Abstract
Some of the most important natural pigments have been produced from fungi and used for coloring in food, cosmetics, textiles, and pharmaceutical products. Forty-seven isolates of endophytic fungi were isolated from Cinnamomum zeylanicum in northern Thailand. Only one isolate, CMU-ZY2045, produced an extracellularly red pigment. This isolate was identified as Nigrospora aurantiaca based on morphological characteristics and the molecular phylogenetic analysis of a combined four loci (large subunit and internal transcribed spacer of ribosomal DNA, β-tubulin, and translation elongation factor 1-alpha genes). The optimum conditions for red pigment production from this fungus were investigated. The results indicated that the highest red pigment yield was observed in the liquid medium containing glucose as a carbon source and yeast extract as a nitrogen source, at a pH value of 5.0 and at 27 °C with shaking for 5 days. The crude red pigment revealed the highest level of solubility in methanol. A fungal red pigment was found to have high stability at temperatures ranging from 20 to 50 °C and pH values at a range of 5.0–6.0. Based on liquid chromatography-mass spectrometry analyses, the red pigment was characterized as bostrycin. The extracted pigment was used for the textile dyeing process. Crude fungal red pigment revealed the highest staining ability in cotton fabrics and displayed excellent fastness to washing, which showing negative cytotoxicity at the concentrations used to cell culture. This is the first report on bostrycin production from N. aurantiaca.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pigments are used for coloring in food, cosmetics, textiles, and pharmaceutical products and can attract the attention of consumers to specific products (Kumar et al. 2015). Nowadays, consumer concern has increased regarding the potential long-term toxicity of synthetic pigments in food processing, cosmetics, pharmaceuticals, and the textile industries due to their carcinogenicity, hyperallergenicity and other potential toxicological problems (Osman et al. 2004; Unagul et al. 2005). Hence, the interest in natural pigments derived from plants and microorganisms continues to increase and many research efforts have been made to replace synthetic pigments with natural pigments (Akilandeswari and Pradeep 2016; Malik et al. 2012). Microbial pigments are advantageous over plants in term of their high availability, stability and yield, low residues, and easy harvesting (Durán et al. 2002; Joshi et al. 2003; Venil and Lakshmanaperumalsamy 2009). Microorganisms including many types of bacteria, fungi, algae, and protozoa are recognized as potential sources for various pigments, e.g., carotenoids, flavins, melamins, quinines, and more specifically monascin, violacein, phycocyanin, or indigo (Dufossé et al. 2005; Torres et al. 2016). Many microbial pigments not only act as coloring agents, but also possess antioxidant, anti-inflammation, and antimicrobial activities (Venil and Lakshmanaperumalsamy 2009). However, the limitations of high-yield production of microbial pigments on a large-scale can be affected by various microbial strains, the fermentation process, cell growth, and nutrition content (Dufossé et al. 2014; Joshi et al. 2003; Panesar et al. 2015). To defeat these constraints, selection of the appropriate strain, mutations or genetic engineering techniques, and the selection of the appropriate fermentation process can all help to significantly improve pigment production yields, as well as the selection of suitable substrates for cultivation (Arumugam et al. 2014; Mapari et al. 2005).
Filamentous fungi have been identified as potential pigment-producing microorganisms (Babitha et al. 2007). Many fungi in the genera Alternaria, Aspergillus, Ashbya, Blaskeslea, Curvularia, Epicoccum, Fusarium, Monascus, Nigrospora, Penicillium, and Trichoderma have been reported as pigment producers (Akilandeswari and Pradeep 2016; Dufossé et al. 2014; Sharma et al. 2012; Torres et al. 2016). Generally, fungal pigments have been used as food colorants, e.g., ankaflavin, monascin, monascorubin, and rubropunctatin, from the Monascus species, Arpinkred™ (anthrachinone-type) from Penicillium oxalicum, β-carotene, and lycopene from Blakeslea trispora, and riboflavin from Ashbya gossypii (Dufossé et al. 2014). The application of fungal pigments in the dyeing of cotton, silk, and wool has been evaluated (Chadni et al. 2017; Mabrouk et al. 2011; Nagia and El-Mohamedy 2007; Perumal et al. 2009; Sharma et al. 2012; Velmurugan et al. 2010; Hinsch et al. 2015; Palomino et al. 2017). In this study, endophytic fungi were isolated from cinnamon plants, and the pigment-producing strains were screened. The appropriate pigment-producing strain was selected and the optimal culture conditions (liquid culture medium, temperature, pH, carbon and nitrogen sources, and cultivation period) for pigment production were determined. The values associated with the solubility, stability, and cytotoxicity of the crude pigment were evaluated. The fungal pigment was purified and identified. Moreover, the crude pigment extract was applied for multifiber dyeing. Therefore, the knowledge acquired from this study could lead to the development of pigments production from endophytic fungi as textile colorants, which may replace the toxic colorants that are currently being used.
Materials and methods
Fungal isolation
Endophytic fungi were isolated from healthy mature leaves and stems of cinnamon plants (Cinnamomum zeylanicum L.) collected from the University of Phayao, Phayao Province, Thailand, in February 2008. The cinnamon leaves and stems were washed in running tap water for 15 min and cut into small pieces (5 mm2 leaf and 1 mm length stem). The cut samples were triple surface-sterilized (70% ethanol for 1 min, 2% sodium hypochlorite for 3 min and 95% ethanol for 30 s) under a laminar flow hood. Twenty-five leaf and stem pieces were placed on potato dextrose agar (PDA) supplemented with 50 ppm of chloramphenicol and 35 ppm of Rose Bengal. The plates were incubated at 25 °C in darkness. The fungi growing out from the tissue samples were transferred to fresh PDA plates. The pure cultures were preserved on PDA slants for the short-term and 20% glycerol at − 20 °C for the long-term preservation.
Identification of pigment-producing endophytic fungus
Morphological studies
Conventional morphological characters were used to tentatively identify the selected pigment-producing endophytic fungi. Colony characteristics including aerial mycelium, density and pigment production were recorded. Micromorphological characteristics were examined using a light microscope (Olympus CX51, Japan). Size data of the anatomical features are based on at least 50 measurements of each structure.
