Abstract
Initial acetic acid can improve the ethanol oxidation rate of acetic acid bacteria for acetic acid fermentation. In this work, Acetobacter pasteurianus was cultured in ethanol-free medium, and energy production was found to increase by 150% through glucose consumption induced by initial acetic acid. However, oxidation of ethanol, instead of glucose, became the main energy production pathway when upon culturing ethanol containing medium. Proteome assay was used to analyze the metabolism change induced by initial acetic acid, which provided insight into carbon metabolic and energy regulation of A. pasteurianus to adapt to acetic acid fermentation conditions. Results were further confirmed by quantitative real-time PCR. In summary, decreased intracellular ATP as a result of initial acetic acid inhibition improved the energy metabolism to produce more energy and thus adapt to the acetic acid fermentation conditions. A. pasteurianus upregulated the expression of enzymes related to TCA and ethanol oxidation to improve the energy metabolism pathway upon the addition of initial acetic acid. However, enzymes involved in the pentose phosphate pathway, the main pathway of glucose metabolism, were downregulated to induce a change in carbon metabolism. Additionally, the enhancement of alcohol dehydrogenase expression promoted ethanol oxidation and strengthened the acetification rate, thereby producing a strong proton motive force that was necessary for energy production and cell tolerance to acetic acid.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acetic acid is a highly important organic acid in food industry. Also, it is the main composition of vinegars. In addition, acetic acid is recognized as an effective antimicrobial compound that prevents the growth of pathogenic and spoilage organisms in fermented foods (Gullo et al. 2014). Acetic acid is produced by acetic acid bacteria (AAB), belonging to the family of Acetobacteraceae (Deppenmeier and Ehrenreich 2009; Gillis and De Ley 1980; Gullo and Giudici 2008). These bacteria are commonly used in the industry due to its remarkable ability to oxidize ethanol to acetic acid and its tolerance to acetic acid (Hattori et al. 2011; Nakano and Fukaya 2008; Saichana et al. 2015). However, acetic acid as the fermentation product is considered as the main inhibitor for getting high acetification rate (Xia et al. 2015). Several mechanisms are responsible for the high tolerance against acetic acid in AAB. Among those mechanisms are (i) alcohol dehydrogenase (ADH) (Chinnawirotpisan et al. 2003; Takemura et al. 1993; Trcek et al. 2006), (ii) acetic acid assimilation (Lasko et al. 1997; Fukaya et al. 1993), (iii) pumping out of acetic acid by the ATP-binding cassette (ABC) transporters and proton motive force-dependent efflux system (Matsushita et al. 2005; Nakano et al. 2006), (iv) changes in cell morphology and membrane composition to adapt to acetic acid (Teitzel and Parsek 2003; Trcek et al. 2007), and (v) improvement in the adaption response to proteins involving acetic acid tolerance (Ishikawa et al. 2010; Okamoto-Kainuma et al. 2011; Okamoto-Kainuma et al. 2004). Recently, the proteomic method has been used to study the different molecular mechanism responsible for acetic acid tolerance in AAB. During acetic acid fermentation, most differentially expressed proteins were functionally related to the TCA cycle, stress response, and other metabolic processes in Komagataeibacter spp. (Andres-Barrao et al. 2016). The proteomic approach was utilized to analyze the proteomic profiles of A. pasteurianus cultured in different acidic titers (Xia et al. 2016). The differentially expressed proteomes of A. pasteurianus grown in glucose with those that underwent acetic acid fermentation were compared using method of 2D-PAGE as previously described (Andres-Barrao et al. 2012). The responses of cellular proteins were found to be related to protein folding, stress response, oxidation-reduction, metabolism, protein biosynthesis, and membrane modification.
The culture conditions are important for AAB acetic acid tolerance and acid production (Giudici et al. 2016; Gullo et al. 2016; Mounir et al. 2016; Shafiei et al. 2017). Oxygen, heat, ethanol stocks, and fermentation modes may contribute to the production and resistance of bacteria to high concentrations of acetic acid (Gullo et al. 2014). The initial acetic acid is important for the productivity in acetic acid fermentation, which can affect the ADH activity (Ory et al. 2002; Xia et al. 2015). Initial acetic acid can increase the acetification rate of acetic acid fermentation under 100 g/L total concentration (Krusong et al. 2015). A start-up protocol with 10 g/L initial acetic acid enhanced acetic acid production compared with other initial acetic acid concentrations (Xia et al. 2015). However, the metabolic mechanism induced by initial acetic acid is not further investigated. The objective of this study is to explore the role of initial acetic acid on cell growth and acetic acid fermentation. Specially, the metabolic change due to the initial acetic acid was researched using the proteomic method and quantitative real-time PCR (qRT-PCR).
