Abstract
Microbial production of solvents like acetone and butanol was a couple of the first industrial fermentation processes to gain global importance. These solvents are important feedstocks for the chemical and biofuel industry. Ralstonia eutropha is a facultatively chemolithoautotrophic bacterium able to grow with organic substrates or H2 and CO2 under aerobic conditions. This bacterium is a natural producer of polyhydroxyalkanoate biopolymers. Recently, with the advances in the development of genetic engineering tools, the range of metabolites R. eutropha can produce has enlarged. Its ability to utilize various carbon sources renders it an interesting candidate host for synthesis of renewable biofuel and solvent production. This review focuses on progress in metabolic engineering of R. eutropha for the production of alcohols, terpenes, methyl ketones, and alka(e)nes using various resources. Biological synthesis of solvents still presents the challenge of high production costs and competition from chemical synthesis. Better understanding of R. eutropha biology will support efforts to engineer and develop superior microbial strains for solvent production. Continued research on multiple fronts is required to engineer R. eutropha for truly sustainable and economical solvent production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ralstonia eutropha (also known as Cupriavidus necator) is a betaproteobacterium that is known for its ability to interconvert carbon and has been considered an ideal microbial chassis for bioconversion of waste carbon to value-added product compounds. R. eutropha was originally isolated from soil and noted for two aspects of its growth: hydrogenase activity and the ease of culturing of the organism (Schatz and Bovell 1952). It was also shown to grow robustly on a wide variety of carbon sources, including carbon dioxide (CO2) (Wilde 1962).
R. eutropha is very well known for its ability to produce intracellular polyhydroxyalkanoates (PHA) from many different carbon sources. PHA synthesis in this case is a stress response and occurs during non-carbon nutrient (e.g., nitrogen) starvation. Intracellular PHA is then mobilized as a carbon source when R. eutropha is present in carbon-deficient conditions (Brigham et al. 2012). Wild-type R. eutropha produces polyhydroxybutyrate (PHB), a homopolymer in the PHA family, under most conditions. PHB homeostasis has been studied for decades in R. eutropha. We currently have a thorough understanding of conditions necessary for polymer synthesis and mobilization (Steinbuchel 1991; Jendrossek 2009; Jendrossek and Handrick 2002), as well as what enzymes and structural proteins are required to maximize PHB synthesis. PHB is synthesized in organelle-like structures called granules in the R. eutropha cytoplasm. There are many different types of granule-associated proteins (PGAP) that are involved in PHB synthesis, containment, and mobilization (Jendrossek 2009; Pfeiffer et al. 2011; Sznaider et al. 2015). Because so much is known about PHA synthesis in R. eutropha, it has been rightly called the model organism for study of PHA homeostasis. Many types of PHA have thermal and mechanical properties that are similar to petroleum-based plastics like polypropylene, polyethylene, or polystyrene. Successful genetic manipulation efforts have constructed strains of R. eutropha capable of synthesizing PHA with a broader range of monomers, resulting in production of biodegradable polymer with the potential for many different applications.
Because of its robust growth, amenability to genetic manipulations and ability to interconvert relatively large amounts of many types of carbon, R. eutropha is often considered an industrially relevant organism. With the advent of metabolic engineering, R. eutropha can be reconstructed to produce more than just PHA. In recent years, production of biofuel molecules has been of interest. The use of R. eutropha as a biofuel-producing organism brings up an interesting consideration: the use of a chemolithoautotrophic (i.e., growing solely on CO2, H2, O2) biocatalyst could convert CO2 to biofuels, suggesting that greenhouse gases can be reclaimed to make value-added products. This review discusses the efforts that have been made to convert R. eutropha from a PHA-producing organism to a solvent (biofuel)-producing organism. Biofuel products examined include alcohols like ethanol and isobutanol, ketones, alkanes, and alkenes, intermediates for which wild-type R. eutropha has been shown to synthesize as intermediates for other pathways. The implications of non-photosynthetic biofuel production from CO2 are a driving factor for many, if not all, the research discussed below.
Alcohol synthesis
Ethanol
The focus of rerouting R. eutropha metabolism to produce biofuel molecules has only infrequently involved the most commonly used and lower energy alcohol, ethanol. In one recent study, genetically engineered R. eutropha has been used to produce ethanol from acetate (Lee et al. 2016). The phaCAB operon encoding PHB metabolic pathway genes was deleted in R. eutropha H16. The heterologous gene adhE, encoding a bi-functional acetaldehyde-CoA dehydrogenase and alcohol dehydrogenase from Escherichia coli (Clark and Cronan 1980), was overexpressed for conversion of acetyl-CoA to ethanol (Fig. 1). When cultivated in minimal media batch cultures under microaerobic conditions supplemented with 5 mg/L of acetate as a sole source of carbon, the engineered strain produced ethanol up to 170 mg/L. When growth conditions were optimized by adjusting nitrogen source (NH4Cl) content and employing fed-batch culturing with periodic feeding of acetate, the engineered R. eutropha strain produced up to 350 mg/L ethanol over 84 h (Lee et al. 2016). Another work examined a R. eutropha strain overexpressing a pyruvate decarboxylase and an alcohol dehydrogenase (Fig. 1). Cells were grown on glucose as the carbon source, and ethanol was produced in an electrochemical reactor, which electrically split water to provide R. eutropha with reducing equivalents (i.e., H2). The molar yield of ethanol on glucose (YPG) was shown to be approximately 1.4 mmol ethanol/mmol glucose consumed, which was a 200% increase compared to conventional (non-electrochemical) batch cultivation (Jeon et al. 2013). Table 1 summarizes the ethanol-producing R. eutropha strains.