Molecular studies
Molecular techniques were used to confirm the identification of pigment-producing endophytic fungi. Genomic DNA was extracted from 1-week-old fungal mycelia on PDA (1–5 mg) using a DNA Extraction Mini Kit (FAVORGEN, Taiwan) following the manufacturer’s protocol. The large subunit (LSU) and internal transcribed spacer (ITS) of ribosomal RNA, β-tubulin (TUB) and translation elongation factor 1-alpha (TEF1) genes were amplified with primers and annealing temperatures following the previous studies (Table 1). Polymerase chain reaction (PCR) was performed in 20 μL reaction containing 1.0 μL DNA template, 1.0 μL of each forward and reverse primers, 10.0 μL 2X Quick Taq HS DyeMix (TOYOBO, Japan), and 7 μL deionized water, and the following thermal conditions: 94 °C for 2 min, followed by 35 cycles of 94 °C for 2 min, the temperatures dependent on the amplified gene (Table 1) for 1 min and 72 °C for 1 min, and a final 72 °C for 10 min on a peqSTAR thermal cycler (PEQLAB Ltd., UK). PCR products were purified using a PCR clean up Gel Extraction NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Germany). The purified PCR products were directly sequenced. Sanger sequencing was carried out by 1ST Base Company (Kembangan, Malaysia) using the PCR primers mentioned above. Sequences were used to query GenBank via BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
For phylogenetic analysis, the sequences obtained from this study and from previous studies along with sequences from GenBank database were used. The multiple sequence alignment was carried out using MUSCLE (Edgar 2004). A maximum likelihood (ML) phylogenetic tree was constructed using RAxML v7.0.3 (Stamatakis 2006), applying the rapid bootstrapping algorithm for 1000 replications using the GTRGAMMA model. Arthrinium arundinis and Ar. sacchari were used as the outgroup. The ML trees were viewed with TreeView32 (Page 2001). Clades with bootstrap values (BS) ≥ 70% were considered as significantly supported (Hillis and Bull 1993). Bayesian phylogenetic analyses were carried out using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method in MrBayes version 3.2 (Ronquist et al. 2012), under a GRT + I + G model. Markov chains were run for 1,000,000 generations, with six chains and random starting trees. The chains were sampled every 100 generations. Among these, the first 2000 trees were discarded as the burn-in phase of each analysis and the resulting trees were used to calculate Bayesian posterior probabilities. Bayesian posterior probabilities (PP) ≥ 0.95 were considered as significant support (Alfaro et al. 2003).
Optimization of fungal pigment production
Fungal cultivation and pigment yield estimation
Three fungal mycelial plugs (5 mm in diameter) obtained from the periphery of the growing colony on PDA at 30 °C for 1 week were transferred into a 25-mL of liquid medium in each 125-mL Erlenmeyer flask after being autoclaved at 121 °C for 15 min. Cultivation was performed in the dark at 30 °C with shaking at 125 rpm on a reciprocal shaker. After incubation, the cultures were centrifuged at 11,000 rpm for 15 min to harvest the supernatant. The fungal mycelia were dried at 30 °C for 48 h and maintained in desiccators for 20 min before being weighed. Only the secreting pigment was considered in this study. Analysis of pigment production was carried out by measurement of the maximum level of absorbance (λmax) of pigments using a spectrophotometer (BOEGO spectrophotometer model S-220 UV/VIS, Germany). Pigment yield in the supernatant was estimated following the method described by Tseng et al. (2000) in which the wavelength of its absorbance maxima was expressed in absorbance unit (AU). Three replications were performed for each treatment.
Effect of culture liquid medium
Five different liquid media were used in this experiment; potato dextrose broth (PDB; Conda®, Spain), Czapek Dox broth (CDB; Signma-Aldrich, USA), glucose yeast extract medium (GY; glucose10 g, yeast extract 2 g), malt extract medium (MM; malt extract 20 g), and oatmeal medium (Difco™, USA). In all media, the volume was adjusted to 1000 mL with distilled water and the pH was adjusted to 6.0 using 1 N HCl and 1 N NaOH. After being inoculated, the culture media were incubated in darkness with shaking at 30 °C for 1 week. The culture liquid medium that presented the highest yield of the pigment was selected for further experiments.
Effect of temperature
In this experiment, a fungal culture was inculcated in the selected liquid medium that had been obtained from previous experiments. A pH value of 6.0 was achieved and the culture was incubated in darkness at 20, 25, 27, 30, 35, 40, and 45 °C for 1 week. The temperature that presented the highest yield of the pigment was selected for further experiments.
Effect of carbon and nitrogen sources
In this experiment, GY medium was selected based on the highest amount of production yield of the pigment. For each treatment glucose was replaced with alternative carbon sources; maltose, fructose, sucrose, soluble starch, carboxy methyl cellulose (CMC), lactose, and mannitol. Beef extract, peptone, malt extract, typtone, yeast extract, urea, (NH4)H2PO4, and KNO3 were assayed as the nitrogen source. The pH of all media was adjusted to 6.0 before being autoclaved. The inoculated flasks were incubated in darkness at a selected temperature. The carbon and nitrogen sources that presented the highest yield of the pigment were selected for further experiments.
Effect of initial of pH
The initial pH of the selected suitable components of the medium that had been obtained from previous experiments was adjusted from 3.0 to 9.0 in each flask before being autoclaving. After inoculation, cultures were incubated at temperature that presented the highest pigment yield for 1 week. The initial pH of culture media that presented the highest yield of the pigment was selected for further experiments.
Effect of cultivation period
The effect of the incubation period on fungal pigment production was determined. The suitable components of the medium and pH that had been obtained from previous experiments were used in this experiment. Cultivation was performed in darkness at a suitable temperature with shaking for 1 week. The fungal biomass was harvested and pigment yield was measured.
Fungal pigment fermentation and extraction
This experiment was conducted in 1000-mL Erlenmeyer flasks containing 300 mL of GY medium and at a pH of 5.0 under optimized conditions. Ten fungal mycelial plugs (5 mm in diameter) obtained from the periphery of the growing colony on PDA at 30 °C for 1 week were transferred into the medium after being autoclaved. The inoculation medium was incubated for up to 1 week at 27 °C and then shaken at 125 rpm on a reciprocal shaker. The same conditions were maintained throughout the experiment.
After fermentation, the culture media were filtrated through Whatman’s No. 1 (Whatman, UK) filter paper. The culture filtrate was extracted using two volumes of ethyl acetate according to the method described by Arumugam et al. (2014). The ethyl acetate fraction was evaporated until dryness using a rotary evaporator (BUCHI Rotaryvapor™ model R-210/215, Switzerland). The crude extracts were kept at − 20 °C before being used in further experiments.
Characterization of crude fungal pigment
Solubility test
The solubility of the pigment was evaluated using various types of solvents; distilled water, absolute methanol, 95% ethanol, 0.1 M HCl, 0.1 M NaOH, acetone, ethyl acetate, and 0.1 M alum. The solubility test was performed following the method described by Ahmad et al. (2012) with some modifications. Ten milliliters of the solvents were added to 15 × 150 mm test tubes containing 1 mg of the pigment extract. The solutions were mixed by vortex and kept in darkness at room temperature (25 ± 2 °C). After 12 h at room temperature, the λmax of each solution was scanned (400–650 nm) by a spectrophotometer and the total pigment yield at 505 nm was recorded. The color names and codes of each solution were recorded using the Methuen handbook of color (Kornerup and Wanscher 1967). Three replications were performed for each treatment.
Stability test
The stability of the fungal pigment toward pH, temperature, light, and being autoclaved was evaluated according to the methods of Velmurugan et al. (2010) and Ahmad et al. (2012) with some modifications. The pH value of the pigment concentration that was used was adjusted to between 4 and 11 by either 1 M NaOH or 1 M HCl at 0.1 mg/mL in methanol with a total volume of 10 mL in 15 × 150 mm test tubes. The effect of pH on the pigment’s stability was recorded after the pH adjustment and the samples were incubated at room temperature for 2 h. With regard to the stability of the pigment solution, a pH value of 6.0 (the highest pigment yield) was incubated in a water bath at 20, 25, 30, 35, 40, 50, 60, and 80 °C for 5 h. The degree of stability under sunlight (11:00 am to 4:00 pm with a light intensity of 12,000 to 55,000 lx), UV light (Philips 15 W, 1 m distance), and fluorescent light (Philips 18 W, 1 m distance, light intensity 2000 lx) for 5 h accompanied by autoclave conditions was investigated. The λmax of each treatment was scanned; the total pigment yield at 505 nm was recorded and compared with the control treatment. The stability percentage of pigment at 505 nm was calculated according to the following formula. The percentage of stability = (As / Ao) × 100, where As is the absorbance of pigment after treatment and Ao is the absorbance of pigment before treatment. Each treatment was performed in triplicate.