Materials and methods
Strains, media, and cultivations
The A. pasteurianus CGMCC 3089 strain that was registered in the Chinese General Microbiological Culture Collection Center was stored at 4 °C on solid medium containing 15 g/L glucose, 10 g/L yeast extract, 20 g/L CaCO3, 28 g/L ethanol, and 17 g/L agar. Seed medium containing 30 g/L glucose, 15 g/L yeast extract, and 28 g/L ethanol and the fermentation medium containing 20 g/L glucose, 20 g/L peptone, and 64 g/L ethanol were used in the acetic acid fermentation process.
GY medium containing 30 g/L glucose and 15 g/L yeast extract; GYA medium containing 30 g/L glucose, 15 g/L yeast extract, and 10 g/L acetic acid; and GYAE medium containing 30 g/L glucose, 15 g/L yeast extract, 10 g/L acetic acid, and 28 g/L ethanol were used for proteome assay.
For acetic acid fermentation, the cells from slant cultures were incubated to 40 mL of seed medium in 250-mL Erlenmeyer flasks, and were cultured at 30 °C and 180 r/min. When the optical density (OD) at 610 nm was approximately 1.2 and acetic acid concentration was approximately 11 g/L, the cells were transferred into the fermentation medium with 10% inoculum. Acetic acid fermentation was performed in the 5-L self-inspiriting fermenter containing 3.5 L of fermentation medium at 30 °C and the aeration rate at 0.15 vvm. The dissolved oxygen (DO) was monitored with an oxygen electrode (Hamilton, Bonaduz, Switzerland). For proteome assay, cells from slant culture were incubated into GY medium. When the OD at 610 nm was approximately 0.7, the cells were transferred into GY, GYA, and GYAE media and cultured at 30 °C and 180 r/min to OD 610 nm was approximately 0.8, respectively.
Protein extraction
The cells were collected by centrifugation at 8000×g for 10 min at 4 °C, and the pellets were washed three times with PBS having pH 7.4 (137 mM of NaCl, 2.7 mM of KCl, 10 mM of Na2HPO4, and 2 mM of KH2PO4). Next, pellets were resuspended in 10 mL of pre-cooled lysis buffer (5 mM of Tris-HCl pH 8.0, 0. 5 M NaCl, 10 μg/mL PMSF, 1% DTT) and sonicated with a circulation of 40 w for 3 s and then pause 5 s until the microbial solutions appeared transparent. Subsequently, the samples were centrifuged at 12,000×g for 20 min at 4 °C. Then, the supernatant was collected and precipitated using 40 mL of cold acetone at −20 °C overnight. The protein was collected by centrifugation at 12,000×g for 20 min at 4 °C and washed three times by cold acetone. The collected protein was vacuum-dried and stored at −80 °C.
Trypsin digestion and LC-MS/MS analysis
The sample was dissolved in solution containing 6 M guanidine hydrochloride and 100 mM of ammonium bicarbonate with pH 8.0–8.5. The final concentration of protein was 2 to 3 μg/μL. A 200 μL solution was mixed with 2 μL of 1 M DTT. The mixture was incubated at 56 °C for 1 h. A 10-μL aliquot of 1 M iodoacetamide was added, and the mixture was incubated at room temperature for 40 min in the dark. The protein mixtures were spun, exchanged into 100 mM of ammonium bicarbonate buffer, and then incubated with trypsin (20:1 to 50:1) at 37 °C for 20 h. The HPLC solvents were 0.1% formic acid (v/v) aqueous (A) and 0.1% formic acid (v/v) acetonitrile (B). A total of 20 μL of the prepared sample was injected and subjected to gradient elution (rate = 200 μL/min) as follows: 5% B for 30 min, from 5% B to 32% B for 60 min, to 90% for 10 min, balance for 5 min, to 5% in 0.1 min, and 5% B for 35 min. The international standard (IS)-optimized mass parameters used in this study were 3.5-kV voltage of capillary and cone, 260 °C source temperature, and 260 °C desolvation temperature. System control and data collection were performed using XCalibur software (version 1.4) (Shen et al. 2015).