Ethanol biosynthesis pathways in engineered R. eutropha strains. Two different pathways have been designed for ethanol production. One pathway (Lee et al. 2016) is fed using acetate as the main carbon source and involves the native acetyl-CoA synthase enzyme (1) and a heterologously expressed, bi-functional acetaldehyde-CoA dehydrogenase and alcohol dehydrogenase enzyme (2, 3). Another pathway (Jeon et al. 2013) converts pyruvate to acetaldehyde using a heterologously expressed pyruvate decarboxylase enzyme (4) and converts acetaldehyde to ethanol by a heterologously expressed alcohol dehydrogenase. The polyhydroxybutyrate biosynthesis pathway (PhaA, PhaB, PhaC) can be a major carbon sink, as it shares a key intermediate, acetyl-CoA, with the ethanol synthesis pathway. Genes encoding the PHB biosynthesis pathway were deleted in (Lee et al. 2016)
Could R. eutropha be engineered to produce large quantities of ethanol biofuel? The studies discussed above suggest the possibility of this. Indeed, R. eutropha is capable of interconverting large amounts of carbon as part of natural metabolism. For example, a R. eutropha strain that is incapable of synthesizing intracellular PHA will overproduce and eventually secrete pyruvate (Steinbüchel and Schlegel 1989), which can be used as a precursor molecule for ethanol synthesis (Fig. 1). However, a model organism like Saccharomyces cerevisiae, which has traditionally produced ethanol in many beverages and, more recently as biofuel from sugarcane juice and other carbon sources (Macedo and Brigham 2014), is such an efficient producer of the alcohol that more engineering and process design efforts would be needed for other organisms to compete with the product yield and efficiency of S. cerevisiae. In recent years, focus has been on the engineering of R. eutropha strains to produce higher alcohols.
Isopropanol
Isopropanol is an interesting biofuel molecule due to its high research octane number (RON = 129). Indeed, isopropanol is already being used as a gasoline and diesel fuel additive (Peralta-Yahya and Keasling 2010). Isopropanol is used as an industrial solvent and blending agent in products like paints and inks. To add to its many uses, isopropanol is also a chemical intermediate and is converted to the useful polymer propylene (Grousseau et al. 2014). Thus, isopropanol produced in a sustainable manner (e.g., fermentatively) is a promising “green” chemical target molecule for multiple industries and applications.
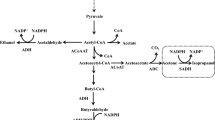
The heterologous expression of engineered isopropanol pathways was evaluated in R. eutropha strain Re2133, which is incapable of producing PHB (Budde et al. 2011). Synthetic production pathways were designed and expressed from plasmids to efficiently divert carbon flow from PHB precursors (acetyl-CoA and acetoacetyl-CoA) towards isopropanol. Among the constructed pathways, strain Re2133/pEG7c (Table 1) overexpressing a native β-ketothiolase (phaA) and CoA-transferase (ctfAB) genes, respectively, and codon-optimized Clostridium acetoacetate decarboxylase and alcohol dehydrogenase genes (Fig. 2) produced up to 3.44 g/L isopropanol in batch culture when the strain was grown on fructose as the sole source of carbon. Examination of carbon distribution in Re2133/pEG7c showed that more carbon went through the engineered isopropanol synthesis pathway in this strain than in any other constructed strain. The overall yield of isopropanol (compared to substrate used) and the overall specific productivity of isopropanol from this strain were higher than the values reported in the literature to date for other engineered isopropanol production strains in batch culture (Grousseau et al. 2014).
Synthesis of isopropanol from acetyl-CoA in engineered R. eutropha strains. Two molecules of acetyl-CoA form one molecule of acetoacetyl-CoA using the β-ketothiolase enzyme (PhaA). The heterologously expressed acetoacetyl-CoA transferase (1) exchanges the Coenzyme A group with succinate to form succinyl-CoA and acetoacetate. The heterologously expressed acetoacetate decarboxylase (2) produces acetone and CO2. Lastly, a heterologously expressed alcohol dehydrogenase (3) converts acetone to isopropanol. The polyhydroxybutyrate biosynthesis pathway (PhaA, PhaB, PhaC) can be a major carbon sink, as it shares key intermediates, acetyl-CoA, and acetoacetyl-CoA, with the isopropanol synthesis pathway. Thus, the phaB and phaC genes were deleted to maximize isopropanol yield (Grousseau et al. 2014)
Conversion of greenhouse gases like CO2 to useful bioproducts has become a goal for many researchers in recent years. The ability of R. eutropha to grow and synthesize products chemolithoautotrophically is of interest because engineered R. eutropha can then be used as a biocatalyst for conversion of CO2, H2, and O2 to polymers, biofuels, and other products, which serves as a method of sustainable production and a potential method for mitigating greenhouse gases. With this in mind, a bioelectrochemical cell has been developed that uses a cobalt phosphate catalyst to split water (2H2O) into O2 and protons (4H+) and then a nickel molybdenum zinc (NiMoZn) or stainless-steel cathode to reduce protons to make H2. The H2 produced in this manner is then used as an energy source for autotrophically growing R. eutropha cultures (Torella et al. 2015). To test the ability of this bioelectrochemical cell to produce biofuel products, engineered R. eutropha cells were used to make isopropanol. The R. eutropha strain Re2133/pEG12 (Table 1) is unable to produce PHB and contains an engineered pathway for isopropanol production (Grousseau et al. 2014). The Re2133/pEG12 strain grew robustly in this bioelectrochemical cell setup, produced up to 216 mg/L isopropanol, which is the highest bioelectrochemical fuel yield reported to date (Torella et al. 2015).