Fungal pigment purification
Sample preparation and column chromatographic separation
The crude extract of the pigment was dissolved in absolute methanol and loaded into Sephadex™ LH-20 (GE Healthcare Bio-Science AB, Sweden) columns. Absolute methanol was used as an eluent. Column eluted fractions that were read through a spectrophotometer between 450 to 550 nm were pooled together. To obtain high purity of the compound, an additional preparative thin layer chromatography (TLC) step was incorporated. The pigment fraction was concentrated and applied to a preparative TLC plate (20 cm × 20 cm, thickness 0.25 mm, Silica gel G F254; Merk, Germany). The running solvent was a mixture of 2% methanol in dichloromethane. The target band of the red pigment was scrapped off, dissolved in methanol, and separated from silica through filtrations with Whatman’s No. 1 filter paper. The samples were concentrated and dried using a rotary evaporator.
Liquid chromatography-mass spectrometry analyses
The purified pigment was analyzed using HPLC (LC-20 AD binary pump system, Shimadzu, Japan) coupled with high-resolution mass spectrometry (LTQ Orbitrap XL; Thermo Fisher Scientific, Waltham, MA). Four microliters of the pigment samples were injected into the L-Column2 ODS (2.1 × 150 mm, 3 μm; Chemicals Evaluation and Research Institute, Japan) and the column was heated to 40 °C. The gradient elution was applied. Solvent A consisted of 0.1% formic acid and solvent B consisted of acetonitrile containing 0.1% formic acid. The mobile phase began with A: B = 80: 20 to 20: 80 for 10 min at a flow rate of 0.2 mL/min. The MS data was obtained in ESI-positive and negative mode using the following parameters: source voltage, 4500 V (positive mode) and 4000 V (negative mode); capillary voltage, 10 V (positive mode) and − 20 V (negative mode); and capillary temperature of 400 °C. The MS spectrums in both the positive and negative modes were used to search for the possible structures as described below.
Identification of the purified fungal pigment was confirmed with a bostrycin standard (AdipoGen Life Science, USA) by LC/MS/MS analysis. LC/MS/MS was performed with 3200 QTRAP (AB Sciex, Framingham, MA) equipped with Prominence UFLC (Shimadzu, Japan). Four microliters of the pigment sample were injected to the Mightysil RP-18GP Aqua (150 × 2.0 mm, 5 μm) column heated at 40 °C. The gradient elution was applied as mentioned above. The retention time was determined and MS/MS data was obtained from ESI in the positive ion mode (ion spray voltage, 5000 V; temperature, 300 °C; N2 as both curtain gas (set to 12 arbitrary units) and collision gas (set to “high”); collision energy, 30 V; scan range, m/z 100–1200; scan speed, 4000 Da/s; declustering potential, 10 V).
Searching of the structure of pigment by MetFrag
MetFrag (http://msbi.ipb-halle.de/MetFrag/), a freely accessible web application, can automatically assign possible structures from a chemical compound database such as KEGG, PubChem, or ChemSpider (Wolf et al. 2010). The possible candidates were ranked by comparison of their in silico fragmentations against the measured MS fragments. The molecular formula and the fragmentation mass spectrum of the pigment were submitted to MetFrag. The search was performed for negative ions with an accuracy of 10 ppm.
Textile dyeing
Textile substrate
The SDC Multifiber SLW (SDC Enterprises Ltd., UK) was used in this study. The multifiber substrate was in the form of a 10-cm wide wrap stripe comprised of a 25 mm wide band of silk, cellulose, cotton, nylon, polyester, acetate, and wool.
Dyeing procedure
The dye solution was prepared by dissolving 5-mg pigment extract in 5 mL absolute methanol and adjusted to 50 mL by distilled water. The initial pH of the dye solution was adjusted to 6.0 using 1 N CH3COOH and 1 N Na2CO3 for the color red. The dyeing process was done following the method of Kramar et al. (2014) and Sharma et al. (2012) with some modifications. Dyeing was carried out by shaking a multifiber substrate in 100-mL steel beakers containing a 50 mL of dye solution at 35 °C for 1 h. After dyeing, samples were thoroughly washed with warm distilled water (35 °C) to remove the unfixed pigment, then soaked in 0.1% CuSO4 solution for 10 min and rinsed with tap water. The samples were air dried at room temperature. The color name and code of each sample was recorded. Five replications were made.
Characterization of dyed textile
Color fastness testing
The color fastness of the dyed multifiber substrate with respect to washing was evaluated according to the standard ISO 105-C01 (1981) by washing the dyed samples in a water bath containing 0.5% (w/v) of standard detergent (SDC Enterprises Ltd., UK) at 40 °C for 30 min. After washing, samples were rinsed for 10 min and dried at 60 °C. The color name and code of each sample was recorded. The surface color of the samples was measured using a Hunter Lab Mini Scan XE Plus colorimeter (Hunter Association Laboratory, Inc., USA).The L* (lightness), a* (red/green), and b* (yellow/blue) values were measured in the CIELab color space. The differences in color value (ΔE*) were determined between the dyed samples and the samples that were subjected to the washing fastness test. The color name and code of the samples were recorded. Experiments involving each sample were performed in five replicates.
In vitro cytotoxic assay
In vitro cytotoxic assay was performed using the human embryonic kidney cell (HEK 293T). The MTT assay was used according to the methods described by Carmichael et al. (1987). Cells were grown in Dulbecco’s Modified Eagle’s medium (DMEM; Thermoscientific, USA) until covering 70% of the T-25 flask area. The cell line was plated overnight in 96-well plates with 1 × 104 cells for each well. The crude red pigment was treated at various concentrations. After 48 h, 10 μL of 5 mg/mL MTT solution (Sigma-Aldrich, USA) was added to each well and incubated for 4 h. To dissolve the formazan crystals, 0.04 N HCl in isopropanol was added and the absorbance at 570 nm was measured using a EZ Read 2000 microplate reader (Biochom, Germany). The percentage of cell viability and the concentration for 50% lethality (LC50) were calculated.
Statistical analysis
Data were analyzed by one-way analysis of variance (ANOVA) by SPSS program version 16.0 for Windows and Tukey’s test was used for significant differences (P < 0.05) between treatments.
Results
Fungal isolation and identification of pigment-producing isolate
Twenty isolates were obtained from stem tissues and 27 isolates were obtained from leaf tissues. Only one fungal isolate (CMU-ZY2045) obtained from leaf tissue could produce an extracellularly red pigment. Colonies of isolate CMU-A109 on PDA grew to 80–85 mm at 25 °C in the darkness after 1 week. Colonies were flat, edge entire, floccose at the center with gray aerial mycelia, becoming black with age in the center and produced a soluble red pigment (Fig. 1A). Hyphae were pale brown, smooth, branched, septate, 1.5–4.5 μm. Conidiophore reduced to conidiogenous cells. Conidiogenous cells were pale brown, monoblastic, discrete, doliform, ovoid, or ampulliform, 7.5–12 × 6–8 μm. Conidia were ellipsoidal, dark brown to black, shiny, smooth surface, 12–15 × 9–15 μm (Fig. 1B). Based on morphological observations, this fungal isolate was identified as N. aurantiaca (Wang et al. 2017). A pure culture was deposited in the Culture Collection of the SDBR Laboratory, Faculty of Science, Chiang Mai University, which a partner of Thailand Bioresource Research Center (TBRC), Thailand, under number SDBR-CMU350.