Protein identification and data analysis
The acquired MS/MS spectra were compared against the NCBI database by using Turbo SEQUEST program in the Proteomics Discovery software suite (version 1.2). For data analysis, protein data with relative expression (R) level of >1.5 and <0.75 and P value of <0.05 were selected to ensure upregulation and downregulation authenticity. The Cluster of Orthologous Groups (COG) database (http://www.ncbi.nlm.nih.gov/COG/) was used to classify and group the identified proteins. The gi numbers of the selected proteins were imported into the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) for biological pathway analysis.
Determination of gene transcription
For qRT-PCR experiments, the A. pasteurianus was cultured under conditions present or absent of 10 g/L initial acetic acid for acetic acid fermentation, and the cells were collected when the OD 610 nm reached about 0.6. The total RNA was isolated using RNA Plus Kit (TaKaRa Biotechnology, Dalian, China) following the manufacturer’s protocol. To remove residual DNA, total RNA was treated with DNase I for 30 min at 37 °C. RNA samples were reverse transcribed with RevertAid™ First Strand cDNA Synthesis Kit (Takara Biotechnology, Dalian, China) according to the manufacturer’s instructions. The 16S rRNA gene was used as internal standard. Primers used in this study are listed in Table 1.
Analytical methods
Viable count was measured using colony-counting method. Cells taken from various sampling points were serially diluted in sterile saline (0.85%, w/v). Next, 0.1 mL of diluted samples was plated on solid medium. Then, colony-forming units were determined after aerobic incubation at 30 °C until the distinct colonies appeared. The acidity of the broth was measured by 0.1 M NaOH with phenolphthalein as an indicator. Acetic acid stoichiometric yield is the conversion rate of acetic acid from ethanol and is calculated in the following equation (Qi et al. 2014):
where Y acid presents the concentration of acetic acid in fermentation broth (g/L), C ethanol presents the concentration of ethanol, and 1.304 presents 1 g/L ethanol completely transformed to obtain 1.304 g/L acetic acid. Concentrations of glucose and ethanol were quantified with a biosensor (ShanDong Academy of Sciences, Jinan, China). Intracellular ATP was quantified using BacTiter-Glo™ (Promega Inc., Madison, USA) (Gengenbacher et al. 2010; Xu et al. 2015). In this study, all experiments were repeated three times. Results were expressed as the mean with a standard error. The analysis of the differences between the categories was calculated with a confidence interval of 95 or 99% using SPSS (least significant differences) analysis.
Results
Effect of initial acetic acid on acetic acid fermentation
As shown in Fig. 1a, the average acetification rate was 1.83 g/L/h by A. pasteurianus CGMCC 3089 with 1% initial acetic acid. Of the substrate ethanol, 64 g/L was almost totally conversed into acetic acid (Fig. 1b), and the stoichiometric yield was 91.1%. However, the fermentation became sluggish without the presence of initial acetic acid. After 42 h of cultivation, the acetic acid was only 20.8 ± 0.05 g/L and the residual ethanol was about 40 g/L. Before 6 h cultivation, the viable cell in fermentation with initial acetic acid was lower compared with that of without initial acetic acid due to the inhibitory effects from acetic acid on cells. However, after 6 h fermentation, the viable cell in fermentation with initial acetic acid became more than that of without initial acetic acid. Moreover, the viable cell in fermentation without initial acetic acid quickly decreased after 15 h of fermentation when the acetic acid levels reached 10 g/L, resulting in the premature termination of fermentation. In addition, glucose consumption rate without initial acetic acid was higher than that 1% initial acetic acid before 25 h, as shown in Fig. 1b.
Effect of initial acetic acid on cell growth and energy product
In analyzing the effect of initial acetic acid on A. pasteurianus, the cell was further cultured in conditions with or without acetic acid and ethanol. As shown in Fig. 2, the lag phase of cell growth in GYA (containing acetic acid) medium was longer than that in GY (no acetic acid) medium due to the acetic acid inhibitory effect. However, the inhibitory effect of acetic acid on cell growth was not obviously in GYAE (containing acetic acid and ethanol) medium due to the present of ethanol. And the most viable cells obtained in GYAE medium were 16.4 and 39.3% higher than those obtained in GY and GYA media, respectively. This result indicated that ethanol reduced the inhibitory effect of acetic acid.
Additionally, the carbon source consumption and energy production were different in three media. Oxidation 1 mol of glucose produces about 2870 kJ of Gibbs free energy. The oxidation 1 mol of ethanol to acetic acid is associated with a Gibbs free energy of 493 kJ (Adler et al. 2014). As listed in Table 2, A. pasteurianus produced more Gibbs free energy when cultured in GYA or GYAE medium than in GY medium. This result is due to the presence of acetic acid, suggesting that the initial acetic acid could improve cell metabolism and produce more energy to resist the acidic environment. Also, initial acetic acid resulted in the similar energy production from glucose (GYA) or combining with ethanol (GYAE). However, the oxidation of ethanol became the main pathway for energy production in GYAE medium and a little of glucose (about 4.7 g/L) was consumed.