Although Grousseau and coworkers demonstrated high titers of isopropanol using an engineered R. eutropha strain (Grousseau et al. 2014), further increase in production titer was not possible for the engineered strain due to isopropanol toxicity and subsequent growth inhibition. Increasing expression of native chaperonin genes has been shown to mitigate alcohol toxicity in production strains (Tomas et al. 2003; Zingaro and Papoutsakis 2013). As such, chaperonin genes groESL were identified in the genome of R. eutropha. The GroEL and GroES complex assist in the folding of proteins and have been implicated in a variety of bacterial stress responses (Vilasi et al. 2018). It was seen that overexpression of native groEL and groES genes in the engineered R. eutropha strain Re2133/pEG23 (also containing the isopropanol synthesis pathway) led to a better tolerance of the strain towards exogenous isopropanol. The final concentration of isopropanol produced by this strain was 9.8 g/L in a fed-batch culture when fructose was the sole carbon source. This new strain overexpressing the chaperonin genes also showed higher enzyme activity levels of the two heterologous isopropanol synthesis enzymes (acetoacetate carboxylase and alcohol dehydrogenase), suggesting a higher specific production rate of isopropanol at the expense of accumulation of the metabolic intermediate acetone. It was observed that increasing expression of the native GroESL chaperonins resulted in a 9–18% increase in the isopropanol yield on fructose (Marc et al. 2017).
C4 alcohols: Butanol and Isobutanol
The alcohols n-butanol and isobutanol have been considered attractive biofuel targets. Both alcohols possess greater energy density than ethanol or isopropanol, and isobutanol has a Research Octane Number (RON) that is similar to that of gasoline and liquefied petroleum gas (LPG) (Fei et al. 2013). n-Butanol is produced by several wild-type strains of Clostridium via the acetone-butanol-ethanol (ABE) process (Richter et al. 2012; Schwarz et al. 2017), and several groups have used E. coli as a microbial chassis for recombinant n-butanol production (Bond-Watts et al. 2011; Shen et al. 2011). The most likely pathway that could be used for n-butanol synthesis in R. eutropha is through 3-hydroxybutyryl-CoA and crotonyl-CoA as intermediate molecules (Lan and Liao 2012; Shen et al. 2011). In a recent work, n-butanol was produced using crotonyl-CoA intermediate. Heterologous genes, including enoyl-CoA hydratase (phaJ) from Aeromonas caviae, trans-2-enoyl-CoA reductase (ter) from Treponema denticola, CoA-acylating aldehyde dehydrogenase (bldh) from Clostridium saccharoperbutyacetonicum, and an aldehyde reductase/alcohol dehydrogenase (yqhD) from E. coli, were introduced into R. eutropha H16 (Fig. 3). The resulting engineered strain was able to produce 30 mg/L n-butanol from fructose as the sole carbon source. The amount of n-butanol was increased to 80 mg/L upon overexpression of the native phaAB genes. Placing the n-butanol biosynthesis operon under a stronger promoter increased production even further to 200 mg/L using fructose as the sole carbon source and allowed for 30 mg/L n-butanol production using formic acid as the sole carbon source (Black et al. 2018).
n-butanol, isobutanol, and 3-methyl-1-butanol (3MB) biosynthesis pathways in engineered R. eutropha strains. Isobutanol produced by the Ehrlich pathway shares intermediates with the native valine biosynthesis pathway (i.e., pyruvate conversion to 2-ketoisovalerate). A heterologous keto acid decarboxylase (1) converts 2-ketoisovalerate to isobutyraldehyde. An alcohol dehydrogenase enzyme (2) converts isobutyraldehyde to isobutanol. The alcohol dehydrogenase could be a constitutively expressed native enzyme (Lu et al. 2012) or a heterologously expressed enzyme (Li et al. 2012). An alternative isobutanol synthesis pathway involves the conversion of acetyl-CoA to crotonyl-CoA, which can be done by native PHA pathway enzymes (Eggers and Steinbuchel 2013; Kawashima et al. 2012). Crotonyl-CoA is converted to butyryl-CoA by a heterologously expressed CoA-acylating aldehyde dehydrogenase (3). Butyryl-CoA is converted to isobutyryl-CoA by an overexpressed, native isobutyryl-CoA mutase (4). Heterologously expressed CoA-acylating aldehyde dehydrogenase and alcohol dehydrogenase enzymes (5) convert isobutyryl-CoA to isobutanol. These enzymes also convert butyryl-CoA to n-butanol
Isobutanol (2-butanol) is an alcohol biofuel that possesses > 90% of the energy density of petroleum-based gasoline, is compatible with the current fuel distribution infrastructure of most countries, and can be used to run vehicles without gasoline blending (Brigham et al. 2013). Isobutanol is one fusel alcohol that can be synthesized via the Ehrlich pathway. In R. eutropha and other organisms, precursors for isobutanol synthesis can come from branched-chain amino acid biosynthesis pathways, specifically the valine synthesis pathway (Lu et al. 2012). In R. eutropha, two additional enzyme activities are needed to produce isobutanol and another fusel alcohol, 3-methyl-1-butanol (3MB), from valine synthetic pathway precursors: ketoisovalerate decarboxylase (KIVD) and alcohol dehydrogenase (ADH) (Brigham et al. 2013; Lu et al. 2012). The kivd gene from Lactococcus lactis was heterologously expressed in R. eutropha to impart the necessary decarboxylase activity. The KIVD gene product has been demonstrated to possess broad-substrate-specificity, including the isobutanol precursor ketoisovalerate (Atsumi et al. 2008; De la Plaza et al. 2004). There are several genes present in the R. eutropha H16 genome that are annotated as “short-chain alcohol dehydrogenase” (Pohlmann et al. 2006). However, assays of extracts from aerobically grown cells demonstrate that no alcohol dehydrogenase is present. Alcohol dehydrogenase activity is observed in anaerobically grown R. eutropha cells, suggesting that ADH enzymes may only be active under anoxic conditions (Brigham et al. 2013). Fortuitously, mutant strains of R. eutropha H16 exhibiting constitutive alcohol dehydrogenase activity were previously characterized (Steinbüchel et al. 1987; Jendrossek et al. 1990). These strains were deleted of PHA synthesis genes and used in combination with the overexpression of native branched-chain amino acid biosynthesis pathway genes and the expression of the heterologous kivd gene for the biosynthesis of isobutanol. A schematic of this isobutanol synthesis pathway is shown in Fig. 3. Several potential pyruvate or ketoisovalerate “sinks” were also removed from the isobutanol production strain in an effort to increase biofuel yield. The resulting engineered strain was shown to produce 270 mg/L isobutanol and 40 mg/L 3MB when cultivated on fructose. In a semi-continuous culture, the engineered strain produced over 14 g/L total branched-chain alcohols over the course of 50 days (Lu et al. 2012). As all alcohols are toxic products for wild-type or engineered R. eutropha strains, one method for increasing yield is the design of a two-stage fermentation strategy where biomass is accumulated to high levels in the first stage, and product is synthesized in the second stage. Fei and coworkers performed two-stage fed batch cultivations where the engineered R. eutropha strain was subjected to nitrogen limitation during the second stage of cultivation. The total alcohol production was shown to increase to 790 mg/L, with a product yield of 0.03 g/g fructose consumed (Fei et al. 2013).