Morphological characteristics and phylogenetic tree of Nigrospora aurantiaca CMU-ZY2045. A. Colony on potato dextrose agar at 25 °C for 1 week. B. Conidiophore and conidia. C. Phylogram derived from maximum likelihood analysis of a combined three-gene loci (LSU, ITS, TUB, and TEF1) from Nigrospora aurantiaca CMU-ZY2045 and related species. Arthrinium arundinis and Arthrinium sacchari were used as the outgroup. The numbers above branches represent maximum likelihood bootstrap percentages (left), Bayesian posterior probabilities (right) and the species described in this study is shown in bold. Bar A = 10 mm, B = 10 μm, C = the number of nucleotide substitutions per site
Molecular methods were used to confirm the identity of the isolate CMU-ZY2045. The LSU, ITS, TUB2, and TEF1 sequences of this fungal isolate were deposited in GenBank database under accession number KY048408, KY048409, MK388224, and MK388225, respectively. The combined LSU, ITS, TUB, and TEF1 dataset consisted 19 isolates with the final alignment dataset comprised 2867 characters including gaps (LSU, 1–853; ITS, 854–1469; TUB, 1467–2301; and TEF, 2302–2867). A phylogram of combined LSU, ITS, TUB, and TEF sequences is showed in Fig. 1C. Our phylogenetic results assigned isolate CMU-ZY2045 to N. aurantiaca. The species was clearly distinguished from other Nigrospora species and formed a sister taxon to N. vesicularis with BS support of 100% and PP value support of 1.0. Therefore, isolate CMU-ZY2045 was identified as N. aurantiaca based on molecular characteristics.
Optimization of fungal pigment production
The wavelength of maximum absorbance was scanned in a range of 300 to 650 nm. The results showed that the wavelength of maximum absorbance of red pigment in the supernatant was observed at 505 nm; therefore, indication of the total pigment yield was estimated at this wavelength. Red pigment and fungal biomass yields of N. aurantiaca CMU-ZY2045 in various liquid media are shown in Fig. 2A. The results indicated that the significantly highest pigment yield (3.652 ± 0.174 AU505) was observed in GY medium followed by PDB (3.217 ± 0.122 AU505) and OM medium (1.550 ± 0.163 AU505), while the highest biomass was found in PDB (4.97 ± 0.78 mg/mL). Temperature affected both pigment production and fungal growth (Fig. 2B). The highest pigment yield (3.591 ± 0.480 AU505) was determined at 27 °C, which was the optimum temperature. However, a fungus could not grow at 40 and 45 °C.
Effects of liquid medium (A), temperature (B), carbon source (C), nitrogen source (D), the initial pH of liquid medium (E), and cultivation period (F) on fungal red pigment and fungal biomass productions by Nigrospora aurantiaca CMU-ZY2045. The results are means of three replicates ± SD. Different letters above each bar in the same parameter indicate the significant difference (P < 0.05)
After 1 week of incubation, the pigment and fungal biomass yield responses to various carbon sources were observed (Fig. 2C). A fungus that produced the highest pigment yield was observed in glucose at 3.598 ± 0.210 AU505, but no significant differences were observed when compared with fructose (3.282 ± 0.307 AU505). Glucose was chosen as a suitable carbon source based on its availability at low cost. Both the lowest pigment and biomass yields were observed when using CMC as substrate. The results showed that the yeast extract was the best nitrogen source for pigment production and fungal growth (Fig. 2D). Our results indicated that the initial pH medium affected pigment production and fungal growth (Fig. 2E). The significantly highest pigment yield (4.042 ± 0.154 AU505) was found in GY medium at pH 5.0, followed by pH 6.0 (3.734 ± 0.098 AU505) and pH 7.0 (3.665 ± 0.213 AU505). The effect of the incubation period on pigment production was investigated for this fungus. The results indicated that the high amount of fungal pigment yield was produced when fungus was grown in the stationary phase, which then slowly decreased (Fig. 2F). The highest pigment yield (4.215 ± 0.148 AU505) was found at 5 days after incubation. Therefore, the use of GY medium (carbon and nitrogen sources were glucose and yeast extract, respectively), at a pH value of 5.0 and incubation at 27 °C for 5 days made up the optimum conditions for the pigment production of this fungus.
Fungal pigment fermentation and extraction
The evaluation of pigment production in 1000-mL Erlenmeyer flasks containing 300 mL of GY medium was observed at the optimal fermentation conditions (pH 5.0, temperature 27 °C). After 1 week of inoculation, the highest pigment yield (4.302 ± 0.238 AU505) was obtained and the liquid culture was filtrated. The extraction of the culture filtrates produced yields of crude pigment at 238.75 ± 42.20 mg/L with five replications.
Characterization of crude fungal pigment
Solubility test
The solubility of the crude fungal pigment extract in different solvents was evaluated and the results are shown in Supplemental Table S1. The solubility of the crude fungal pigment extract varied and appeared in red color in absolute methanol, 95% ethanol and acetone with a maximum absorption level at 505 nm. It was found that both the highest solubility ability was found in absolute methanol. The solubilization in distilled water, 0.1 M HCl and ethyl acetate showed an orange-red color. However, the solubilization in 0.1 M NaOH showed a bluish-violet color with a maximum absorption value of 600 nm, while a pink color was observed in 0.1 M alum showing a maximum absorption value at 565 nm.
Stability test
The crude fungal pigment extract was subjected to various treatments to test its stability and the results are presented in Supplemental Table S2. For the pH stability test, the initial color of this pigment was red (pH 6.4, λmax 505 nm, yield 0.984 ± 0.030 AU505), and different shades of colors resulted when the pigment extract was adjusted to different pH values. At pH values of 3, 4, 5, 6, and 7, the red color was observed with a maximum wavelength of 505 nm, except for the pH value of 7. The highest percentage of stability was found at pH 6.0 (99.6 ± 0.4%), followed by pH 5.0 (98.3 ± 1.1%) and pH 7.0 (97.3 ± 0.5%). At all other pH values, different colors such as bluish-red (pH 8.0 and 9.0), purplish-red (pH 10.0), and reddish-violet (pH 11.0) were observed with the maximum wavelength values at 540, 545, and 590 nm, respectively. The pigment stability was tested at different temperatures ranging from 20 to 80 °C, and the pigments that were subjected to all tested temperatures showed no changes in color. A high level of stability at low temperatures (20–50 °C) was observed. The pigment subjected to sunlight, UV, fluorescent lights, and autoclaved conditions showed no changes in color. The precipitation of the pigment was not observed at the bottom of the tubes after all stability tests. The red pigment from this fungus revealed high percentage values of stability at various temperatures and under sunlight. Our study indicates that the red pigment produced by N. aurantiaca CMU-ZY2045 can be used as a textile colorant.