Those results suggested that initial acetic acid in GYA media resulted in increased glucose consumption to release more energy and resist the acetic acid stress. Specially, A. pasteurianus can adjust the energy metabolism and ethanol oxidation became the main energy source in acetic acid fermentation conditions (GYAE).
Proteome analysis of A. pasteurianus in different media
Acetic acid can induce the changes in the energy metabolism pathway. To further understand the whole metabolism change caused by initial acetic acid, the researchers compared the proteome grown under different media. The 1.5-fold upregulation and 0.75-fold downregulation were considered as the minimum level for differential expression, and the identified proteins were assigned by COG, which were classified into 18 categories (as shown in Fig. S1). In the presence or absence of initial acetic acid or ethanol conditions, the different expression proteins were mainly categorized into several classes of carbohydrate and energy metabolism, translation, ribosomal structure and biogenesis, amino acid transport and metabolism, etc. Because of the influence of initial acetic acid and ethanol on the glucose or ethanol consumption and energy production, the carbohydrate and energy metabolism of A. pasteurianus became the focus of this study.
The protein differentially expressed under GYA and GY media was compared (shown in Table S1). The following are various carbohydrate metabolic enzymes. In the pentose phosphate pathway (PPP), they were 6-phosphogluconate dehydrogenase, transaldolase, and ribose 5-phosphate isomerase. The differential enzymes in the glycolysis (EMP) pathway were glucose-6-phosphate isomerase, glyceraldehyde 3-phosphate dehydrogenase, phosphoglycerate kinase, phosphoglyceromutase, phosphopyruvate hydratase, and pyruvate kinase. The enzymes involved in the TCA cycle are pyruvate dehydrogenase, citrate synthase, isocitrate dehydrogenase, succinate dehydrogenase, fumarate hydratase, and malate:quinone oxidoreductase. These enzymes were upregulated due to initial acetic acid in GYA medium compared with those of the GY medium. The upregulation of those enzymes can strengthen PPP, EMP, and TCA cycle. The acetyl-CoA hydrolase and phosphotransacetylase can feed a new molecule of acetyl-CoA for TCA cycle by assimilating a molecule acetic acid. These particular enzymes were upregulated, suggesting that the more acetyl-CoA was produced and flow into the TCA cycle (Mullins et al. 2008; Wang et al. 2015b). Malic enzyme (malate dehydrogenase) involving pyruvate metabolism is known as a supply shunt of the TCA cycle as it can supply malate from pyruvate (Sakurai et al. 2013). The downregulation of malic enzyme may result in the enhancement of TCA cycle to produce more intermediate metabolites of TCA cycle due to the initial acetic acid in GYA medium. Also, the membrane-bound ADH large subunit involved in ethanol oxidation was upregulated due to the initial acetic acid.
The protein expression in the presence of ethanol (GYAE medium) was compared with that of the expression levels in the absence of ethanol (GYA medium). We observed that enzymes involved in the PPP (i.e., 6-phosphogluconate dehydrogenase, transaldolase, and ribose 5-phosphate isomerase) and in the TCA cycle (i.e., pyruvate dehydrogenase, citrate synthase, isocitrate dehydrogenase, fumarate hydratase, and malate:quinone oxidoreductase) were downregulated due to ethanol addition. Interestingly, malic enzyme was upregulated due to the presence of ethanol in the GYA medium. On the contrary, it was downregulated due to addition of initial acetic acid in the GY medium. Ethanol addition resulted in decreased PPP and TCA cycle compared with that in GYA medium, which suggested decreased glucose metabolism and energy production. The upregulation of the malic enzyme that supplies oxaloacetate was due to the decreased of TCA cycle. Aaldehyde dehydrogenase (ALDH) was upregulated by about 6.6-fold in GYAE medium compared with that in GYA medium, indicating enhanced ethanol oxidation pathway. However, the glucose metabolism was decreased due to the presence of ethanol, as shown in Table 2. Thus, the ethanol oxidation became the main energy source for cell growth and metabolism in the GYAE medium.
