing wblI gene. The positive control of ssgR by AveI is same as the findings that AtrA activates the transcription of ssgR, which in turn activates ssgA transcription in S. coelicolor (Kim et al. 2015).
The regulatory role of AveI in morphology. a Binding of His6-AveI to the promoter regions of genes involved in morphology by EMSA. EMSA conditions as described for Fig. 2. b qRT-PCR analysis of the related genes in DaveI and WT. RNAs were prepared from cells grown on YMS for 2 and 6 days. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant
Prediction and verification of the AveI regulon
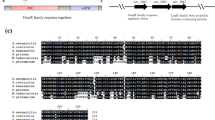
To fully understand the regulatory role of AveI in S. avermitlis, we used the palindromic consensus sequence (5′-GGAAT-n5-ATTCC-3′) of ArtA (Wang et al. 2015) to scan the S. avermitilis genome, through the PREDetector web-based application (http://predetector.hedera22.com/login) to search for putative AveI target genes. The genome-wide search revealed 440 putative AveI target genes with score > 6.5 (Table S2). About half of the target genes (203) are unknown or unclassified genes, the others are involved in regulatory function (58), transport (44), amino acids and related molecules metabolism (28), carbohydrate metabolism (17), protein synthesis, folding, and modification (13), DNA synthesis, repair, recombination, modification, and packaging (9), secondary metabolism (9), fatty acid and lipid metabolism (7), differentiation (4), and other metabolisms. Nine of them (aveA1, aveD, aveI, melC1, olmC, olmRII, ssgD, ssgR, and wblI) have been confirmed to be directly regulated by AveI. To test whether AveI binds to the promoter regions of other putative target genes, we selected 42 genes with predicted gene function associated with regulation function, secondary metabolism, carbon metabolism, energy metabolism, and protein folding for EMSAs (Table 1).
EMSA results showed that His6-AveI bound specifically to the promoter regions of 25 putative AveI target genes (aspC2, avaA, avaL1, ctaC, fadS3, fahA, galE6, gltI2 groES1, ilvk3, leuA1, leuA2, maeB1, mcr, nrpS5, pepA, pepD1, pitH2, pks9-4, ppc, SAV577, SAV1671, SAV5600, SAV6046, and SAV6634) (Fig. 4), but not to the promoter regions of aceE1, bkdF, cdh, dnaK2, echA6, eno, fadA4, glmS1, glnR, gpmA1, idnO, ilvE, maeB2, pckA, pstS, and rhaT. qRT-PCR analysis demonstrated that the transcriptional levels of 14 genes (avaA, avaL1, ctaC, fadS3, galE6, groES1, maeB1, mcr, pepA, pitH2, ppc, SAV577, SAV6046, and SAV6634) from 25 newly identified AveI target genes were decreased in DaveI (Fig. 5), indicating the positive control of AveI on these genes. The transcriptional levels of aspC2, fahA, gltI2, leuA1, leuA2, ilvK3, nrpS5, pepD1, pks9-4, SAV1671, and SAV5600 were increased in DaveI (Fig. 5), suggesting the negative regulatory role of AveI on these genes. The findings indicate that AveI plays a pleiotropic role in primary metabolism, secondary metabolism, and morphological differentiation in S. avermitilis and acts as dual role of a repressor and an activator.