Fungal pigment purification and identification
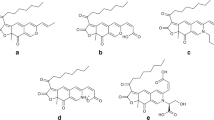
The scheme for purification of the red pigment is shown in Fig. 3. One band was observed in the TLC having Rf values of 0.65. A final yield of the purified fungal red pigment was 1.6 mg from 25 mg of the red pigment fraction. LC/MS analysis of the pigment was performed. The MS spectrum in a positive mode showed a peak at m/z 337.0925, which corresponded to C16H17O8 ([M + H]+, calcd 337.0918), in the negative mode, m/z 335.0753 (C16H15O8, [M − H]−, calcd 335.0761) as it had appeared. Therefore, the molecular formula was confirmed as C16H16O8. In the negative mode, MS/MS analysis of m/z 335.0753 [M − H]− showed nineteen product ions (Supplemental Table S3). To search for the structure of red pigment in the chemical database, in silico fragmenter MetFrag was performed. The molecular formula and the fragmentation mass spectrum of the red pigment were submitted to MetFrag. When applying KEGG, PubChem, and ChemSpider as the chosen database, the MetFrag search resulted in 2 hits, 81 hits, and 63 hits, respectively. The most probable forms of red pigment with chemical structures were altersolanol A, altersolanol D, or bostrycin.
Previous studies reported that altersolanol A, altersolanol D, and bostrycin resulted in yellow, orange-yellow, and red pigments, respectively (Fuoillaud et al. 2016; Suemitsu and Nakamura 1981; van Eijk 1975; Yagi et al. 1993). Therefore, a fungal red pigment in this study was compared with the bostrycin standard using the LC/MS/MS technique. Under relevant conditions, the retention times of a purified fungal red pigment showed a peak at 14.0 min, which corresponded to a bostrycin standard with a maximum absorption value of 504 nm (Fig. 4A and C). The identification of the fungal red pigment was confirmed by co-injection with a bostrycin standard. Moreover, it was found that the fungal red pigment and bostrycin standard showed the same fragmentation of mass spectrum (Figs. 4B and D). Therefore, the fungal red pigment was considered as bostrycin based on the comparisons of MS profile and a retention time with bostrycin standard.
Characterization of dyed textile
Color fastness testing
The color name, code, fastness, and total color difference values of the dyed multifiber substrates before and after being washed are presented in Supplemental Table S4. The results revealed that the differences in the stained colors varied for the different types of fiber substrates (Fig. 5). Silk, cotton, and nylon fabrics displayed a pastel red color, cellulose fabric displayed a pale red, color and the acetate fabric was pink. However, both the polyester and wool fabrics presented a pinkish color with slight staining. After the washing test, silk and cellulose fabrics showed a pale red color, whereas cotton and nylon fabrics showed a pastel red color and the acetate fabric appeared pink. The polyester and wool fabrics were white indicating no staining. The irregular staining in the cellulose, cotton and acetate fabrics was not changed after being washed. It was found that all the dyed fabrics after washing were lighter than the dyed fabrics before being washed because an increase in the L* value in the washed fabrics was observed. The A* value was decreased in the dyed fabrics after being washed. Consequently, the red shade of the washed fabrics was lighter than the unwashed fabrics. The ΔE* values ranged from 1.32–4.80, while the ΔE* values in the cotton fabrics were lower than the other fabrics. Our study indicated that the red pigment produced by N. aurantiaca CMU-ZY2045 can produce a higher degree of staining in cotton fabrics than in other fabrics.
In vitro cytotoxic assay
The viability percentage of HEK 293T in each selected concentration is shown in Supplemental Fig. S1. The results indicated that cell viability percentage was decreased when the concentration of the crude red pigment was increased. The viability percentage of HEK 293T cell at a concentration of 0.1 mg/mL in methanol (a concentration used in textile dyeing) was 89.06 ± 7.50%, which was considered non-significant with regard to methanol and the untreated cell treatments. It could presumptively indicate that it is a non-toxic concentration by comparing control as the viability was above 85%. The LC50 of the crude red pigment was observed at 2.34 mg/mL.
Discussion
Endophytic fungi are natural sources in the production of secondary metabolites, e.g., antibiotics and pigments (Babitha et al. 2007; Qui et al. 2010; Torres et al. 2016). In the present study, an endophytic fungus, N. aurantiaca CMU-ZY2045 isolated from cinnamon trees produced an extracellularly red pigment. The results were similar to those of previous studies which found that pure cultures of fungal genera, e.g., Blakeslea, Eurotium, Fusarium, Isaria, Monascus, Nigrospora, Paecilomyces, Penicillium, and Talaromyces could secrete the red pigment in both solid and liquid media (Arumugam et al. 2014; Chadni et al. 2017; Cho et al. 2002; Lebeau et al. 2017; Méndez et al. 2011; Qui et al. 2010; Torres et al. 2016; Unagul et al. 2005; Velmurugan et al. 2010).
In this study, the optimum conditions for the highest yield of red pigment production from N. aurantiaca CMU-ZY2045 was obtained at 5 days of cultivation in GY medium used with carbon and nitrogen sources as glucose and yeast extract, respectively, at a pH of 5.0 and incubation at 27 °C. This result was supported by the findings of several previous studies, which reported that fungal growth and pigment production yields were greatly influenced by cultivation conditions (e.g., cultivation medium, temperature, pH, carbon and nitrogen sources, mineral, aeration, and type of fermentation) and the optimum conditions for pigment production were not homologous for the fungal species and strains (Chadni et al. 2017; Cho et al. 2002; Kumar et al. 2015; Méndez et al. 2011; Pandey et al. 2018; Pradeep et al. 2013; Sharma et al. 2012; Souza et al. 2016; Unagul et al. 2005). Notably, the optimum conditions for red pigment production from Nigrospora sp. NIOT was found at 6 days of cultivation in PDB using soluble starch as carbon source and peptone as nitrogen source, a pH of 6.0 and incubation at 30 °C (Arumugam et al. 2014). Zhou et al. (2015) found that the cultivation of Monascus ruber in liquid medium at a pH of 5.4 using corn flour and NH4Cl as the carbon and nitrogen sources, respectively and being incubated at 30 °C for 1 week gave the highest yield of red pigment production. In addition, previous studies suggested that the optimal physical conditions and suitable nutrients for fungal pigment fermentation could improve the pigment production yields significantly on an industrial production scale (Arumugam et al. 2014; Kumar et al. 2015; Mapari et al. 2005). Additionally, Tudor et al. (2013) reported that pH values did not only affect the fungal growth and pigment production, but also the intensity of the pigment production.
The solubility of crude red pigment of N. aurantiaca CMU-ZY2045 varied in different solvent types, and the highest solubility was found in methanol. This result is similar to those of previous reports where the solubilization ability of the crude microbial pigment was dependent upon various solvent types (Narendrababu and Shishupala 2017; Rokade and Pethe 2016). Souza et al. (2016) reported that pigment production from Epicoccum nigrum, Lecanicillium aphanocladii, and Pe. flavigenum was soluble in ethyl acetate, but pigments production from As. aureolatus, As. sydowii, As. keveii, and Pe. chermesinum were insoluble in ethyl acetate. Orange-red pigment production occurred from Aspergillus showed the highest solubility in ethyl acetate (Narendrababu and Shishupala 2017). In addition, the solubilization ability was influenced by the composition of the polarity substances in the crude pigment, for which polar substances tend to dissolve in polar solvents and non-polar substances dissolve in non-polar solvents (Latha and Jeevaratnam 2010; Robinson et al. 2014; Rokade and Pethe 2016; Souza et al. 2016).