Li and coworkers also designed a strain capable of producing isobutanol and 3MB. A heterologous ketoisovalerate supply pathway, consisting of an acetohydroxyacid synthase gene (alsS) from Bacillus subtilis and acetohydroxyacid isomeroreductase and dihydroxyacid dehydratase genes (ilvC and ilvD, respectively) from E. coli, was introduced into the R. eutropha genome. In addition, the kivd gene from L. lactis and the yqhD gene from E. coli were added to complete the fusel alcohol production pathway. This engineered R. eutropha strain produced 846 mg/L isobutanol and 570 mg/L 3MB using formic acid as the main carbon and energy source. Cell growth and biofuel production were also examined in an electrobioreactor. At the cathode of the electrobioreactor CO2 was converted to formate, which was then used as a carbon source for growth and production. Using this electrochemical generation system to produce formate for R. eutropha growth and production, over 140 mg/L combined biofuels were produced (Li et al. 2012).
Black and coworkers demonstrated a novel isobutanol biosynthesis pathway in R. eutropha (Fig. 3). Using the heterologous n-butanol biosynthesis pathway discussed above, a native, novel isobutyryl-CoA mutase, Sbm1, was overexpressed to produce 32 mg/L isobutanol using fructose as the sole carbon source. The Sbm1 enzyme activity is highly dependent on the presence of vitamin B12 in the cytoplasm, and the highest levels of isobutanol production were observed when 1 mM of vitamin B12 was added to the culture media. Concomitantly, n-butanol was also produced by this engineered R. eutropha strain (Black et al. 2018). Table 1 summarizes the n-butanol and isobutanol-producing R. eutropha strains.
Synthesis of non-alcohol fuel molecules
Methyl ketones/terpenes
Methyl ketones have captured interest of biofuel researchers for their potential relevance as a fuel molecule. The highly reduced, aliphatic character of methyl ketones, coupled with the ability to engineer organisms to produce these molecules, makes them ideal candidates for biofuels. Methyl ketones can be produced by incomplete β-oxidation of fatty acids (Goh et al. 2012). R. eutropha has been engineered for the production of fatty acid-derived methyl ketones. Using a strain that was deficient in fatty acid β-oxidation and PHB synthesis, overexpression of a cytoplasmic version of the TesA thioesterase in R. eutropha led to > 150-fold increase in fatty acid synthesis in defined media conditions. With an increase in precursor supply demonstrated, the heterologous expression of three heterologous genes (acyl coenzyme A oxidase gene from Micrococcus luteus and fadB and fadM from E. coli) was expressed, which resulted in the production of 50–65 mg/L methyl ketones under heterotrophic growth conditions and 50–180 mg/L methyl ketones under chemolithoautotrophic growth conditions (Muller et al. 2013).
Recently, a R. eutropha-based process was developed to produce terpenes, specifically the sesquiterpene α-humulene. This was achieved by heterologous expression of the mevalonate pathway (from Myxococcus xanthus) and α-humulene synthase (from Zingiber zerumbet). Humulene yields of up to 10 mg/g cell dry weight (CDW) were produced under heterotrophic conditions. Under chemolithoautotrophic conditions in a bioelectrochemical system, humulene yields were shown to be 17 mg/g CDW. This is the first reported chemolithoautotrophic production of a terpene from CO2, H2, and O2 and represents a promising starting point for high-value terpene production for different biological functions and applications (Krieg et al. 2018).
Alkanes/alkenes
Biologically produced alkanes and alkenes can serve as drop-in alternatives to petroleum-based fuels and have been of recent interest as metabolic pathways have been designed to allow for engineering of microorganisms to produce these molecules. Bi and coworkers developed a synthetic biology toolkit for bioproduction and demonstrated the synthesis of the hydrocarbons pentadecane and heptadecane by overexpressing synthetic acyl-ACP reductase (aar) and aldehyde decarbonylase (adc, Fig. 4) genes in engineered R. eutropha. Resulting strains were able to produce up to 6 mg/L of total hydrocarbons (Bi et al. 2013). Recently, a cyanobacterial alkane synthesis pathway was expressed in R. eutropha in the form of acyl-ACP reductase and aldehyde deformylating oxygenase (ADO, Fig. 4). Tridecane (C13), pentadecane (C15), heptadecane (C17), heptadecane (C17:1), and hexadecanal (C16) were produced in cultures grown on fructose, yielding 12 mg total hydrocarbon/g CDW. Growth and production were also tested in autotrophic cultures, with pentadecane, heptadecane, and heptadecane being produced with a yield of ~ 1 mg total hydrocarbon/g CDW. This is the first know attempt at autotrophic alkane/alkene production using engineered R. eutropha (Crepin et al. 2016).
Aldehyde decarbonylases (ADs) are key enzymes for heterologous alkane synthesis in engineered R. eutropha strains. ADs, such as the ADO enzyme (Crepin et al. 2014) and Adc enzyme (Bi et al. 2013), convert aldehydes to equimolar amounts of alkane and formate. The alkanes can then be collected for use as biofuels
Overcoming biofuel product toxicity to engineered R. eutropha strains
R. eutropha can be engineered to produce many different types of biofuel molecules. This is advantageous because R. eutropha is proficient at converting carbon into value-added products, in part because of its amenability to genetic engineering. However, one caveat is the toxicity of even low levels (< 2.0% v/v) of biofuel molecules, especially alcohols, to R. eutropha (Bernardi et al. 2016). Engineering a stable R. eutropha biofuel production platform will most definitely involve overcoming this toxicity, either by metabolic engineering or by fermentation and recovery strategy, or a combination of both. Experimental evolution of R. eutropha cells in the presence of increasing concentrations of isobutanol has been examined. A prior, similar study had been performed using E. coli strains, which identified some genetic determinants for isobutanol tolerance (Minty et al. 2011). Mutations in two genes, acrA and acrA6, were shown to affect tolerance to isobutanol in R. eutropha. In-frame deletions of these genes were shown to improve survival of R. eutropha cells in the presence of the alcohol (Bernardi et al. 2016).
Another option for mitigation of biofuel molecule toxicity to R. eutropha is the removal of the molecule from the production media concomitant with its production. Keeping levels of alcohol or other biofuel molecules below the toxic level would allow for increased cell survival and provide more optimal conditions for continued biofuel production. In a semi-continuous culture experiment, engineered R. eutropha cells were recovered from the production medium and placed in fresh medium every 24 h. Under these conditions, isobutanol was produced by the culture continuously for 50 days (Lu et al. 2012). In fermenter cultures, simultaneous fermentation and biofuel product removal has been examined using engineered E. coli. Recovery techniques like liquid-liquid extraction and pervaporation were shown to increase biofuel molecule production in E. coli cultures (see (Zheng et al. 2009) and references therein). To the best of our knowledge, there have been no published works on the effect of simultaneous fermentation and recovery in R. eutropha cultures.
Chemolithoautotrophic growth and production by R. eutropha
One of the principal attractions of R. eutropha as a microbial chassis for production of biofuel is the ability of the organism to grow and synthesize product chemolithoautotrophically. This suggests that CO2, a greenhouse gas, could be collected and used as a source of carbon to produce biomass and a value-added compound, thus potentially producing a fuel with a diminished “carbon footprint.” Indeed, wild-type R. eutropha has been shown to synthesize PHB under chemolithoautotrophic conditions (Ishizaki et al. 2001; Volova and Kalacheva 2005). R. eutropha uses the Calvin-Benson-Bassham cycle to grow using CO2 as a carbon source, H2 as a source of reducing equivalents and O2 as a terminal electron acceptor. The genome of R. eutropha contains two cbb operons that contain the genes necessary for chemolithoautotrophic growth. Both operons, one present on the chromosome and the other on the megaplasmid pHG1, are under the strict control of the regulator CbbR, and are expressed during growth on CO2, H2, and O2 (Schwartz et al. 2009; Bowien and Kusian 2002).
R. eutropha also expresses O2-tolerant hydrogenase enzymes, which allow robust autotrophic growth and production, due to their ability to keep the NAD+/NADH ratios balanced in the presence of O2. There are two O2-tolerant hydrogenases in R. eutropha, each with a critical role to play in hydrogen metabolism. The membrane-bound hydrogenase feeds electrons derived from the oxidation of H2 into the respiratory chain, and the cytoplasmic soluble hydrogenase connects H2 oxidation directly to the reduction of NAD+ to form NADH (Burgdorf et al. 2005; Lenz et al. 2010). Because these hydrogenase enzyme complexes are resistant to inhibition by oxygen, H2 oxidation can be coupled to the reduction of O2, which allows for bioproduct synthesis by engineered R. eutropha using H2 and O2 produced from electrically split water along with CO2 as the carbon source (Brigham et al. 2013).
Carbonic anhydrase enzymes are responsible for the interconversion of CO2 to bicarbonate ion (HCO3−), which can be instrumental for CO2 transport in a chemolithoautotrophically growing cell. R. eutropha is unique in that it expresses multiple carbonic anhydrase enzymes, each with its own specific role in cellular metabolism. One of the four carbonic anhydrases, the periplasm-located Caa, converts CO2 to bicarbonate, which is then transported into the cell cytoplasm (Gai et al. 2014). The role of Caa could be critical if R. eutropha is growing and synthesizing bioproduct chemolithoautotrophically, using CO2 as the sole carbon source. The carbonic anhydrase enzyme Can has been demonstrated to be required for growth of R. eutropha under ambient CO2 conditions. Can could be responsible for providing CO2 to key carboxylation reactions that take place in the R. eutropha cell under heterotrophic conditions (Kusian et al. 2002). Another carbonic anhydrase, Cag, could be playing a similar role to Can, perhaps supplying CO2 to the enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), a key enzyme in the Calvin-Benson-Bassham cycle. It is thought that the carbonic anhydrase Can2 could use the interconversion of CO2 to bicarbonate to regulate cytoplasmic pH (Gai et al. 2014). The interplay of these four enzymes can give us a glimpse of the importance of CO2 metabolism in R. eutropha.
Outlook
It has been said that, while microbial conversion of CO2 is possible, “will it make a difference?” (E. Toone, personal communication). The rationale behind this question is that we have shown that R. eutropha can produce a variety of solvent molecules that can be used as biofuels, but if the productivity (space time yield) of these fuel molecules is not significant (i.e., > 2.0 g product/L/h), then biofuels made in this way will not be competitive with petroleum-based fuels. To increase biofuel productivity of R. eutropha strains, further strain engineering must be performed. RuBisCO assimilates CO2 and is the key enzyme in the Calvin-Benson-Bassham cycle. Unfortunately, O2 is a competing substrate for active sites in RuBisCO (Klein et al. 2009), which is a problem for growth and production using CO2, H2, and O2. Carboxysomes are multi-protein microcompartment structures found in many cyanobacteria that can act as an “air lock” to selectively exclude certain molecules, such as O2. These microcompartments contain RuBisCO and carbonic anhydrase, and key steps in carbon fixation take place there in relatively high-CO2, low-O2 environments (Bonacci et al. 2012; Klein et al. 2009). This serves to make carbon fixation by RuBisCO more efficient by decreasing the likelihood of the enzyme encountering a competing substrate. A heterologous carboxysome operon from Halothiobacillus neapolitanus was expressed under an arabinose-inducible promoter in R. eutropha. Induction of gene expression demonstrated increase in overall RuBisCO and carbonic anhydrase activities (Li et al. 2015). However, the assembly of carboxysome-like microstructures in the R. eutropha cytoplasm has yet to be observed. Expression of functioning carboxysomes in R. eutropha and demonstration of O2 exclusion could increase efficiency of carbon fixation for growth and biofuel production.
One concern about chemolithoautotrophic fermentation is the use of H2 and O2 gases, which, in certain conditions, represents an explosion hazard. To mitigate this issue, a novel bioreactor has been designed that separates the two incompatible gases, decreasing the risk of an explosion. R. eutropha cells would grow on the wall of the hollow fiber and have access to H2 and O2 for growth and production, one gas transported through the lumen of the hollow fiber, the other will diffuse across the fiber wall to reach the cells. This type of reactor setup also has the added benefit to be able to transport any secreted biofuel product away from the cells, thus also mitigating product toxicity (Brigham et al. 2013). Successful scale-up of this type of bioreactor could help revolutionize the CO2-to-biofuel space.
At this point in time, biofuels remain a conscious choice of alternative fuel, as opposed to a necessity. The positive aspects of biofuels make for a viable and exciting field of study. Robust biofuel research and development adds needed diversity to the overall fuel portfolio of companies and nations. R. eutropha, as a versatile organism able to interconvert and reclaim “waste” carbon, is an attractive biocatalyst.
References
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451(7174):86–90. https://doi.org/10.1038/nature06450
Bernardi AC, Gai CS, Lu J, Sinskey AJ, Brigham CJ (2016) Experimental evolution and gene knockout studies reveal AcrA-mediated isobutanol tolerance in Ralstonia eutropha. J Biosci Bioeng 122(1):64–69. https://doi.org/10.1016/j.jbiosc.2015.12.015
Bi C, Su P, Müller J, Yeh YC, Chhabra SR, Beller HR, Singer SW, Hillson NJ (2013) Development of a broad-host synthetic biology toolbox for Ralstonia eutropha and its application to engineering hydrocarbon biofuel production. Microb Cell Factories 12:107. https://doi.org/10.1186/1475-2859-12-107
Black WB, Zhang L, Kamoku C, Liao JC, Li H (2018) Rearrangement of coenzyme A-acetylated carbon chain enables synthesis of isobutanol via a novel pathway in Ralstonia eutropha. ACS Synth Biol 7:794–800. https://doi.org/10.1021/acssynbio.7b00409
Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF (2012) Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci U S A 109(2):478–483
Bond-Watts BB, Bellerose RJ, Chang MC (2011) Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol 7(4):222–227. https://doi.org/10.1038/nchembio.537
Bowien B, Kusian B (2002) Genetics and control of CO2 assimilation in the chemoautotroph Ralstonia eutropha. Arch Microbiol 178(2):85–93. https://doi.org/10.1007/s00203-002-0441-3
Brigham CJ, Gai CS, Lu J, Speth D, Worden RM, Sinskey AJ (2013) Engineering Ralstonia eutropha for production of isobutanol from CO2, H2, and O2. In: Lee JW (Ed.) Advanced Biofuels and Bioproducts. Springer. New York 1065–1090. doi: https://doi.org/10.1007/978-1-4614-3348-4_39
Brigham CJ, Zhila N, Shishatskaya E, Volova TG, Sinskey AJ (2012) Manipulation of Ralstonia eutropha carbon storage pathways to produce useful bio-based products. Subcell Biochem 64:343–366. https://doi.org/10.1007/978-94-007-5055-5_17.
Budde CF, Riedel SL, Willis LB, Rha CK, Sinskey AJ (2011) Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from plant oil by engineered Ralstonia eutropha strains. Appl Environ Microbiol 77(9):2847–2854. https://doi.org/10.1128/AEM.02429-10
Burgdorf T, Lenz O, Buhrke T, van der Linden E, Jones AK, Albracht SPJ, Friedrich B (2005) [NiFe]-hydrogenases of Ralstonia eutropha H16: modular enzymes for oxygen-tolerant biological hydrogen oxidation. J Mol Microbiol Biotechnol 10(2–4):181–196. https://doi.org/10.1159/000091564
Clark D, Cronan J (1980) Escherichia coli mutants with altered control of alcohol dehydrogenase and nitrate reductase. J Bacteriol 141(1):177–183
Crepin L, Lombard E, Guillouet SE (2016) Metabolic engineering of Cupriavidus necator for heterotrophic and autotrophic alka(e)ne production. Metab Eng 37:92–101. https://doi.org/10.1016/j.ymben.2016.05.002
De la Plaza M, de Palencia PF, Pelaez C, Requena T (2004) Biochemical and molecular characterization of alpha-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol Lett 238(2):367–374
Eggers J, Steinbüchel A (2013) Poly(3-hydroxybutyrate) degradation in Ralstonia eutropha H16 is mediated stereoselectively to (S)-3-hydroxybutyryl coenzyme A (CoA) via crotonyl-CoA. J Bacteriol 195(14):3213–3223. https://doi.org/10.1128/JB.00358-13
Fei Q, Brigham CJ, Lu J, Fu R, Sinskey AJ (2013) Production of branched-chain alcohols by recombinant Ralstonia eutropha in fed-batch cultivation. Biomass Bioenergy 56:334–341
Gai CS, Lu J, Brigham CJ, Bernardi AC, Sinskey AJ (2014) Insights into bacterial CO2 metabolism revealed by the characterization of four carbonic anhydrases in Ralstonia eutropha H16. AMB Express 4:2. https://doi.org/10.1186/2191-0855-4-2
Goh EB, Baidoo EEK, Keasling J, Beller H (2012) Engineering of bacterial methyl ketone synthesis for biofuels. Appl Environ Microbiol 78(1):70–80. https://doi.org/10.1128/AEM.06785-11
Grousseau E, Lu J, Gorret N, Guillouet SE, Sinskey AJ (2014) Isopropanol production with engineered Cupriavidus necator as bioproduction platform. Appl Microbiol Biotechnol 98(9):4277–4290. https://doi.org/10.1007/s00253-014-5591-0
Ishizaki A, Tanaka K, Taga N (2001) Microbial production of poly-D-3-hydroxybutyrate from CO2. Appl Microbiol Biotechnol 57(1–2):6–12
Jendrossek D (2009) Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191(10):3195–3202. https://doi.org/10.1128/JB.01723-08
Jendrossek D, Handrick R (2002) Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol 56:403–432. https://doi.org/10.1146/annurev.micro.56.012302.160838
Jendrossek D, Kruger N, Steinbüchel A (1990) Characterization of alcohol dehydrogenase genes of derepressible wild-type Alcaligenes eutrophus H16 and constitutive mutants. J Bacteriol 172(9):4844–4851. https://doi.org/10.1128/jb.172.9.4844-4851.1990
Jeon BY, Yi JY, Jung IL, Park DH (2013) Activation of ethanol production by combination of recombinant Ralstonia eutropha and electrochemical reducing power. Adv Microbiol 3(1):42–45. https://doi.org/10.4236/aim.2013.31006
Kawashima Y, Chang W, Mifune J, Orita I, Nakamura S, Fukui T (2012) Characterization and functional analyses of R-specific enoyl coenzyme A hydratases in polyhydroxyalkanoate-producing Ralstonia eutropha. Appl Environ Miocrbiol 78(2):493–502. https://doi.org/10.1128/AEM.06937-11
Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA (2009) Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol 392(2):319–333. https://doi.org/10.1016/j.jmb.2009.03.056
Krieg T, Sydow A, Faust S, Huth I, Holtmann D (2018) CO2 to terpenes: autotrophic and electroautotrophic α-humulene production with Cupriavidus necator. Angew Chem Int Ed 57:1–5. https://doi.org/10.1002/anie.201711302
Kusian B, Sueltmeyer D, Bowein B (2002) Carbonic anhydrase is essential for growth of Ralstonia eutropha at ambient CO2 concentrations. J Bacteriol 184(18):5018–5026. https://doi.org/10.1128/JB.184.18.5018-5026.2002
Lan EI, Liao JC (2012) Microbial synthesis of n-butanol, isobutanol, and other higher alcohols from diverse resources. Bioresour Technol 135:339–349. https://doi.org/10.1016/j.biortech.2012.09.104
Lee H, Jeon B, Oh M (2016) Microbial production of ethanol from acetate by engineered Ralstonia eutropha. Biotechnol Bioprocess Eng 21: 402–407 (2016) DOI https://doi.org/10.1007/s12257-016-0197-2
Lenz O, Ludwig M, Schubert T, Buerstel I, Ganskow S, Goris T, Schwarze A, Friedrich B (2010) H2 conversion in the presence of O2 as performed by the membrane-bound [NiFe]-hydrogenase of Ralstonia eutropha. ChemPhysChem 11(6):1107–1119. https://doi.org/10.1002/cphc.200901002
Li S, Lu J, Brigham CJ, Sinskey AJ (2015) Improving the efficiency of carbon fixation in Ralstonia eutropha with carbon-concentrating microcompartments. MURJ 29:36–42
Li H, Opgenorth PH, Wernick DG, Rogers S, Wu TY, Higashide W, Malati P, Huo YX, Cho KM, Liao JC (2012) Integrated electromicrobial conversion of CO2 to higher alcohols. Science 335(6076):1596. https://doi.org/10.1126/science.1217643
Lu J, Brigham CJ, Gai CS, Sinskey AJ (2012) Studies on the production of branched-chain alcohols in engineered Ralstonia eutropha. Appl Microbiol Biotechnol 96(1):283–297. https://doi.org/10.1007/s00253-012-4320-9
Macedo N, Brigham CJ (2014) From beverages to biofuels: journeys of ethanol-producing microorganisms. Int J Biotechnol Wellness Ind 3(3):79–87. https://doi.org/10.6000/1927-3037.2014.03.03.1
Marc J, Grousseau E, Lombard E, Sinskey AJ, Gorret N (2017) Over expression of GroESL in Cupriavidus necator for heterotrophic and autotrophic isopropanol production. Metabolic Eng 42:74–84. https://doi.org/10.1016/j.ymben.2017.05.007
Minty JJ, Lesnefsky AA, Lin F, Chen Y, Zaroff TA, Veloso AB, Xie B, McConnell CA, Ward RJ, Schwartz DR, Rouillard JM, Gao Y, Gulari E, Lin XN (2011) Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb Cell Factories 10:18. https://doi.org/10.1186/1475-2859-10-18
Müller J, Maceachran D, Burd H, Sathitsuksanoh N, Bi C, Yeh Y, Lee TS, Hillson NJ, Chhabra SR, Singer SW, Beller HR (2013) Engineering of Ralstonia eutropha H16 for autotrophic and heterotrophic production of methyl ketones. Appl Environ Microbiol 79(14):4433–4439. https://doi.org/10.1128/AEM.00973-13
Peralta-Yahya PP, Keasling JD (2010) Advanced biofuel production in microbes. Biotechnol J 5(2):147–162. https://doi.org/10.1002/biot.200900220
Pfeiffer D, Wahl A, Jendrossek D (2011) Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82(4):936–951. https://doi.org/10.1111/j.1365-2958.2011.07869.x
Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, Cramm R, Eitinger T, Ewring C, Potter M, Schwartz E, Strittmatter A, Voss I, Gottschalk G, Steinbüchel A, Friedrich B, Bowein B (2006) Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat Biotechnol 24(10):1257–1262
Richter H, Qureshi N, Heger S, Dien B, Cotta MA, Angenent LT (2012) Prolonged conversion of n-butyrate to n-butanol with Clostridium saccharoperbutylacetonicum in a two-stage continuous culture with in-situ product removal. Biotechnol Bioeng 109:913–921. https://doi.org/10.1002/bit.24380
Schatz A, Bovell C Jr (1952) Growth and hydrogenase activity of a new bacterium, Hydrogenomonas facilis. J Bacteriol 63(1):87–98
Schwartz E, Voigt B, Zuehlke D, Pohlmann A, Lenz O, Albrecht D, Schwarze A, Kohlmann Y, Krause C, Hecker M, Friedrich B (2009) A proteomic view of the chemolithoautotrophic lifestyle of Ralstonia eutropha H16. Proteomics 9(22):5132–5142. https://doi.org/10.1002/pmic.200900333
Schwarz KM, Grosse-Honebrink A, Derecka K, Rotta C, Zhang Y, Minton NP (2017) Towards improved butanol production through targeted genetic modification of Clostridium pasteurianum. Metab Eng 40:124–137. https://doi.org/10.1016/j.ymben.2017.01.009
Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, Liao JC (2011) Driving forces enable high-titer anaerobic 1-butanol synthesis in E. coli. Appl Environ Microbiol 77(9):2901–2915. https://doi.org/10.1128/AEM.03034-10
Steinbüchel A (1991) Polyhydroxyalkanoic acids. In: Byrom D (Ed.) Biomaterials—novel materials from biological sources, Palgrave MacMillan, UK pp. 123–213. doi: https://doi.org/10.1007/978-1-349-11167-1_3
Steinbüchel A, Frund C, Jendrossek D, Schlegel HG (1987) Isolation of mutants of Alcaligenes eutrophus unable to derepress the fermentative alcohol-dehydrogenase. Arch Microbiol 148(3):178–186. https://doi.org/10.1007/BF00414809
Steinbüchel A, Schlegel HG (1989) Excretion of pyruvate by mutants of Alcaligenes eutrophus which are impaired in the accumulation of poly-([beta]-hydroxybutyric acid) (PHB), under conditions permitting synthesis of PHB. Appl Microbiol Biotechnol 31(2):168–175 Doi.org/10.1007/BF00262457
Sznajder A, Pfeiffer D, Jendrossek D (2015) Comparative proteome analysis reveals four novel polyhydroxybutyrate (PHB) granule-associated proteins in Ralstonia eutropha H16. Appl Environ Microbiol 81(5):1847–1858. https://doi.org/10.1128/AEM.03791-14
Tomas CA, Welker NE, Papoutsakis ET (2003) Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell’s transcriptional program. Appl Environ Microbiol 69(8):4951–4965. https://doi.org/10.1128/AEM.69.8.4951-4965.2003
Torella JP, Gagliardi CJ, Chen JS, Bediako K, Colon B, Way JC, Silver PA, Nocera DG (2015) Efficient solar-to-fuels production from a hybrid microbial-water-splitting catalyst system. Proc Natl Acad Sci U S A 112(8):2337–2342. https://doi.org/10.1073/pnas.1424872112
Vilasi S, Bulone D, Bavisotto CC, Campanella C, Gammazza AM, San Biagio PL, Cappello F, de Macario EC, Macario AJL (2018) Chaperonin of group I: oligomeric spectrum and biochemical and biological implications. Front Mol Biosci 4(99):1–14. https://doi.org/10.3389/fmolb.2017.00099
Volova TG, Kalacheva GS (2005) The synthesis of hydroxybutyrate and hydroxyvalerate copolymers by the bacterium Ralstonia eutropha. Microbiology 74(1):63–69. https://doi.org/10.1007/s11021-005-0028-5
Wilde E (1962) Studies on growth and storage synthesis of Hydrogenomonas. Arch Mikrobiol 43(2):109–137. https://doi.org/10.1007/s00253-011-3102-0
Zheng YN, Li LZ, Xian M, Mia YJ, Yang JM, Xu X, He DZ (2009) Problems with the microbial production of butanol. J Ind Microbiol Biotechnol 36:1127–1138
Zingaro KA, Papoutsakis ET (2013) GroESL overexpression imparts Escherichia coli tolerance to i-, n-, and 2-butanol, 1,2,4-butanetriol and ethanol with complex and unpredictable patterns. Metab Eng 15:196–205. https://doi.org/10.1016/j.ymben.2012.07.009
Acknowledgements
We thank Prof. Alexander Steinbüchel and the editorial team of Applied Microbiology and Biotechnology for the opportunity to write and publish this work. CJB thanks Prof. Anthony Sinskey of Massachusetts Institute of Technology for the opportunity to work on a biofuel production project, which serves as the inspiration for continued interest in this topic.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Chakravarty, J., Brigham, C.J. Solvent production by engineered Ralstonia eutropha: channeling carbon to biofuel. Appl Microbiol Biotechnol 102, 5021–5031 (2018). https://doi.org/10.1007/s00253-018-9026-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9026-1