The protein expressions under GYAE and GY media were compared. The proteins involved in PPP (6-phosphogluconate dehydrogenase, transaldolase, and ribose 5-phosphate isomerase) were downregulated. However, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, phosphoglyceromutase, and phosphopyruvate hydratase involved in EMP were upregulated. Glucose-6-phosphate isomerase and pyruvate kinase were downregulated. Meanwhile, fructose 1, 6-bisphosphatase, and pyruvate phosphate dikinase involved in gluconeogenesis pathway were upregulated due to the presence of initial acetic acid and ethanol. The results were compared with those in GY medium. In addition, the enzymes involved in the TCA cycle, including pyruvate dehydrogenase, citrate synthase, isocitrate dehydrogenase, fumarate hydratase, and malate:quinone oxidoreductase, were downregulated. The downregulation of PPP enzymes indicated the weakening of glucose metabolism. The upregulation of enzymes involving EMP and gluconeogenesis implied that the pyruvate was anaplerosis for EMP by gluconeogenesis (Sakurai et al. 2011).
Therefore, the proteomics showed that the initial acetic acid can promote energy metabolism such as TCA cycle. Additionally, the presence of ethanol enhanced the ethanol oxidation pathway. Pyruvate metabolism could supply the EMP and TCA cycle when ethanol became the main energy source.
Assay of the gene transcription in acetic acid fermentation
Changes in gene transcriptions due to presence of initial acetic acid were analyzed. Genes were selected based on the results of proteomic analysis, including PPP, pyruvate metabolism, TCA cycle, ethanol oxidation, and acetic acid tolerance. As shown in Fig. 3, the improved acetic acid fermentation with initial acetic acid could be explained by the enhancement of ADH expression (about 2-fold). The transcription of gnd that encodes 6-phosphogluconate dehydrogenase as the key protein of PPP was downregulated due to the presence of initial acetic acid. This finding suggests that the glucose consumption decreased due to the addition of initial acetic acid as shown in Fig. 1b. The gene transcriptions, such as cs, aarC, and mqo involved in the TCA cycle, were upregulated. This result indicates that the TCA cycle was enhanced by initial acetic acid. Pyruvate kinase encoded by pyk supplies pyruvate from phosphoenolpyruvate and produces ATP. Pyruvate phosphate dikinase encoded by gene ppdK and involved in gluconeogenesis can convert pyruvate to phosphoenolpyruvate. The upregulation of pyk transcription and downregulation of ppdk suggested that more pyruvate was produced in acetic acid fermentation with initial acetic acid than that of without initial acetic acid. Pyruvate can be converted to malate by malate dehydrogenase, an enzyme encoded by gene mae, to enter the TCA cycle. Downregulation of mae implied that the TCA cycle was decreased. The protein DnaK was also downregulated by initial acetic acid, which was a response protein to protect other proteins from denaturation and aggregation caused by environmental stress (Ishikawa et al. 2010; Okamoto-Kainuma et al. 2004). The results of qRT-PCR indicated that initial acetic acid induced the metabolic pathway change by decreasing the PPP and enhancing the energy metabolism.
Discussion
PPP is the main glucose metabolic pathway of A. pasteurianus due to the absence of the phosphofructokinase (Illeghems et al. 2013; Azuma et al. 2009). Upregulation of PPP enzymes increased the glucose consumption and metabolism due to the presence of initial acetic acid in GYA compared with that of GY medium. In addition, the EMP could supply pyruvate for TCA cycle that was also upregulation with TCA cycle. The enzymes involved in the TCA cycle were observed to be upregulated due to the production of acetic acid in AAB (A. pasteurianus or Komagataeibacter spp.) (Andres-Barrao et al. 2016; Andres-Barrao et al. 2012; Sakurai et al. 2012). The enzymes included citrate synthase, aconitate hydrolase and succinate dehydrogenase, isocitrate dehydrogenase, 2-oxoglutatare dehydrogenase, and fumarate hydratase. Moreover, acetyl-CoA hydrolase and phosphotransacetylase can feed a new molecule of acetyl-CoA into the TCA cycle by assimilating a molecule of acetic acid that diffused into the cell (Lasko et al. 1997; Fukaya et al. 1993; Fukaya et al. 1990; Mullins et al. 2008). The TCA cycle was enhanced to ensure the efficient supply of oxaloacetate and other metabolite and maintain the normal metabolism for AAB growth. The results of the present study indicate that TCA cycle is important in fermentation process and tolerance of AAB to live in a naturally aggressive environment. In the absence of ethanol, AAB enhanced the PPP, EMP, and TCA cycle by utilizing glucose as the main carbon and energy source to increase energy metabolism and improve cell tolerance against acetic acid.
As one of the most important enzymes in ethanol oxidation, membrane-bound ADH contributes greatly to acetic acid tolerance in AAB (Steiner and Sauer 2001; Wang et al. 2015c). Loss of membrane-bound ADH, A. aceti, and A. pasteurianus displayed a loss of acetic acid tolerance and exhibited a growth reduction in the presence of acetic acid (Chinnawirotpisan et al. 2003; Takemura et al. 1993). Some strains of K. europaeus with a higher ADH activity than A. pasteurianus exhibited higher acetic acid tolerance than A. pasteurianus (Trcek et al. 2006). Expression of ADH was upregulated in GYA medium compared with that in GY medium, implying that initial acetic acid can improve the ADH expression. Therefore, upregulation expression of ADH will lead to the increased tolerance of A. pasteurianus against acetic acid that was according to the previous report (Xia et al. 2015). The expression of ALDH that is the other enzyme for ethanol oxidation was upregulated in GYAE medium compared with that in GYA medium (as listed in Table S1) because ALDH is more sensitive to ethanol than ADH (Mamlouk and Gullo 2013). Ethanol oxidation is the main energy producing pathway in acetic acid fermentation (GYAE medium) since ethanol is the preferred substrate over glucose for AAB (Sakurai et al. 2011). To perform the oxidation of ethanol to acetic acid ADH and ALDH transfers electron via ubiquinone (UQ). UQ gains one electron from the oxidation of ethanol and acetaldehyde and then transmits electrons to O2 when it is oxidized by ubiquinone oxidase in the respiratory chain. During the transmittance of electrons, 4 H+ in the cell react with O2 that gained 4 e− from the UQ to produce 2 H2O. While, 4 H+ from the oxidized UQ are released into the periplasm to result in a higher concentration of H+ out of the cells than that inside of the cells. Thus, a proton motive force necessary for energy production is obtained (Mamlouk and Gullo 2013; Wang et al. 2015a). The proton motive force as the energy source is used for pumping out acetic acid in a proton uncoupler-sensitive manner that accompanied by H+ passively penetrate into the cell (Matsushita et al. 2005; Wang et al. 2015a). The enhancement of the ethanol oxidation pathway promotes the produce of energy from a proton motive force used for the acetic acid transfer to improve the tolerance of AAB. Thus, the metabolism change induced by initial acetic acid and ethanol improved the cell growth in GYAE media, which is brought about by increase in viable cells, as shown in Fig. 2.
As shown in Fig. 4, the assay of gene transcription in acetic acid fermentation demonstrated that the TCA and ethanol oxidation as the important energy metabolism pathway were enhanced, whereas the PPP as main pathway of glucose metabolism was decreased. The acetic acid tolerance was improved as energy production was important for AAB to pump out acetic acid and maintain the suitable intracellular environment (Matsushita et al. 2005; Nakano et al. 2006). Also, TCA cycle can assimilate intracellular acetic acid to supply the cycle and produce more ATP to adapt to the acetic acid fermentation conditions. Thus, more viable count was obtained and acetic acid fermentation was processed as shown in Fig. 1.
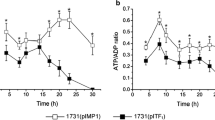
To further study the effects of initial acetic acid on energy metabolism, the DO and the intracellular ATP concentration were compared. As shown in Fig. 5, DO was decreased due to the cell growth and the ethanol oxidation. However, the DO without initial acetic acid was higher than that of the 1% initial acetic acid. Specially, after 15 h of fermentation, the DO increased accompanied with the viable cell decrease. Thus, the acetification process was stopped (Fig. 1). For acetic acid fermentation, oxygen is the final electron acceptor to form H2O and the proton motive force that is necessary for energy production. In addition, the lower DO indicated that initial acetic acid can promote cell metabolism resulting in more oxygen consumption and energy production necessary for cell growth and tolerance against sharp environment. The ethanol oxidation and the TCA cycle were linked to the respiratory chain and produce energy that contains a part of ATP (Adler et al. 2014). In acetic acid fermentation, ATP is necessary for cell growth by improving the acetic acid tolerance. Some studies also discovered that intracellular ATP is related to the cell’s response to acid shocks in Bacillus cereus and Salmonella enterica (Mols et al. 2010; Tan et al. 2015). As shown in Fig. 5, intracellular ATP was lower before 21 h due to the presence of initial acetic acid compared with that without initial acetic acid. In AAB, acetic acid is pumped out by the ATP-dependent ABC transporters when acetic acid diffused into the cell (Nakano et al. 2006). Similar to GrpE and DnaJ, DnaK as a molecular chaperone consumes ATP to ensure proper folding of the protein under acid pressure (Harrison et al. 1997; Ishikawa et al. 2010; Okamoto-Kainuma et al. 2004). The lower ATP was brought about by the increased consumption of ATP to resist the acid.
In summary, decreased intracellular ATP as a result of initial acetic acid inhibition can change the energy metabolism (TCA cycle and ethanol oxidation). This situation induces the AAB to produce more energy to resist acetic acid and promote the cell growth in initial period of acetic acid fermentation. Additionally, enhanced ADH expression promotes the ethanol oxidation and strengthens the acetification rate, meanwhile producing a strong proton motive force that is necessary for energy production and acetic acid transfer. Also, expression of some tolerance protein including Dnak is induced by initial acetic acid to improving the acetic acid tolerance.
This study provided a global metabolic change of AAB induced by initial acetic acid according to proteome assay, which highlighted insight carbon metabolism and energy regulation to adapt acetic acid fermentation conditions. However, future study will focus on the metabolic control to improve the acetic acid fermentation considering the energy production.
References
Adler P, Frey LJ, Berger A, Bolten CJ, Hansen CE, Wittmann C (2014) The key to acetate: metabolic fluxes of acetic acid bacteria under cocoa pulp fermentation-simulating conditions. Appl Environ Microbiol l80(15):4702–4716. doi:10.1128/AEM.01048-14
Andres-Barrao C, Saad MM, Chappuis ML, Boffa M, Perret X, Ortega Perez R, Barja F (2012) Proteome analysis of Acetobacter pasteurianus during acetic acid fermentation. J Proteome 75:1701–1717. doi:10.1016/j.jprot.2011.11.027
Andres-Barrao C, Saad MM, Ferrete EC, Bravo D, Chappuis ML, Perez RO, Junier P, Perret X, Barja F (2016) Metaproteomics and ultrastructure characterization of Komagataeibacter spp. involved in high-acid spirit vinegar production. Food Microbiol 55:112–122. doi:10.1016/j.fm.2015.10.012
Azuma Y, Hosoyama A, Matsutani M, Furuya N, Horikawa H, Harada T, Shirai M (2009) Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus. Nucleic Acids Res 37:5768–5783. doi:10.1093/nar/gkp612
Chinnawirotpisan P, Theeragool G, Limtong S, Toyama H, Adachi OO, Matsushita K (2003) Quinoprotein alcohol dehydrogenase is involved in catabolic acetate production, while NAD-dependent alcohol dehydrogenase in ethanol assimilation in Acetobacter pasteurianus SKU1108. J Biosci Bioeng 96:564–571. doi:10.1016/s1389-1723(04)70150-4
Deppenmeier U, Ehrenreich A (2009) Physiology of acetic acid bacteria in light of the genome sequence of Gluconobacter oxydans. J Mol Microbiol Biotechnol 16(1–2):69–80. doi:10.1159/000142895
Fukaya M, Takemura H, Okumura H, Kawamura Y, Horinouchi S, Beppu T (1990) Cloning of genes responsible for acetic acid resistance in Acetobacter aceti. J Bacteriol 172(4):2096–2104. doi:10.1128/jb.172.4.2096-2104.1990
Fukaya M, Takemura H, Tayama K, Okumura H, Kawamura Y, Horinouchi S, Beppu T (1993) The aarC gene responsible for acetic acid assimilation confers acetic acid resistance on Acetobacter aceti. Ferment Bioeng 76(4):270–275. doi:10.1016/0922-338X(93)90192-B
Gengenbacher M, Rao SP, Pethe K, Dick T (2010) Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology 156:81–87. doi:10.1099/mic.0.033084-0
Gillis M, De Ley J (1980) Intra-and intergeneric similarities of the ribosomal ribonucleic acid cistrons of Acetobacter and Gluconobacter. Int J Syst Bacteriol 30(1):7–27. doi:10.1099/00207713-30-1-7
Giudici P, Vero L D, Gullo M, Solieri L, Lemmetti F (2016) Fermentation strategy to produce high gluconate vinegar. Acetic Acid Bact 5(1). doi:10.4081/aab.2016.6067
Gullo M, Giudici P (2008) Acetic acid bacteria in traditional balsamic vinegar: phenotypic traits relevant for starter cultures selection. Int J Food Microbiol 125(1):46–53. doi:10.1016/j.ijfoodmicro.2007.11.076
Gullo M, Verzelloni E, Canonico M (2014) Aerobic submerged fermentation by acetic acid bacteria for vinegar production: process and biotechnological aspects. Process Biochem 49:1571–1579. doi:10.1016/j.procbio.2014.07.003
Gullo M, Zanichelli G, Verzelloni E, Lemmetti F, Giudici P (2016) Feasible acetic acid fermentations of alcoholic and sugary substrates in combined operation mode. Process Biochem 51(9):1129–1139. doi:10.1016/j.procbio.2016.05.018
Harrison CJ, Hayer-Hartl M, Liberto MD, Hartl FU, Kuriyan J (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276(5311):431–435. doi:10.1126/science.276.5311.431
Hattori R, Yamada K, Kikuchi M, Hirano S, Yoshida N (2011) Intramolecular carbon isotope distribution of acetic acid in vinegar. J Agric Food Chem 59:9049–9053. doi:10.1021/jf200227e
Illeghems K, De Vuyst L, Weckx S (2013) Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genomics 14:526. doi:10.1186/1471-2164-14-526
Ishikawa M, Okamoto-Kainuma A, Jochi T, Suzuki I, Matsui K, Kaga T, Koizumi Y (2010) Cloning and characterization of grpE in Acetobacter pasteurianus NBRC 3283. J Biosci Bioeng 109:25–31. doi:10.1016/j.jbiosc.2009.07.008
Krusong W, Yaiyen S, Pornpukdeewatana S (2015) Impact of high initial concentrations of acetic acid and ethanol on acetification rate in an internal venturi injector bioreactor. J Appl Microbiol 118(3):629. doi:10.1111/jam.12715
Lasko DR, Schwerdel C, Bailey JE, Sauer U (1997) Acetate-specific stress response in acetate-resistant bacteria an analysis of protein patterns. Biotechnol Prog 13(5):519–523. doi:10.1021/bp970075f
Mamlouk D, Gullo M (2013) Acetic acid bacteria: physiology and carbon sources oxidation. Indian J Microbiol 53:377–384. doi:10.1007/s12088-013-0414-z
Matsushita K, Inoue T, Adachi O, Toyama H (2005) Acetobacter aceti possesses a proton motive force-dependent efflux system for acetic acid. J Bacteriol 187:4346–4352. doi:10.1128/JB.187.13.4346-4352.2005
Mols M, van Kranenburg R, Tempelaars MH, van Schaik W, Moezelaar R, Abee T (2010) Comparative analysis of transcriptional and physiological responses of Bacillus cereus to organic and inorganic acid shocks. Int J Food Microbiol 137:13–21. doi:10.1016/j.ijfoodmicro.2009.09.027
Mounir M, Shafiei R, Zarmehrkhorshid R, Hamouda A, Alaoui MI, Thonart P (2016) Simultaneous production of acetic and gluconic acids by a thermotolerant Acetobacter strain during acetous fermentation in a bioreactor. J Biosci Bioeng 121(2):166–171. doi:10.1016/j.jbiosc.2015.06.005
Mullins EA, Francois JA, Kappock TJ (2008) A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti. J Bacteriol 190:4933–4940. doi:10.1128/JB.00405-08
Nakano S, Fukaya M (2008) Analysis of proteins responsive to acetic acid in Acetobacter: molecular mechanisms conferring acetic acid resistance in acetic acid bacteria. Int J Food Microbiol 125:54–59. doi:10.1016/j.ijfoodmicro.2007.05.015
Nakano S, Fukaya M, Horinouchi S (2006) Putative ABC transporter responsible for acetic acid resistance in Acetobacter aceti. Appl Environ Microbiol 72:497–505. doi:10.1128/AEM.72.1.497-505.2006
Okamoto-Kainuma A, Yan W, Fukaya M, Tukamoto Y, Ishikawa M, Koizumi Y (2004) Cloning and characterization of the dnaKJ operon in Acetobacter aceti. J Biosci Bioeng 97:339–342. doi:10.1016/s1389-1723(04)70216-9
Okamoto-Kainuma A, Ishikawa M, Nakamura H, Fukazawa S, Tanaka N, Yamagami K, Koizumi Y (2011) Characterization of rpoH in Acetobacter pasteurianus NBRC3283. J Biosci Bioeng 111:429–432. doi:10.1016/j.jbiosc.2010.12.016
Ory ID, Romero LE, Cantero D (2002) Optimum starting-up protocol of a pilot plant scale acetifier for vinegar production. J Food Eng 52(1):31–37. doi:10.1016/S0260-8774(01)00082-6
Qi Z, Yang H, