Abstract
Δ1-Dehydrogenation is one of the most important reactions for steroid drug modification. Numerous 3-ketosteroid-Δ1-dehydrogenases (KstDs) catalyzing this reaction were observed in various organisms. However, only a few have been characterized and used for substrate conversion. In this study, a promising enzyme (KstD2) from Mycobacterium neoaurum DSM 1381 was purified and characterized. Interestingly, KstD2 displayed a high activity on a range of substrates, including 17α-hydroxypregn-4-ene-3,20-dione (17α-OH-P); androsta-4,9(11)-diene-3,17-dione (NSC 44826); and 4-androstene-3,17-dione (AD). These reactions were performed under optimal conditions at 40 °C and pH 8.0. Noteworthy, both the activity and stability of the enzyme were sensitive to various metal ions. After optimizing the expression and biocatalyst conditions, up to 1586 U mg−1 intracellular KstD activity on AD could be produced. Furthermore, the associated conversion rate was 99% with 30 g L−1 AD after 8 h. On the other hand, we obtained 99%, 90%, and over 80% of conversion with 20 g L−1 NSC 44826; 10 g L−1 16,17α-epoxyprogesterone; and 20 g L−1 17α-OH-P or canrenone, respectively, after 24 h. Sequence homology and structural analyses indicated that the residue R178 located in a unique short loop among cluster 2 is crucial for substrate recognition which was confirmed by mutagenesis. In summary, this study reports on the first purification and characterization of a KstD from cluster 2. Its remarkable properties deserve more attention to potentially lead to further industrial applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Steroid drugs are particularly important in the prevention and treatment of various diseases (Fernandez-Cabezon et al. 2018; García et al. 2012). Moreover, seeking for more active steroid drugs can be achieved by modifying the structure of the steroid nuclei (Donova and Egorova 2012; Wu et al. 2015). However, traditional chemical methods are difficult to implement in whole processes in order to modify steroid intermediates, because of the complex steroid structure (Bhatti and Khera 2012). Therefore, microbial transformation has caught the attention of medicinal chemists due to the associated mild reaction conditions as well as their regio-selectivity and stereo-selectivity (Fernandes et al. 2003). Among these reactions, Δ1-dehydrogenation was one of the most frequently reported (Donova and Egorova 2012).
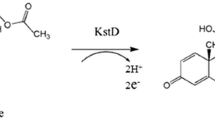
3-Ketosteroid-Δ1-dehydrogenase (KstD) catalyzes Δ1-dehydrogenation (Fig. 1) and was well studied for its prominent role in steroid catabolism (Guevara et al. 2017; Wang et al. 2017; Yao et al. 2014). On the one hand, KstD inactivation was achieved in many actinomycetes including Rhodococcus erythropolis, Rhodococcus ruber, and Mycobacterium neoaurum, leading to the production of important intermediates such as 22-hydroxy-23; 24-bisnorchol-4-ene-3-one (4HP); 4-androstene-3,17-dione (AD); and 9α-hydroxy-4-androsten-3,17-dione (9α-OH-AD) (Guevara et al. 2017; Knol et al. 2008; Yao et al. 2014; Zhang et al. 2018). This resulted from altered degradation of the steroid nuclei. On the other hand, some strains, including Arthrobacter and Mycobacterium, were mutated, selected, and used for catalyzing the Δ1-dehydrogenation of steroids. For instance, the Arthrobacter simplex was employed to transform cortisone acetate to prednisone acetate (Song et al. 2018).
Promising strains were also produced by heterologous expression of KstDs to convert various valuable steroid intermediates (Shao et al. 2017a; Shao et al. 2017b). This approach drew more and more attention because it is time-saving and does not lead to by-product accumulation (Wang et al. 2017). Some of the recent studies are summarized in Table 2. KSDD from M. neoaurum JC-12 was successfully expressed in Bacillus subtilis to produce (1,4-androstene-3,17-dione) ADD from AD (Zhang et al. 2013). Moreover, the heterologous expression of KSDD in Escherichia coli or Corynebacterium crenatum was gradually improved allowing enzyme purification and characterization (Shao et al. 2017a; Zhang et al. 2016). Recently, a number of KstDs from a variety of bacteria were identified and a series of steroids were characterized as preferential substrates. For example, MsKstD1 from M. smegmatis mc2155 allowed the almost complete conversion of 6 g L−1 hydrocortisone (Wang et al. 2017). Furthermore, site-directed mutagenesis of KsdD3 from A. simplex was reported to improve the yields of the corresponding reactions (Mao et al. 2018). In short, the growth conditions and the intrinsic KstD activity are the largest contributors to the bioconversion efficiency.
The crystal structure of KstD1 from R. erythropolis SQ1 (SQ1-KstD1) was resolved previously and gave a deep insight into its catalytic mechanism (Rohman et al. 2013). In addition, several mutations were performed on various KstDs leading to a better understanding of substrate recognition. Such as, the W299A mutation of KsdD3 from A. simplex (KsdD3W299A) improved its specific activity towards substrates including AD and testosterone (Mao et al. 2018). Moreover, the candidate sites reported in M. neoaurum KsdD were proved to be also essential in KsdD3 from A. simplex, although the preferred residues differ. For example, S138 is beneficial to the activity of KsdD from M. neoaurum, but the corresponding residue is H134 in KsdD3 from A. simplex. Furthermore, the H134S mutation leads to the complete inactivation of A. simplex KsdD3 (Mao et al. 2018; Shao et al. 2016). These studies indicate that substrate recognition among the KstD family is similar but involves a number of differences.