Confirmation of putative AveI target genes by EMSA. EMSA conditions as described in Fig. 2
qRT-PCR analysis of putative AveI target genes in DaveI and WT. RNAs were the same ones used in Fig. 2. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant
Determination of the precise AveI-binding sequence
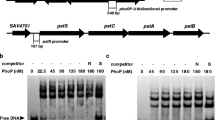
To identify the precise AveI-binding site, we selected the promoter regions of mcr and galE6 which have high affinity with His6-AveI for DNase I footprinting analysis. DNase I footprinting assays were performed on 5′-end fluorescein-labeled probes of mcr (264 bp) and galE6 (296 bp) in the presence of His6-AveI. A 26-nt protected region (5′-ACGTCGGTGGAATGTCCCACTCCGGT-3′) was found in the mcr promoter region, extending from − 67 to − 42 relative to the predicted transcriptional start site of mcr (Fig. 6a), containing a 15-nt palindromic sequence (5′-GGAATGTCCCACTCC-′), similar to the consensus sequence of ArtA. Combined with the qRT-PCR results that AveI positively regulates mcr expression, AveI may help to recruit RNA polymerase to the promoter of mcr through binding to the site adjacent to the promoter and activate its transcription. A 15-nt palindromic ArtA consensus sequence (5′-GGAACCTGGCATTCC-3′) was also found in the 31-nt AveI-protected region (5′-CACACCATCCCGGAACCTGGCATTCCAGGCA-3′) in the galE6 promoter region (Fig. 6b). To confirm that the 15-nt palindromic sequence is the recognition sequence of AveI, we introduced mutation to the palindromic sequence in the mcr promoter region (probe A) to produce probe AM (Fig. 6c). EMSAs showed that His6-AveI bound to probe A, but not to probe AM (Fig. 6c); therefore, the palindromic sequence (5′-GGAAT-n5-ATTCC-3′) is the AveI-binding site. The findings indicate that AveI has very similar binding sites as AtrA, suggesting the conserved regulatory role of AtrA homolog in Streptomyces.
Determination of AveI-binding sites. a and b DNase I footprinting assays of the promoter regions of mcr (a) and galE6 (b) with His6-AveI. Upper fluorograms: control reaction. Protection fluorograms were obtained with increasing amounts of His6-AveI. Nucleotide sequences of mcr and galE6 promoter regions are shown below fluorograms. Underlines: AveI-protected sequences. Arrows: palindromic sequences. Boxes: predicted − 35 and − 10 elements of promoters. Black bent arrows and boldface letters: translational start codon. c EMSAs of probe A of mcr promoter region and mutated probe AM to identify the AveI-binding site. Mutation was introduced into the palindromic sequence of protected region in probe A to produce mutated probe AM. Concentrations of His6-AveI for probes: 0.075, 0.15, 0.225, and 0.3 μM
Deletion of aveI increases avermectin production in an avermectin high-producer
Deletion of aveI could enhance avermectin production in S. avermitilis WT strain. We also tested the possibility of further improving avermectin production in an avermectin high-producer by aveI deletion. Compared to the avermectin B1a production (5470 μg/mL) of the parental strain CAU69, the production level of CAU69/DaveI was increased by ~ 14.3% and reached 6252 μg/mL (Fig. 7), indicating that deletion of aveI may provide an effective strategy to improve avermectin production in the avermectin high-producing strains.
Discussion
In this investigation, we demonstrated that AveI acts as a repressor regulating avermectin and oligomycin production, and also as an activator controlling melanogenesis and morphological differentiation (Fig. 8). At least 35 genes have been confirmed to be AveI targets by EMSAs, most of them are associated with primary metabolism. pepA, pepD1, SAV5600, and SAV6634 encode proteases or peptidases, which digest proteins or peptides to produce amino acids, and aspC2, SAV6046, fahA, leuA1, and leuA2 are involved in the metabolism and biosynthesis of amino acids. fadS3 and mcr, encoding a fatty acid desaturase and racemase, are associated with metabolism of lipids. galE6 (encoding a UDP-glucose 4-epimerase), maeB1 (encoding a malate dehydrogenase), ppc (encoding a phosphoenolpyruvate carboxylase), and SAV1671 (encoding an oxidoreductase) are associated with carbohydrate metabolism. ctaC (encoding a cytochrome c oxidase subunit II) is involved in energy metabolism. gltI2, ilvK3, and pitH2, encoding putative ABC transporter substrate-binding proteins and a low-affinity inorganic phosphate transporter, are involved in substrates transport. These findings were consistent with the transcriptomics analysis results of the aveI mutant vs. WT that aveI deletion affects a variety of genes in both primary and secondary metabolic pathways (Chen et al. 2009). Therefore, AveI acts as a global regulator in S. avermitilis, controls not only secondary metabolism and morphological differentiation, but also primary metabolism. Although TetR-family regulators usually function as repressors, about half of the identified target genes are under the positive control of AveI. The mechanism of activation by AveI probably involves competition with repressors in the promoter regions of target genes or allowing activators to bind, as observed for AtrA-gr in S. griseus which activates streptomycin biosynthesis, probably through facilitating the AdpA-dependent transcriptional activation of strR (Hirano et al. 2008).
Similar to its homolog AtrA in other Streptomycetes, AveI regulates the biosynthesis of several secondary metabolites in S. avermitilis. AveI negatively regulates avermectin production by repressing the transcription of CSR gene aveR and structural genes aveA1, aveA3, and aveD. AveI also negatively regulates oligomycin production by repressing the CSR gene olmRII and the structural gene olmC. AveI positively regulates melanin biosynthesis by activating the expression of melC1C2 operon. nrpS5 (SAV6633, encoding a non-ribosomal peptide synthetase) and pks9-4(SAV2376, encoding putative 3-oxoacyl-ACP synthase I of PKS9) are also under the direct negative control of AveI. Besides, among the AveI target genes, avaA (SAV2269, encoding a γ-butyrolactone biosynthesis protein, homolog of AfsA) and avaL1 (SAV2270, encoding a γ-butyrolactone-dependent transcriptional regulator) belongs to γ-butyrolactone regulatory system, which triggers antibiotic production and morphological differentiation in Streptomyces. SAV577 (encoding a TetR-family transcriptional regulator) downregulates avermectin biosynthesis indirectly (Guo et al. 2014). Therefore, AveI may also regulate secondary metabolism through the cascaded regulation of other regulators. Genes associated with several primary metabolic pathways, such as fatty acid metabolism and protein synthesis, were found to be under positive control of AveI. Therefore, AveI is also possibly involved in directing the carbon flux from primary to secondary metabolism in S. avermitilis.
The SsgA-like proteins are a family of proteins that control cell division and sporulation in actinobacteria (Noens et al. 2005, 2007). In S. coelicolor A3(2), the transcription of ssgA is activated by the regulator SsgR (Traag et al. 2004). AtrA-c activates the transcription of ssgR, which in turn activates the transcription of ssgA (Kim et al. 2015). In S. avermitilis, ssgR is also under the positive control of AveI. Another ssgA-like gene, ssgD, is also under the positive control of AveI. WhiB-like proteins (Wbl) are small transcription factor-like proteins essential for sporulation in actinobacteria (Fowler-Goldsworthy et al. 2011; Molle et al. 2000). The expression of wblI is under the negative control of AveI. Therefore, AveI controls morphological differentiation by regulating the expression of ssgR, ssgD, and wblI. It is interesting to mention that some targets of AveI are conserved in Streptomyces. Besides ssgR, γ-butyrolactone receptor protein encoding gene spbR in S. pristinaespiralis and avaA in S. avermitilis are also the targets of AtrA homolog (Wang et al. 2015). SsgR and γ-butyrolactone receptor protein are involved in morphological differentiation of Streptomyces, suggesting that AtrA homologs may play a conserved role in development.
AtrA (the AveI homolog) has been intensively studied to regulate secondary metabolism in several Streptomyces strains (Chen et al. 2008; Hirano et al. 2008; Li et al. 2015; Mao et al. 2015; Uguru et al. 2005; Wang et al. 2015). AtrA-c and AtrA-gr activate actinorhodin and streptomycin biosynthesis through activating the expression of the CSR genes actII-ORF4 in S. coelicolor and strR in S. griseus, respectively (Hirano et al. 2008; Uguru et al. 2005). AtrA-p activates pristinamycin production through activating the expression of two CSR genes spbR and papR5 (Wang et al. 2015). S. globisporus AtrA activates lidamycin production by activating one of the CSR genes, sgcR1 (Li et al. 2015). AtrA-r positively regulates daptomycin production via activating the structure gene dptE in S. roseosporus (Mao et al. 2015). AveI activates melanin biosynthesis by activating the expression of structure genes melC1C2 operon. However, AveI represses avermectin production and oligomycin production through repressing the transcription of CSR genes aveR and olmRII and structural genes aveA1, aveA3, aveD, and olmC in S. avermitilis. Although the regulatory role of AtrA homolog in antibiotic biosynthesis is conserved in various Streptomyces, the molecular mechanism for each secondary metabolite is relatively diverse.
References
Chen L, Lu Y, Chen J, Zhang W, Shu D, Qin Z, Yang S, Jiang W (2008) Characterization of a negative regulator AveI for avermectin biosynthesis in Streptomyces avermitilis NRRL8165. Appl Microbiol Biotechnol 80(2):277–286
Chen L, Chen J, Jiang Y, Zhang W, Jiang W, Lu Y (2009) Transcriptomics analyses reveal global roles of the regulator AveI in Streptomyces avermitilis. FEMS Microbiol Lett 298(2):199–207
Choudoir MJ, Pepe-Ranney C, Buckley DH (2018) Diversification of secondary metabolite biosynthetic gene clusters coincides with lineage divergence in Streptomyces. Antibiotics (Basel) 7(1):12
Craney A, Ahmed S, Nodwell J (2013) Towards a new science of secondary metabolism. J Antibiot 66(7):387–400
Duong CT, Lee HN, Choi SS, Lee SY, Kim ES (2009) Functional expression of SAV3818, a putative TetR-family transcriptional regulatory gene from Streptomyces avermitilis, stimulates antibiotic production in Streptomyces species. J Microbiol Biotechnol 19(2):136–139
Fowler-Goldsworthy K, Gust B, Mouz S, Chandra G, Findlay KC, Chater KF (2011) The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiol 157(Pt 5):1312–1328
Guo J, Zhao J, Li L, Chen Z, Wen Y, Li J (2010) The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol Gen Genet 283(2):123–133
Guo J, Zhang X, Luo S, He F, Chen Z, Wen Y, Li J (2013) A novel TetR family transcriptional regulator, SAV576, negatively controls avermectin biosynthesis in Streptomyces avermitilis. PLoS One 8(8):e71330
Guo J, Zhang X, Chen Z, Wen Y, Li J (2014) Two adjacent and similar TetR family transcriptional regulator genes, SAV577 and SAV576, co-regulate avermectin production in Streptomyces avermitilis. PLoS One 9(6):e99224
He JM, Zhu H, Zheng GS, Liu PP, Wang J, Zhao GP, Zhu GQ, Jiang WH, Lu YH (2016) Direct involvement of the master nitrogen metabolism regulator GlnR in antibiotic biosynthesis in Streptomyces. J Biol Chem 291(51):26443–26454
Hirano S, Tanaka K, Ohnishi Y, Horinouchi S (2008) Conditionally positive effect of the TetR-family transcriptional regulator AtrA on streptomycin production by Streptomyces griseus. Microbiol 154(Pt 3):905–914
Ikeda H, Omura S (1997) Avermectin biosynthesis. Chem Rev 97(7):2591–2610
Ikeda H, Kazuo SY, Omura S (2014) Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol 41(2):233–250
Jiang L, Liu Y, Wang P, Wen Y, Song Y, Chen Z, Li J (2011) Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis. Biotechnol Lett 33(10):1955–1961
Kim SH, Traag BA, Hasan AH, McDowall KJ, Kim BG, van Wezel GP (2015) Transcriptional analysis of the cell division-related ssg genes in Streptomyces coelicolor reveals direct control of ssgR by AtrA. Antonie Van Leeuwenhoek 108(1):201–213
Kitani S, Ikeda H, Sakamoto T, Noguchi S, Nihira T (2009) Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl Microbiol Biotechnol 82(6):1089–1096
Li X, Yu T, He Q, McDowall KJ, Jiang B, Jiang Z, Wu L, Li G, Li Q, Wang S, Shi Y, Wang L, Hong B (2015) Binding of a biosynthetic intermediate to AtrA modulates the production of lidamycin by Streptomyces globisporus. Mol Microbiol 96(6):1257–1271
Liu G, Chater KF, Chandra G, Niu G, Tan H (2013a) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77(1):112–143
Liu Y, Yan T, Jiang L, Wen Y, Song Y, Chen Z, Li J (2013b) Characterization of SAV7471, a TetR-family transcriptional regulator involved in the regulation of coenzyme A metabolism in Streptomyces avermitilis. J Bacteriol 195(19):4365–4372
Liu W, Zhang Q, Guo J, Chen Z, Li J, Wen Y (2015) Increasing avermectin production in Streptomyces avermitilis by manipulating the expression of a novel TetR-family regulator and its target gene product. Appl Environ Microbiol 81(15):5157–5173
Liu X, Cheng Y, Lyu M, Wen Y, Song Y, Chen Z, Li J (2017) Redox-sensing regulator Rex regulates aerobic metabolism, morphological differentiation, and avermectin production in Streptomyces avermitilis. Sci Rep 7:44567
Luo S, Sun D, Zhu J, Chen Z, Wen Y, Li J (2014) An extracytoplasmic function sigma factor, sigma(25), differentially regulates avermectin and oligomycin biosynthesis in Streptomyces avermitilis. Appl Microbiol Biotechnol 98(16):7097–7112
Macneil DJ, Klapko LM (1987) Transformation of Streptomyces avermitilis by plasmid DNA. J Ind Microbiol 2(4):209–218
Mao XM, Luo S, Zhou RC, Wang F, Yu P, Sun N, Chen XX, Tang Y, Li YQ (2015) Transcriptional regulation of the daptomycin gene cluster in Streptomyces roseosporus by an autoregulator, AtrA. J Biol Chem 290(12):7992–8001
Molle V, Palframan WJ, Findlay KC, Buttner MJ (2000) WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J Bacteriol 182(5):1286–1295
Noens EE, Mersinias V, Traag BA, Smith CP, Koerten HK, van Wezel GP (2005) SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol Microbiol 58(4):929–944
Noens EE, Mersinias V, Willemse J, Traag BA, Laing E, Chater KF, Smith CP, Koerten HK, van Wezel GP (2007) Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol Microbiol 64(5):1244–1259
Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, Titgemeyer F (2010) The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol Microbiol 75(5):1133–1144
Sun D, Zhu J, Chen Z, Li J, Wen Y (2016) SAV742, a novel AraC-family regulator from Streptomyces avermitilis, controls avermectin biosynthesis, cell growth and development. Sci Rep 6:36915
Traag BA, Kelemen GH, Van Wezel GP (2004) Transcription of the sporulation gene ssgA is activated by the IclR-type regulator SsgR in a whi-independent manner in Streptomyces coelicolor A3(2). Mol Microbiol 53(3):985–1000
Uguru GC, Stephens KE, Stead JA, Towle JE, Baumberg S, McDowall KJ (2005) Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol Microbiol 58(1):131–150
van Wezel GP, McDowall KJ (2011) The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28(7):1311–1333
Wang W, Tian J, Li L, Ge M, Zhu H, Zheng G, Huang H, Ruan L, Jiang W, Lu Y (2015) Identification of two novel regulatory genes involved in pristinamycin biosynthesis and elucidation of the mechanism for AtrA-p-mediated regulation in Streptomyces pristinaespiralis. Appl Microbiol Biotechnol 99(17):7151–7164
Xu LH, Fushinobu S, Takamatsu S, Wakagi T, Ikeda H, Shoun H (2010) Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis. J Biol Chem 285(22):16844–16853
Yang R, Liu X, Wen Y, Song Y, Chen Z, Li J (2015) The PhoP transcription factor negatively regulates avermectin biosynthesis in Streptomyces avermitilis. Appl Microbiol Biotechnol 99(24):10547–10557
Zhu J, Sun D, Liu W, Chen Z, Li J, Wen Y (2016) AvaR2, a pseudo gamma-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol Microbiol 102(4):562–578
Zhu J, Chen Z, Li J, Wen Y (2017) AvaR1, a butenolide-type autoregulator receptor in Streptomyces avermitilis, directly represses avenolide and avermectin biosynthesis and multiple physiological responses. Front Microbiol 8:2577
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 31470190) and the project for Extramural Scientists of the State Key Laboratory of Agrobiotechnology (No. 2019SKLAB6-5).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 755 kb)
Rights and permissions
About this article
Cite this article
Liu, L., Cheng, Y., Lyu, M. et al. AveI, an AtrA homolog of Streptomyces avermitilis, controls avermectin and oligomycin production, melanogenesis, and morphological differentiation. Appl Microbiol Biotechnol 103, 8459–8472 (2019). https://doi.org/10.1007/s00253-019-10062-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10062-3