This study found that the color stability of fungal red pigment is highly stable at low temperatures (20–50 °C) and in a more acidic pH range than the basic pH (pH 5.0–6.0). This result was supported by several previous studies that reported that the stability of the fungal pigment was influenced by different conditions such as pH, heat, light, and chemical additives (Kaur et al. 2017; Poorniammal et al. 2010; Torres et al. 2016). Sastrawidana et al. (2016) found that red pigment produced from Pe. purpurogenum was stable at 30–80 °C and at pH values in a range of 4.0–8.0. A red pigment produced from Penicillium sp. HSD07B was stable between pH values of 2 and 10, and temperatures of 10–100 °C (Hailei et al. 2011). The stability of fungal pigments toward different processing conditions, additives, and pH values make up the major factors affecting the selection and application of fungal pigments (Robinson et al. 2013; Sharma et al. 2012; Torres et al. 2016; Wongjewboot and Kongruang 2011).
The fungal red pigment characterized in this study that was identified as bostrycin was originally isolated from the fungus, Alternaria and Ar. phaeospermum (van Eijk 1975). It has also been found in other fungi, e.g., Al. eichorniae, Al. porri, Al. solani, Bostrychonema alpestre, Dermocybe splendida, Fusarium sp. PSU-F14, Fusarium sp. PSU-F15, Nigrospora oryzae, Nigrospora sp. 1403, Nigrospora sp. 407, Nigrospora sp. MA75, Phomopsis juniperovara, Stemphylium botryosum, and Xylaria sp. 2508 (Deshmukh et al. 2014; Huang et al. 2014; Thomson 1987; Trisuwan et al. 2010; Wheeler et al. 1975; Xia et al. 2011; Yang et al. 2012). However, this is the first report on bostrycin produced by N. aurantiaca. Bostrycin has important biological activities, including antibacterial and phytotoxic activities (Charudattan and Rao 1982; van Eijk 1975; Wheeler et al. 1975). This compound has also been shown to possess anticancer activities by the inhibition of the growth of lung, prostate, and gastric cancer cells (Chen et al. 2010, 2011; Lin et al. 2008). Moreover, bostrycin has the potential to be used in food processing as a safe color fixative agent (Huang et al. 2017).
The present study revealed that the red pigment produced by N. aurantiaca CMU-ZY2045 could be used as a textile dye. The red pigment showed the highest strain in cotton fabric, which was expressed by color fastness after being washed. Similarly, previous studies indicated that the fungal pigment can be exploited as one of the natural sources of textile dyes (Babitha et al. 2007; Hernández et al. 2019; Nagia and El-Mohamedy 2007; Sharma et al. 2012; Hinsch et al. 2015; Palomino et al. 2017). In addition, the resulting color of the fabrics after staining and washing varied with both different fabric types and fungal pigments (Kramar et al. 2014; Mabrouk et al. 2011; Poorniammal et al. 2010; Sharma et al. 2012). For examples, the reddish-brown and red pigments produced by Al. alternata, As. niger, Talaromyces verruculosus and Trichoderma virens displayed the highest degree of staining and excellent fastness to washing in cotton fabrics (Anchana devi 2014; Chadni et al. 2017; Devi and Karuppan 2015). However, the pigments from Curvularia lunata, Pe. chtysogenum, Pe. italicum, and Pe. regulosum showed the highest degree of staining in wool fabric and revealed effective color washing fastness (Mabrouk et al. 2011; Sharma et al. 2012).
In conclusion, an endophytic fungus, N. aurantiaca. CMU-ZY2045 isolated from the leaf tissues of the cinnamon plant effectively produced red pigment in vitro. The highest red pigment yield was observed in the glucose yeast extract liquid medium with glucose as a carbon source, yeast extract as a nitrogen source, a pH value of 5.0, and at 27 °C with shaking for 5 days. Red pigment extract was stable in temperatures of 20–50 °C and pH values of 5.0–6.0. The staining of cotton fabrics using this fungal red pigment extract has excellent fastness to washing and the concentration used revealed a negative level of cytotoxicity. The fungal red pigment was identified as bostrycin. Thus, our results suggest that the red fungal pigment extract has the potential to be used in textile processing as a safe natural dye.
References
Ahmad WA, Yusof NZ, Nordin N, Zakaria ZA, Rezali MF (2012) Production and characterization of violacein by locally isolated Chromobacterium violaceum grown in agricultural wastes. Appl Biochem Biotechnol 167:1220–1234. https://doi.org/10.1007/s12010-012-9553-7
Akilandeswari P, Pradeep BV (2016) Exploration of industrially important pigment from soil fungi. Appl Microbiol Biotechnol 100:1631–1643. https://doi.org/10.1007/s00253-015-7231-8
Alfaro ME, Zoller S, Lutzoni F (2003) Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov Chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol Evol 20:255–266
Anchana devi A (2014) Extraction of natural dyes from fungus-an alternate for textile dyeing. J Nat Sci Res 4:1–6
Arumugam GK, Srinivasan SK, Joshi G, Gopal D, Ramalingam K (2014) Production and characterization of bioactive metabolites from piezotolerant deep sea fungus Nigrospora sp. in submerged fermentation. J Appl Microbiol 118:99–111. https://doi.org/10.1111/jam.12693
Babitha S, Soccol CR, Pandey A (2007) Solid-state fermentation for the production of Monascus pigment from jack fruit seed. Bioresour Technol 98:1554–1560. https://doi.org/10.1016/j.biortech.2006.06.005
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556
Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB (1987) Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res 47:936–942
Chadni Z, Rahaman MH, Jerin I, Hoque KMF, Reza MA (2017) Extraction and optimization of red pigment production as secondary metabolites from Talaromyces verruculosus and potential use in textile industries. Mycology 8:48–57. https://doi.org/10.1080/21501203.2017.1302013
Charudattan R, Rao KV (1982) Bostrycin and 4-deoxybostrycin: two nonspecific phytotoxins produced by Alternaria eichhorniae. Appl Environ Microbiol 43:846–849
Chen CQ, Fang LK, Liu JW, Zhang JW, Yang GG, Yang W (2010) Effects of marine fungal metabolites (1386A) from the South China Sea on proliferation, apoptosis and membrane potential of gastric cancer cell line MCG-803. Chin J Pathophysiol 26:1908–1912
Chen WS, Hou JN, Guo YB, Yang HL, Xie CM, Lin YC, She ZG (2011) Bostrycin inhibits proliferation of human lung carcinoma A549 cells via downregulation of the Pl3K/Akt pathway. J Exp Clin Cancer Res 30:17. https://doi.org/10.1186/1756-9966-30-17
Cho YJ, Park JP, Hwang HJ, Kim SW, Choi JW, Yun JW (2002) Production of red pigment by submerged culture of Paecilomyces sinclairii. Lett Appl Microbiol 35:195–202. https://doi.org/10.1046/j.1472-765X.2002.01168.x
Deshmukh SK, Verekar SA, Bhave SV (2014) Endophytic fungi: a reservoir of antibacterials. Front Microbiol 5:715. https://doi.org/10.3389/fmicb.2014.00715
Devi S, Karuppan P (2015) Reddish brown pigment from Alternaria alternata for textile dyeing and printing. Indian J Fibre Text Res 40:315–319
Dufossé L, Galaup P, Yaron A, Arad SM, Blanc P, Murthy KNC, Ravishankar GA (2005) Microorganism and microalgae as source of pigments for food use: a scientific oddity or an industrial reality. Trends Food Sci Technol 16:389–406. https://doi.org/10.1016/j.tifs.2005.02.006
Dufossé L, Fouille M, Caro Y, Mapari SAS, Sutthiwong N (2014) Filamentous fungi are large-scale producers of pigments and colorants for the food industry. Curr Opin Biotechnol 26:56–61. https://doi.org/10.1016/j.copbio.2013.09.007
Durán N, Teixeira MFS, Conti RD, Esposito E (2002) Ecological-friendly pigmentts from fungi. Crit Rev Food Sci Nutr 42:53–66. https://doi.org/10.1080/10408690290825457
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Fuoillaud M, Venkatachalam M, Girard-Valenciennes E, Caro Y, Dufossé L (2016) Anthraquinones and derivatives from marine-derived fungi: structure diversity and selected biological activities. Mar Drugs 14:64. https://doi.org/10.3390/md14040064
Glass LN, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330
Hailei W, Zhifang R, Ping L, Yanchang G, Guosheng L, Jianming Y (2011) Improvement of the production of a red pigment in Penicillium sp. HSD07B synthesized during co-culture with Candida tropicalis. Bioresour Technol 102:6082–6087. https://doi.org/10.1016/j.biortech.2011.01.040
Hernández VA, Galleguillos F, Thibaut R, Müller A (2019) Fungal dyes for textile applications: testing of industrial conditions for wool fabrics dyeing. J Text Inst 110:61–66. https://doi.org/10.1080/00405000.2018.1460037
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192
Hinsch EM, Chen HL, Weber G, Robinson SC (2015) Colorfastness of extracted wood-staining fungal pigments on fabrics: a new potential of textile dyes. J Text Appar Technol Manag 9:1–11
Huang X, Sun X, Lin S, Xiao Z, Li H, Bo D, She Z (2014) Xylanthraquinone, a new anthraquinone from the fungus Xylaria sp. 2508 from the South China Sea. Nat Prod Res 28:111–114. https://doi.org/10.1080/14786419.2013.850687
Huang YH, Yang WJ, Cheng CY, Sung HM, Lin SF (2017) Bostrycin production by agro-industrial residues and its potential for food processing. Food Sci Biotechnol 26:715–721. https://doi.org/10.1007/s10068-017-0082-6
Joshi V, Attri D, Bala A, Bhushan S (2003) Microbial pigments. Indian J Biotechnol 2:362–369
Kaur S, Arora N, Kaur S (2017) Characterization of yellow pigments produced by Pencillium sp. under solid state cultivation. J Biotechnol Biomater 7(259). https://doi.org/10.4172/2155-952X.1000259
Kornerup A, Wanscher JH (1967) Methuen handbook of colour, 2nd edn. Eyre Methuen, London
Kramar A, Ilic-Tomic T, Petkovic M, Radulović N, Kostic M, Jocic D, Nikodinovic-Runic J (2014) Crude bacterial extracts of two new Streptomyces sp. isolates as bio-colorants for textile dyeing. World J Mirobiol Biotechnol 30:2231–2240. https://doi.org/10.1007/s11274-014-1644-x
Kumar A, Vishwakarma HS, Singh J, Dwivedi S, Kumar M (2015) Microbial pigments: production and their application in various industries. IJPCBS 5:203–212
Latha BV, Jeevaratnam K (2010) Purification and characterization of the pigments from Rhodotorula glutinis DFR-PDY isolated from natural source. Global J Biotechnol Biochem 5:166–174
Lebeau J, Venkatachlam M, Fouillaud M, Petit T, Vinale F, Dufossé CY (2017) Production and new extraction method of polyketide red pigments produced by ascomycetous fungi from terrestrial and marine habitats. J Fungi 3:34. https://doi.org/10.3390/jof3030034
Lin W, Fang LK, Liu JW, Cheng WQ, Yun M, Yang HL (2008) Inhibitory effects of marine fungal metabolites from the South China Sea on prostate cancer cell line DU-145. Int J Intern Med 35:562–564
Mabrouk AM, El-Khrisy EAM, Youssef YA, Asem MA (2011) Production of textile reddish brown dyes by fungi. Malaysian J Microbiol 7:33–40
Malik K, Tokkas J, Goyal S (2012) Microbial pigment: a review. Int J Microbial Resour Technol 1:361–365
Mapari SAS, Nielsen KF, Larsen TO, Frisvad JC, Meyer AS, Thrane U (2005) Exploring fungal biodiversity for the production of water-soluble pigments as potential natural food colorants. Curr Opin Biotechnol 16:231–238. https://doi.org/10.1016/j.copbio.2005.03.004
Méndez A, Pérez C, Montanéz JC, Martínez G, Aguilar CN (2011) Red pigment production by Penicillium purpurogenum GH2 is influenced by pH and temperature. J Zhejiang Univ Sci B 12:961–968. https://doi.org/10.1631/jzus.B1100039
Nagia FA, El-Mohamedy RSR (2007) Dyeing of wool with natural anthraquinone dyes from Fusarium oxysporum. Dyes Pigments 75:550–555. https://doi.org/10.1016/j.dyepig.2006.07.002
Narendrababu BN, Shishupala S (2017) Spectrophotometric detection of pigments from Aspergillus and Penicillium isolates. J Appl Biol Biotechnol 5:53–58. https://doi.org/10.7324/JABB.2017.50109
O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci 95:2044–2049. https://doi.org/10.1073/pnas.95.5.2044
Osman MY, Sharaf IA, Osman HM, El-Khouly ZA, Ahmed EI (2004) Synthetic organic food colouring agents and their degraded products: effects on human and rat cholinesterases. Br J Biomed Sci 61:128–132. https://doi.org/10.1080/09674845.11732657
Page RD (2001) TreeView. Glasgow University, Glasgow
Palomino AME, Vega GSM, Chen HL, Robinson SC (2017) Wood-rotting fungal pigments as colorant coating on oil-based textile dyes. Coatings 7:152. https://doi.org/10.3390/coatings7100152
Pandey N, Jain R, Pandey A, Tamta S (2018) Optimisation and characterisation of the orange pigment produced by a cold adapted strain of Penicillium sp. (GBPI_P155) isolated from mountain ecosystem. Mycology 9:81–92. https://doi.org/10.1080/21501203.2017.1423127
Panesar R, Kaur S, Panesar PS (2015) Production of microbial pigments utilizing agro-industrial waste: a review. Curr Opin Food Sci 1:70–76. https://doi.org/10.1016/j.cofs.2014.12.002
Perumal K, Stalin V, Chandrasekarenthiran S, Sumathi E, Saravanakumar A (2009) Extraction and characterization of pigment from Sclerotinia sp. and its use in dyeing cotton. Text Res J 79:1178–1187. https://doi.org/10.1177/0040517508087680
Poorniammal R, Manickam P, Gunasekaran S, Murugesan R, Thilagavathi G (2010) Natural dye production from Thermomyces sp. fungi for textile application. Indian J Fibre Text Res 38:276–279
Pradeep FS, Begam MS, Palaniswamy M, Pradeep BV (2013) Influence of culture media on growth and pigment production by Fusarium moniliforme KUMBF1201 isolated from paddy field soil. World Appl Sci J 22:70–77. https://doi.org/10.5829/idosi.wasj.2013.22.01.7265
Qui M, Xie R, Shi Y, Chen H, Wen Y, Gao Y, Hu X (2010) Isolation and identification of endophytic fungus SX01, a red pigment producer from Ginkgo biloba L. World J Microbiol Biotechnol 26:993–998. https://doi.org/10.1007/s11274-009-0261-6
Robinson SC, Tudor D, Zhang WR, Ng S, Cooper PA (2013) Ability of three yellow pigment producing fungi to colour wood under controlled conditions. Int Wood Prod J 5:103–107. https://doi.org/10.1179/2042645313Y.0000000060
Robinson SC, Hinsch E, Weber G, Freitas S (2014) Method of extraction and resolubilisation of pigments from Chlorociboria aeruginosa and Scytalidium cuboideum, two prolific spalting fungi. Color Technol 130:221–225. https://doi.org/10.1111/cote.12080
Rokade MT, Pethe DAS (2016) Isolation and identification of chromogenic bacteria from various sources. EJPMR 3:295–299
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Sastrawidana IDK, Maryam SY, Sukarta IN (2016) Natural dyeing of silk and cotton fabric with red pigment from Penicillium purpurogenus which is isolated from goat milk contaminated soil. J Nat Sci Res 6:32–37
Sharma D, Gupta C, Aggarwal S, Nagpal N (2012) Pigment extraction from fungus for textile dyeing. Indian J Fiber Text Res 37:68–73
Souza PNC, Grigoletto TLB, de Moraes LAB, Abreu LM, Guimarães LHS, Santos C, Galvão LR, Cardoso PG (2016) Production and chemical characterization of pigments in filamentous fungi. Microbiology 162:12–22. https://doi.org/10.1099/mic.0.000168
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Suemitsu R, Nakamura A (1981) Isolation and identification of altersolanol a from the culture liquid of Alternaria porri (Ellis) Ciferri. Agric Biol Chem 45:2363–2364. https://doi.org/10.1080/00021369.1981.10864889
Thomson RH (1987) Naturally occurring Quinones III: recent advances. Chapman and Hall, London
Torres FAE, Zaccarim BR, Novaes LCL, Jozala AF, Santos CA, Teixeira MFS, Santos-Ebinuma VC (2016) Natural colorants from filamentous fungi. Appl Microbial Biotechnol 100:2511–2521. https://doi.org/10.1007/s00253-015-7274-x
Trisuwan K, Khamthong N, Rukachaisirikul V, Phongpaichit S, Preedanon S, Sakayaroj J (2010) Anthraquinone cyclopentanone and naphthoquinone derivatives from the sae fan-derived fungi Fusarium spp. PSU-F14 and PSU-F135. J Nat Prod 73:1507–1511. https://doi.org/10.1021/np100282k
Tseng YY, Chen MT, Lin CF (2000) Growth, pigment production and protease activity of Monascus purpureus as affected by salt, sodium nitrite, polyphosphate and various sugars. J Appl Microbiol 88:31–37. https://doi.org/10.1046/j.1365-2672.2000.00821.x
Tudor D, Robinson SC, Cooper PA (2013) The influence of pH on pigment formation by lignicolous fungi. Int Biodeterior Biodegradation 80:22–28. https://doi.org/10.1016/j.ibiod.2012.09.013
Unagul P, Wongsa P, Kittakoop P, Intamas S, Srikitikulchai P, Tanticharoen M (2005) Production of red pigments by the insect pathogenic fungus Cordyceps unilateralis BCC1869. J Ind Microbiol Biotechnol 32:135–140. https://doi.org/10.1007/s10295-005-0213-6
van Eijk GW (1975) Bostrycin, a tetrahydroanthraquinone pigment and some other metabolites from the fungus Arthrinium phaeospermum. Experientia 31:783–784
Velmurugan P, Kamala-Kannan S, Balachandar V, Lakshmanaperumalsamy P, Chae JC, Oh BT (2010) Natural pigment extraction from five filamentous fungi for industrial applications and dyeing of leather. Carbohydr Polym 79:262–268. https://doi.org/10.1016/j.carbpol.2009.07.058
Venil CK, Lakshmanaperumalsamy P (2009) An insightful overview on microbial pigment, prodigiosin. Electron J Biol 5:49–61
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Wang M, Lui F, Crous PW, Cai L (2017) Phylogenitic reassessment of Nigrospora: ubiquitous endophytes, plant and human pathogens. Persoonia 39:118–142. https://doi.org/10.3767/persoonia.2017.39.06
Wheeler MM, Wheeler DMS, Peterson GW (1975) Anthraquinone pigments from the phytopathogen Phomopsis juniperovora hahn. Phytochemistry 14:288–289. https://doi.org/10.1016/0031-9422(75)85060-6
White TJ, Burns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. in: Innis MA, Gelfand DH, Sninsky JJ, Whitish TJ (Eds.) PCR protocols, a guide to methods and applications. Academic Press, San Diego. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Wolf S, Schmidt S, Müller-Hannemann M, Neumann S (2010) In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics 11:148. https://doi.org/10.1186/1471-2105-11-148
Wongjewboot I, Kongruang S (2011) pH stability of ultrasonic Thai isolated Monascus purpureus pigments. IJBBB 1:79–83
Xia X, Li Q, Li J, Shao C, Zhang J, Zhang Y, Liu X, Lin Y, Liu C, She Z (2011) Two new derivatives of griseofulvin from the mangrove endophytic fungus Nigrospora sp. (strain no. 1403) from Kandelia candel (L.) Druce. Planta Med 77:1735–1738. https://doi.org/10.1055/s-0030-1271040
Yagi A, Okamura N, Haraguchi H, Abo T, Hashimoto K (1993) Antimicrobial tetrahydroanthraquinones from a strain of Alternaria silani. Phytochemistry 33:87–91. https://doi.org/10.1016/0031-9422(93)85401-C
Yang WJ, Yang CS, Huang CJ, Chen KS, Lin SF (2012) Bostrycin, a novel coupling agent protein immobilization and prevention of biomaterial-cantered infection produced by Nigrospora sp. Enzym Microb Technol 50(407):287–292. https://doi.org/10.1016/j.enzmictec.2012.02.002
Zhou Z, Guo H, Xie C (2015) Effect of culture conditions on production of red pigment and citrinin by fermentation of Monascus ruber. Chem Eng Trans 46:1363–1368. https://doi.org/10.3303/CET1546228
Acknowledgments
We are grateful to Mr. Russell K. Hollis for English proofreading.
Funding
This work was supported by grants from the Chiang Mai University, Center of Excellence on Biodiversity (BDC), Office of Higher Education Commission (BDC-PG2–159010), National Research Council of Thailand (NRCT) and Japan Society for the Promotion of Science (JSPS) in Core-to-Core Program (Establishment of an international research core for new bio-research fields with microbes from tropical area).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 301 kb)
Rights and permissions
About this article
Cite this article
Suwannarach, N., Kumla, J., Nishizaki, Y. et al. Optimization and characterization of red pigment production from an endophytic fungus, Nigrospora aurantiaca CMU-ZY2045, and its potential source of natural dye for use in textile dyeing. Appl Microbiol Biotechnol 103, 6973–6987 (2019). https://doi.org/10.1007/s00253-019-09926-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09926-5