In a previous work, we identified KstD2 from M. neoaurum DSM 1381 as a high potential candidate enzyme with yet unknown properties (Zhang et al. 2018). Moreover, according to the latter study, optimizing codon usage and bacterial transformation as well as regulating protein expression greatly improved the bioconversion (Shao et al. 2017a, b). Therefore, in the present work, we investigated the optimal temperature and pH and the impact of metal ions on the reaction as well as the substrate specificity of the enzyme. The best conversion processes were explored and suggested possible application of KstD2 on other valuable steroid substrates. Finally, KstD2 structural modelization was achieved providing some insights in the mechanism of substrate recognition.
Materials and methods
Chemicals, strains, and media
2,6-Dichlorophenolindophenol (DCPIP); phenazine methosulfate (PMS); FAD; hydrocortisone; 17α-OH-P; canrenone; hydrocortisone acetate (HA); 16,17α-epoxyprogesterone; and AD were bought from Shanghai Macklin Biochemical Co., Ltd. (China). 17α-Hydroxypregna-4-ene-3; 20-dione-21-acetate (RSA); androsta-4,9(11)-diene-3,17-dione (NSC 44826); and 9α-hydroxy-4-androstene-3,17-dione (9α-OH-AD) were obtained from Zhejiang Xianju Junye Pharmaceutical Co., Ltd. (China). ClonExpress® II One Step Cloning Kit (Vazyme Biotech Co., Ltd. Nanjing, China) was used for plasmid construction. The other reagents were obtained from Thermo Fisher Scientific.
E. coli DH5α was cultivated in Luria–Bertani medium (LB medium) at 37 °C and 200 rpm for kstD gene cloning. E. coli BL21 (DE3) was used as a host for heterologous expression of KstD2 and was grown in Terrific Broth medium (TB medium). Isopropyl β-d-1-thiogalactopyranoside (IPTG) and kanamycin 50 μg mL−1 were added when necessary.
Cloning and expression
The kstD2 gene (GenBank accession number MG251736) was amplified and cloned into pET28a (Novagen) as described previously (Zhang et al. 2018). The corresponding codon-optimized gene (kstD2opt) was designed for expression in E. coli and synthesized by Sangon Biotech (Shanghai) Co., Ltd. (China). The latter was subsequently inserted into pET28a between the BamHI and HindIII sites with an N-terminal His6 tag. All the primers are listed in Table S1. To express the recombinant KstD2 with a His6 tag at both its C- and N-terminals, the primers kstd2opt-F/kstd2optCH6-R were used to amplify kstD2opt and the product was inserted into the BamHI site of pET28a. The resulting plasmid was called pET28a-kstD2optCH6. Using pET28a-kstD2optCH6 as a template, the primers kstd2opt-F/kstd2optCH9-R were used for PCR amplification prior to cloning into pET28a digested by BamHI and XhoI. pET28a-kstD2optCH9 which has a his9 tag instead of a his6 at the C-terminal was finally obtained. Plasmid sequences were confirmed by Sanger sequencing prior to transformation of E. coli BL21 (DE3) and kanamycin selection.
Site-directed mutagenesis of KstD2 was performed by PCR using the plasmid pET28a-kstD2optCH9 as a template and the primers are listed in Table S1. The PCR products were treated with DpnI and used for transformation of E. coli BL21 (DE3). Mutants were then confirmed by Sanger sequencing.
The recombinant strains were cultivated overnight in 3 mL LB medium containing kanamycin at 37 °C under agitation (200 rpm). The strains were subcultured in 50 mL of TB medium supplemented with kanamycin at 37 °C and 200 rpm until OD600 reached 1.5–2.5. IPTG was then added at a concentration of 0.1 mM for inducing KstD2 expression at 30 °C and 200 rpm during 20 h. Incubation temperature (10–37 °C), IPTG concentration (0–1.0 mM), and induction time (0–24 h) were optimized for efficient protein production. Optimal conditions were used for further experiments unless explicitly stated.
Purification of KstD2
The recombinant KstD2 protein was expressed from pET28a-kstD2optCH9 and purified by affinity chromatography on a Ni-TED Sefinose (TM) Column (Sangon Biotech (Shanghai) Co., Ltd.). Whole cells were collected, resuspended in 100 mL lysis buffer (50 mM Tris-HCl, pH 8.0, 25 μM FAD), and lysed by sonication. Cell extracts were centrifuged at 12,000 rpm for 30 min at 4 °C. The column was subsequently washed with buffer B (50 mM Tris-HCl, pH 8.0, 30 mM imidazole). KstD2 elution was carried out using buffer C (50 mM Tris-HCl, pH 8.0, 250 mM imidazole), and the fraction containing the protein was exchanged into buffer D (20 mM Tris-HCl, pH 8.0,10 μM FAD) using Millipore Amicon® Ultra-15 centrifugal filter concentrators. The samples were stored at − 20 °C after 10–20% glycerol was added. The purified enzyme was subsequently assayed by SDS-PAGE with the procedure described previously (Zhang et al. 2018).
KstD enzymatic assay
The activity of KstD in cell-free extracts as well as the purified KstD2 was measured at 600 nm (ε600 = 18.7 × 103 cm−1 M−1) with a Nano Drop 2000 spectrophotometer (Thermo Scientific) at 40 °C. The reaction mixture (1 mL) contained 50 mM Tris-HCl pH 8.0, 1.5 mM PMS, 0.12 mM DCPIP, diluted cell-free extracts or purified KstD2, and 500 μM steroid substrate in methanol (2%). Three replicates were analyzed. The Bradford method was employed to determine protein contents. One unit of enzyme activity is defined as the reduction of 1 μmol of DCPIP per minute. Specific activities are defined as micromoles per milligrams per minute (U mg−1).
Characterization of the recombinant KstD2
Optimal temperature for KstD2 activity was determined at pH 8.0 by performing 15-min reactions at various temperatures (25–60 °C). The assay was started by the addition of 500 μM AD and 1.5 mM PMS to the reaction mixture. KstD2 activity at 40 °C was defined as 100%. Thermostability of the enzyme was characterized at 40 °C by measuring the residual activity after incubation at temperatures ranging from 25 to 60 °C during 2 h in Tris-HCl buffer (50 mM, pH 8.0). Residual KstD2 activity at 25 °C was defined as 100%.c
The optimal pH of KstD2 was evaluated at 40 °C at varying pH values in 50 mM buffers (3.0–6.0, citric acid buffer; 6.0–8.0, sodium phosphate buffer; 8.0–9.0, Tris-HCl buffer; 9.0–10.0, Gly-NaOH buffer). pH stability was determined by incubating the enzyme at 4 °C during 2 h and the residual activity was measured at pH 8.0 and 40 °C. KstD2 activity with Tris-HCl buffer at pH 8.0 was defined as 100%.
The effect of NaCl, KCl, and MgCl was determined by adding the different compounds at final concentrations ranging from 0 to 50 mM. In order to measure the KstD2 stability, the purified enzyme was incubated with various concentrations of NaCl (0–500 mM) at different temperatures (4 °C, 25 °C, 35 °C, 45 °C) during 2 h. The relative enzyme activity was determined under standard assay conditions.
Effects of metal ions on KstD activity
To investigate the effect of metal ions (K+, Na+, Mg2+, Ca2+, Mn2+, Cu2+, Ni2+, Fe3+) and ethylenediaminetetraacetic acid (EDTA) on the activity of the purified KstD2, 1 mM of the corresponding chloride salts was added to the reaction. The activity of the control experiment (without salt) was defined as 100%.
Substrate specificity of purified KstD2
The substrate specificity of KstD2 was determined by measuring enzyme activity with 500 μM AD; hydrocortisone; 17α-OH-P; canrenone; HA; 16,17α-epoxyprogesterone; RSA; NSC 44826; or 9α-OH-AD as substrates and using the corresponding optimal pH and temperature. Kinetic parameters of the Michaelis–Menten equation were determined with the use of origin 8.0 by employing Hill function with n = 1. All measurements were performed in triplicate.
Analytical methods
One milliliter of samples was extracted with 2 mL of ethyl acetate twice. The supernatants were then mixed and dried under vacuum. Samples were dissolved in methanol prior to high-performance liquid chromatography (HPLC) analysis. Separation was performed on an Agilent XDB-C18 column (4.6 × 250 mm; 40 °C) and a UV/visible detector (254 nm) was employed to detect the steroid substrate conversion rates with methanol/water (70:30, v/v). The flow rate was 0.6 mL min−1.
Biocatalytic conditions with resting cells of E. coli
The recombinant E. coli BL21/pET28a-kstD2optCH9 strain was grown and submitted to induction under optimal conditions during 12 h. Cells were harvested by centrifugation at 7000 rpm for 10 min and resuspended in 15–20 mL of buffers (50 mM) containing AD and emulsified by hydroxypropyl-β-cyclodextrin (HP-β-CD, molar ratio to AD was 1:1) and Tween 80 (0.1%, v/v). Final cell and AD concentrations were 10 g L−1 and 5 g L−1, respectively. The optimal temperature of bioconversions was carried out with Tris-HCl buffer (pH 8.0) in 250-mL shake flasks at 200 rpm and cells were sampled after 2 h. For determining optimal pH, the same biocatalytic processes were implemented at various pH values and 40 °C. The influence of the dosage of wet cells (5, 10, 20, 50, 80, and 100 g L−1) on the bioconversion of 10 g L−1 AD was investigated at optimal pH and temperature. Fifty grams per liter of wet cells were used to measure the effect of different AD concentrations (20, 30, 40, 50 g L−1) on the conversion rate. Finally, the performances of the recombinant E. coli strain on selected steroid substrates were also investigated.
Structure modeling of KstD2
The sequence similarity network (SSN) for 3-ketosteroid-Δ1-dehydrogenase (KstD) was created using EFI-EST (https://efi.igb.illinois.edu/efi-est/). The input sequences were from FAD_binding_2 InterPro family (IPR003953). The SSN was displayed with an e-value threshold of 10−150. Cytoscape v3.6.1 (Shannon et al. 2003) was used for SSN visualization and analysis.
KstD2 (Uniprot: A0A2P1IUZ5) model was built based on that of KstD1(PDB: 4C3Y) using Schrödinger Suite 2018-2. The structure of 4C3Y was processed by Protein Preparation Wizard. The model of KstD2 was also optimized by Protein Preparation Wizard for analysis.
Multiple sequence alignment was performed by Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), and weblogo (https://weblogo.berkeley.edu/logo.cgi) was used to generate sequence logos.
Accession number
The nucleic acid sequence of kstD2opt has been deposited in the GeneBank database with the accession number MK040595. The accession number of the corresponding amino acid sequence is AVN89960.1.
Results
Purification and characterization of the recombinant KstD2
In order to purify KstD2, its expression was enhanced according to previous reports (Shao et al. 2017a). Using codon optimization, low expression temperature (25 °C), optimum IPTG concentration (0.1 mM), and proper induction time (12 h), the intracellular KstD2 activity reached up to 1585.5 U mg−1 (Fig. S1). Purification was then performed by affinity chromatography using the his6 and his9 tags present at the N- and C-terminal, respectively (Fig. S2). The impact of temperature, pH, NaCl concentration, and metal ions on KstD2 activity was investigated.
This revealed that the optimal temperature of the enzyme is 40 °C, while it also displayed relatively high activities between 25 °C and 45 °C, corresponding to more than 90% activity compared with that observed at 40 °C (Fig. 2a). Thermal stability of the recombinant KstD2 slightly decreased with temperature increase from 25 °C to 40 °C. Meanwhile, the stability was seriously impaired when the temperature reached 45 °C, and enzyme activity was completely inhibited after incubation at 50 °C or higher during 2 h (Fig. 2b).
Effect of the temperature, pH, and metal ions on the activity and stability of recombinant KstD2. Effect of temperature on recombinant KstD2 activity (a) and stability (b). Effects of pH on recombinant KstD2 activity (c) and stability (d). c Effect of metal ions and EDTA on KstD2 activity (e) and influence of Na+ concentrations on recombinant KstD2 stability (f)
The optimum pH was 8.0 (Fig. 2c). The enzyme exhibited about 50% and 42% activity at pH 6.0 and pH 9.0, respectively. Moreover, KstD2 was almost completely deactivated at pH 3.0, pH 4.0, and pH 10.0. On the other hand, the enzyme was relatively stable at pH ranging from 6.0 to 9.0. It retained only 21.9% and 33.6% activity compared with pH 8.0 with Tris-HCl buffer after 2-h incubation at pH 5.0 and 10.0, respectively. Furthermore, Tris-HCl appeared as a better buffer compared with sodium phosphate (Fig. 2d).
Metal ions showed opposite effects on the activity of KstD2. The latter was slightly affected by 1 mM Na+, K+, Mg2+, or EDTA (Fig. 2e). It was almost completely inhibited by Ca2+, Mn2+, and Cu2+ and relatively less inhibited by Fe3+ and Ni2+ with 40.6–59.1% of remaining activity, respectively.
In addition, phosphate-buffered saline (PBS) or Tris-HCl buffer containing high NaCl concentration was found not suitable for the efficient production of active recombinant KstD2 during purification. Thus, the effect of different concentrations of NaCl was also investigated. This analysis showed KstD2 activity was slightly influenced by NaCl with final concentration ranging from 20 to 50 mM (Fig. S3a). Nevertheless, the presence of high NaCl concentration impaired KstD2 stability, especially at high temperature. At 20 mM, the remaining activity of KstD2 was hardly decreased after 2 h of incubation at the four selected different temperature (4, 25, 35, 45 °C) (Fig. 2f). Enzyme stability was more seriously altered when the final NaCl concentration was increased. The relative activity of the enzyme dropped down to 59.8% at 4 °C and 500 mM and was completely inhibited at 35 °C and 500 mM after 2 h. The same was observed with KCl or MgCl (Fig. S3 and Fig. S4).
Substrate specificity
The Km (Michaelis–Menten constant) and kcat (catalytic rate constant towards various substrates) of KstD2 are displayed in Table 1. These parameters allowed to estimate the catalytic efficiency (kcat/Km). The kcat of KstD2 was much higher than those described previously for the other isoenzymes indicating the excellent property of KstD2.
The catalytic efficiency (kcat/Km) favored 17α-OH-P (470.91 × 106 M−1 s−1) as a substrate. High kcat/Km value towards 16,17α-epoxyprogesterone was also observed. Lower kcat (8882.95 s−1) and kcat/Km (67.13 × 106 M−1 s−1) values were measured using AD as a substrate. KstD2 had the lowest affinity (832.72 μM) and kcat (1862.9 s−1) toward hydrocortisone and 9α-OH-AD, respectively. Furthermore, when the hydroxyl group at C21 of hydrocortisone was substituted by an acetyl group, the Δ1-dehydrogenation reaction was largely promoted. Indeed KstD2 displayed a much higher catalytic efficiency with HA compared with hydrocortisone. The hydroxyl group at C11 hindered the activity of KstD2 since the activity was higher on RSA than on HA. Moreover, the activity of KstD2 on NSC 44826 was lower than that observed with AD indicating that the C=C structure at C9–C11 hampers the reaction. The Δ1-dehydrogenation reaction catalyzed by KstD2 on different substrates varied depending on the structure of the compound.
Optimization of the conditions for improving AD conversion rate
The conversion rates using 5 g L−1 of AD and 10 g L−1 of wet cells were measured at different temperatures and pH. The highest conversion rate was 85.2 % after 2 h and was obtained at 40 °C and pH 8.0. The conversion rate decreased when the temperature was shifted away from the optimum. To investigate the influence of the biomass on the conversion rate, various concentrations of wet cells were used and the bioconversion was carried out with 10 g L−1 of AD under optimal conditions. As shown in Fig. 3, both the conversion speed and rate rose with the increase of E. coli cell weight. Fifty grams per liter of wet cells were used in the subsequent experiments as the conversion rate was up to 99.6% at this concentration after 2.5 h. To further determine the potential of E. coli/pet28a-kstD2CH9, the AD concentration was increased to 20, 30, 40, and 50 g L−1. After 8 h, AD (20 g L−1) was almost completely converted to ADD (99.32%). Furthermore, when using a concentration of 30 g L−1, 99.0% of AD was transformed to ADD. Forty grams per liter and 50 g L−1 of AD were also used, and conversion rates were 86.5% and 77.7%, respectively.
Bioconversion of steroids by resting cells
The bioconversion of another four valuable steroids, including canrenone; 16,17α-epoxyprogesterone; 17α-OH-P; and NSC 44826, were investigated over 24 h using E. coli resting cells (Fig. 4). About 99% of NSC 44826 (10 g L−1 and 20 g L−1) were transformed in 8 h. 89.3% of NSC 44826 could be converted when using a concentration of 30 g L−1 and the conversion rate hardly increased after 8 h. The performances of resting cells on 17α-OH-P and canrenone were quite similar at the three concentrations. The conversion rate reached approximately 77.8–80.6% and 73.0–78.2% at 8 h, respectively, and increased slightly to 83.9–86.7% and 81.6–88.2% at 24 h, respectively. For 16,17α-epoxyprogesterone, the conversion rate declined sharply with 46.6%, 72.1%, and 89.6% for 10 g L−1, 15 g L−1, and 20 g L−1, respectively, after 8 h. On the other hand, it hardly increased from 8 h to 24 h. Even though the purified KstD2 showed much higher activities on 17α-OH-P and 16,17α-epoxyprogesterone compared with AD, the performance of E. coli BL21 (DE3) harboring KstD2 was barely satisfactory. We speculate that the reasons may be the lower solubilities under the given conditions compared with AD and NSC 44826.
Sequence and structural analysis of the KstD2 homology model
In order to further the study of KstD2 protein, the sequence similarity network (SSN) of FAD_binding_2 family (IPR003953) was constructed and displayed with an e-value cutoff of 10−150 (share ~ 50% sequence identity inside each cluster). The 3-ketosteroid-Δ1-dehydrogenases with clear function from different organisms were located into four clusters (Fig. 5a, Fig. S5). Moreover, KstD2 and SQ1-KstD1 (for which the crystal structure was resolved, PDB: 4C3Y) belong to clusters 2 and 1, respectively. According to previous studies (Mao et al. 2018; Shao et al. 2016; Xie et al. 2015), the amino acid residues of the catalytic pocket are involved in KstD activity. So, to further understand the function of KstD2, we used 4C3Y as a template to build a homology model of KstD2. The residues of KstD2 and SQ1-KstD1 within a 4 Å distance from the AD molecule were used to define the substrate binding site. From the overall structure (Fig. 5b), most of the sequence was well aligned apart from two short loops close to the substrate binding site in KstD2. The latter was highlighted in light blue (Fig. 5c). We performed multiple sequence alignment for cluster 1 and cluster 2 and used the Weblogo to generate sequence logo graphically representing the amino acid conservation (positions) in the substrate binding site (Fig. 5d).
Sequence and structural analysis of the KstD2 homology model. a The sequence similarity network (SSN) of KstDs. b Ribbon representation of the predicted KstD2 overall structure (light blue) and its model template (4C3Y, pink). FAD and AD are shown as orange and red stick. c The residues of KstD2 and 4C3Y within 4 Å to AD. AD was colored as orange and red stick, and the predicted unique sites for KstD2 within this area were highlighted in light blue and labeled in red. d Comparison of substrate binding motifs of cluster 1 and cluster 2 which contain SQ1-KstD1 and KstD2, respectively. And the unique sites for cluster 2 were also labeled in red corresponding to c
It was reported that Y119, Y359, Y459, and G491 are essential for the dehydrogenation reaction in SQ1-KstD1 and the counterpart residues in KstD2 are Y121, Y359, Y459, and G536 (Rohman et al. 2013). In addition to the four key residues, the G50, L492 and Y532 were also conserved, which corroborate their significant role in substrate binding. Besides, in a previous study, the mutant Y541F (Y532 in KstD2) of M. neoaurum obviously displayed reduced KstD1 activity (Qin et al. 2017).
Several residues were conserved and unique in cluster 2. This includes E136, L334, Y463, and T538. In addition, the positions from M176 to P180 were located into a loop which was not conserved in cluster 2. This excluded R178, which was close to the C17 position of the steroid substrates and is mainly occupied by basic amino acids. Consequently, we mutated the R178 to various amino acids as shown in Fig. S6. As expected, the R178K mutant retained most of the activity on AD compared with wild type KstD2. On the contrary, when it was substituted by a Y which is aromatic amino acid, the activity of KstD2 on AD was completely inhibited. This result, on the one hand, confirms the importance of R178 in the catalysis. Moreover, it proves the reliability of our homology-based structural analysis. Thus, the residues mentioned above probably contribute to the substrate recognition and catalytic reaction of KstDs of cluster 2.
To summarize, the high activity and substrate specificity of KstD2 is probably conferred by unique residues of the substrate binding site. As the activity of KstD2 on AD is much higher than SQ1-KstD1, the unique five residues which were labeled in red attracted much attention, especially R178. Overall, this is the first time that a KstD from cluster 2 was purified and characterized.
Discussion
The properties of numerous KstDs from various hosts were investigated in recent years (Mao et al. 2018; Wang et al. 2017; Zhang et al. 2015, 2016). These enzymes play important functions in steroid degradation and significant applications can be implemented in the production of steroid drugs (Donova and Egorova 2012). However, none of the KstDs from cluster 2 was purified and characterized. Moreover, there is an important need for KstDs with higher activity and interesting properties. Thus, the KstD2 from M. neoaurum DSM 1381 that has been previously reported as a good candidate was purified, characterized, and used to transform various steroids.
KstD2 could easily be expressed at high levels in E. coli, but the enzyme was mainly accumulated in the insoluble fraction (Fig. S2). Both codon-optimization and improvement of the induction conditions were implemented to enhance soluble expression of the enzyme. During protein purification, KstD2 slightly binds to the Ni2+-column if the protein only contains a his6 tag at the N-terminal. Purification was much more efficient when a his6 tag was added at the C-terminal and further improved by lengthening the his6 tag to a his9 tag (Fig. S7).
The optimal pH of KstD2 was pH 8.0, which is also the best pH value for storing the protein. KstD2 stability was best in Tris-HCl buffer. We speculated that Na+ (present in the sodium phosphate buffer) was the causal compound since the stability of KstD2 reduced when NaCl was supplied to the Tris-HCl buffer. Furthermore, the effect of NaCl increased with the temperature and this phenomenon was first observed with KstD2, since the KsdD3 from A. simplex was reported to be stored with 0.2 M NaCl (Mao et al. 2018). Surprisingly, most of the selected metal ions inhibited KstD2. This does not correlate with the observation that Ca2+ strongly stimulates the activity of KSDD from M. neoaurum JC-12 (Zhang et al. 2016). Another trait of KstD2 is the relatively high optimal temperature of 40 °C and that it keeps high activity over a broad temperature ranging from 25 to 45 °C. By contrast, most of the well-studied KstDs showed highest activity under 30 °C (Mao et al. 2018; Wang et al. 2017; Zhang et al. 2016). This trait is strongly conducive to its industrial application.
Besides the specificities mentioned above, another more remarkable feature is its high catalytic rate (kcat) and catalytic efficiency (kcat/Km) towards various substrates, especially AD, 17α-OH-P, and 16,17α-epoxyprogesterone. The enzyme catalytic efficiency (kcat/Km) for AD was 67.13 × 106 M−1 s−1, which was almost 20 times higher than that of the KsdD3W299A reported recently (Mao et al. 2018). The catalytic efficiency (kcat/Km) for17α-OH-P was also much higher than that of MsKstD1 reported by Wang et al. (2017). Moreover, the substrate profile of KstD2 was also different from MsKstD1 and ReKstD. For example, MsKstD preferred hydrocortisone and 9α-OH-AD rather than AD and the kcat/Km of ReKstD towards the three substrates was similar (Wang et al. 2017). However, for KstD2 from M. neoaurum DSM 1381, the kcat/Km for AD was much higher than for hydrocortisone and 9α-OH-AD. Besides, both the C=C structure at C9–C11 and hydroxyl groups at C9 and C11 reduced the efficiency of the Δ1-dehydrogenation reaction catalyzed by KstD2.
In the recent years, several studies of the Δ1-dehydrogenation of steroid substrates employed heterologously expressed KstDs from various organisms (Table 2). This is only related to cluster 3 and cluster 4 KstDs. No KstD of cluster 2 was purified and applied in this process. In addition to the excellent properties mentioned above, the recombinant E. coli BL21 (DE3) harboring KstD2 showed impressive performance when used for Δ1-dehydrogenation of steroid substrates. It also displayed the highest feedstock of AD (30 g L−1) ever reported with the achievement of a 99% conversion rate. Another valuable steroid named NSC 44826 was also completely catalyzed when supplied with 20 g L−1 precursors. This performance was much better than that of KstD3gor expressed in E. coli which was able to transform 2 g L−1 of NSC 44826 with a 96% conversion rate. Nevertheless, only 90.0% of the 30 g L−1 of NSC 44826 were transformed, which implies that KstD2 prefers steroid substrates without C=C structure at C9–C11. Considering the catalytic efficiency (kcat/Km) towards 16,17α-epoxyprogesterone and 17α-OH-P is much higher than towards AD; the poor performances on the 16,17α-epoxyprogesterone and 17α-OH-P compared with AD may be due to the inappropriate conversion conditions and may probably be improved by increasing substrates’ solubility (Table 1). On the other hand, the main reason for the relatively low conversion rate on canrenone is the low catalytic efficiency of KstD2. All in all, KstD2 was proved as a high potential KstD performing much better than previously reported enzymes (Table 2), and KstDs of cluster 2 deserve more attention.
Then, we attempted to understand the high KstD2 activity by structural analysis and multiple sequence alignment of cluster 1 and cluster 2 enzymes. As shown in Fig. 5, KstD2 shares a lot of conserved residues with its homologs in cluster 2 at the substrate binding sites. This may explain the similar properties between these proteins. For example, the Km values of KstD2s from M. neoaurum DSM 1381, R. erythropolis SQ1, and R. ruber strain Chol-4 on AD are much lower than on 9α-OH-AD (Guevara et al. 2017; Knol et al. 2008; Zhang et al. 2018). Besides, R178 was identified as an important residue responsible for KstD2 activity. This residue is conserved and unique in cluster 2. Some other residues were also specific to KstD2 among the cluster 2. These residues may be potential sites for developing a more efficient KstD based on the structure of KstD2. Thus, we hope that more attention will be paid to cluster 2 KstDs which may lead to a deeper insight in the KstD family.
In short, the high activity KstD isoform from M. neoaurum DSM 1381 belongs to cluster 2. In the present study, the enzyme was purified and characterized for the first time. It displayed high specific activities on various substrates, especially AD, NSC 44826, 17α-OH-P, and 16,17α-epoxyprogesterone. Moreover, its relatively high optimal temperature of 40 °C also benefits to its industrial application. However, it is sensitive to many metal ions which should capture our attention. Furthermore, its high conversion efficiency on selected steroid substrates further proved its excellent properties. The analysis of its structure and the cluster 2 KstD sequences revealed that two short loops, which are close to the substrate binding site, are specific to cluster 2. Finally, a conserved residue, R178, is located in one of these loops and indeed plays an important role for the activity of KstD2.
References
Bhatti HN, Khera RA (2012) Biological transformations of steroidal compounds: a review. Steroids 77(12):1267–1290. https://doi.org/10.1016/j.steroids.2012.07.018
Donova MV, Egorova OV (2012) Microbial steroid transformations: current state and prospects. Appl Microbiol Biotechnol 94(6):1423–1447. https://doi.org/10.1007/s00253-012-4078-0
Fernandes P, Cruz A, Angelova B, Pinheiro HM, Cabral JMS (2003) Microbial conversion of steroid compounds: recent developments. Enzym Microb Technol 32(6):688–705. https://doi.org/10.1016/s0141-0229(03)00029-2
Fernandez-Cabezon L, Galan B, Garcia JL (2018) New insights on steroid biotechnology. Front Microbiol 9:958. https://doi.org/10.3389/fmicb.2018.00958
García J, Uhía I, Galán B (2012) Catabolism and biotechnological applications of cholesterol degrading bacteria. Microb Biotechnol 5(6):679–699. https://doi.org/10.1111/j.1751-7915.2012.00331.x
Guevara G, Fernandez de Las Heras L, Perera J, Navarro Llorens JM (2017) Functional differentiation of 3-ketosteroid Δ1-dehydrogenase isozymes in Rhodococcus ruber strain Chol-4. Microb Cell Factories 16(1):42. https://doi.org/10.1186/s12934-017-0657-1
Knol J, Bodewits K, Hessels GI, Dijkhuizen L, Van der Geize R (2008) 3-Keto-5 alpha-steroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem J 410:339–346. https://doi.org/10.1042/bj20071130
Mao S, Wang JW, Liu F, Zhu Z, Gao D, Guo Q, Xu P, Ma Z, Hou Y, Cheng X, Sun D, Lu F, Qin HM (2018) Engineering of 3-ketosteroid-(1)-dehydrogenase based site-directed saturation mutagenesis for efficient biotransformation of steroidal substrates. Microb Cell Factories 17(1):141. https://doi.org/10.1186/s12934-018-0981-0
Qin N, Shen Y, Yang X, Su L, Tang R, Li W, Wang M (2017) Site-directed mutagenesis under the direction of in silico protein docking modeling reveals the active site residues of 3-ketosteroid-Delta(1)-dehydrogenase from Mycobacterium neoaurum. World J Microbiol Biotechnol 33(7):146. https://doi.org/10.1007/s11274-017-2310-x
Rohman A, van Oosterwijk N, Thunnissen A-MWH, Dijkstra BW (2013) Crystal structure and site-directed mutagenesis of 3-ketosteroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 explain its catalytic mechanism. J Biol Chem 288(49):35559–35568. https://doi.org/10.1074/jbc.M113.522771
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Shao ML, Zhang X, Rao ZM, Xu MJ, Yang TW, Li H, Xu ZH, Yang ST (2016) A mutant form of 3-ketosteroid-Δ(1)-dehydrogenase gives altered androst-1,4-diene-3, 17-dione/androst-4-ene-3,17-dione molar ratios in steroid biotransformations by Mycobacterium neoaurum ST-095. J Ind Microbiol Biotechnol 43(5):691–701. https://doi.org/10.1007/s10295-016-1743-9
Shao M, Chen Y, Zhang X, Rao Z, Xu M, Yang T, Li H, Xu Z, Yang S (2017a) Enhanced intracellular soluble production of 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum in Escherichia coli and its application in the androst-1,4-diene-3,17-dione production. J Chem Technol Biotechnol 92(2):350–357. https://doi.org/10.1002/jctb.5012
Shao M, Sha Z, Zhang X, Rao Z, Xu M, Yang T, Xu Z, Yang S (2017b) Efficient androst-1,4-diene-3,17-dione production by co-expressing 3-ketosteroid-Δ1-dehydrogenase and catalase in Bacillus subtilis. J Appl Microbiol 122(1):119–128. https://doi.org/10.1111/jam.13336
Song B, Zhou Q, Xue HJ, Liu JJ, Zheng YY, Shen YB, Wang M, Luo JM (2018) IrrE Improves organic solvent tolerance and Δ(1)-dehydrogenation productivity of Arthrobacter simplex. J Agric Food Chem 66(20):5210–5220. https://doi.org/10.1021/acs.jafc.8b01311
Wang X, Feng J, Zhang D, Wu Q, Zhu D, Ma Y (2017) Characterization of new recombinant 3-ketosteroid-Δ1-dehydrogenases for the biotransformation of steroids. Appl Microbiol Biotechnol 101(15):6049–6060. https://doi.org/10.1007/s00253-017-8378-2
Wu Y, Li H, Zhang X-M, Gong J-S, Rao Z-M, Shi J-S, Zhang X-J, Xu Z-H (2015) Efficient hydroxylation of functionalized steroids by Colletotrichum lini ST-1. J Mol Catal B-Enzym 120:111–118. https://doi.org/10.1016/j.molcatb.2015.07.003
Xie R, Shen Y, Qin N, Wang Y, Su L, Wang M (2015) Genetic differences in ksdD influence on the ADD/AD ratio of Mycobacterium neoaurum. J Ind Microbiol Biotechnol 42(4):507–513. https://doi.org/10.1007/s10295-014-1577-2
Yao K, Xu LQ, Wang FQ, Wei DZ (2014) Characterization and engineering of 3-ketosteroid- big up tri, open1-dehydrogenase and 3-ketosteroid-9alpha-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9alpha-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab Eng 24:181–191. https://doi.org/10.1016/j.ymben.2014.05.005
Zhang W, Shao M, Rao Z, Xu M, Zhang X, Yang T, Li H, Xu Z (2013) Bioconversion of 4-androstene-3,17-dione to androst-1,4-diene-3,17-dione by recombinant Bacillus subtilis expressing ksdd gene encoding 3-ketosteroid-Delta(1)-dehydrogenase from Mycobacterium neoaurum JC-12. J Steroid Biochem Mol Biol 135:36–42. https://doi.org/10.1016/j.jsbmb.2012.12.016
Zhang Q, Ren Y, He J, Cheng S, Yuan J, Ge F, Li W, Zhang Y, Xie G (2015) Multiplicity of 3-ketosteroid Δ1-dehydrogenase enzymes in Gordonia neofelifaecis NRRL B-59395 with preferences for different steroids. Ann Microbiol 65(4):1961–1971. https://doi.org/10.1007/s13213-015-1034-0
Zhang X, Wu D, Yang TW, Xu MJ, Rao ZM (2016) Over-expression of Mycobacterium neoaurum 3-ketosteroid-Δ1-dehydrogenase in Corynebacterium crenatum for efficient bioconversion of 4-androstene-3,17-dione to androst-1,4-diene-3,17-dione. Electron J Biotechnol 24:84–90. https://doi.org/10.1016/j.ejbt.2016.10.004
Zhang RJ, Liu XC, Wang YS, Han YC, Sun JS, Shi JP, Zhang BG (2018) Identification, function, and application of 3-ketosteroid Δl -dehydrogenase isozymes in Mycobacterium neoaurum DSM 1381 for the production of steroidic synthons. Microb Cell Factories 17:16. https://doi.org/10.1186/s12934-018-0916-9
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
This research was funded by the grants from the State Key Project of Research and Development Plan (grant number, 2017YFE0112700).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This article does not contain any studies with human participants or animals performed by the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 690 kb)
Rights and permissions
About this article
Cite this article
Zhang, R., Xu, X., Cao, H. et al. Purification, characterization, and application of a high activity 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum DSM 1381. Appl Microbiol Biotechnol 103, 6605–6616 (2019). https://doi.org/10.1007/s00253-019-09988-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09988-5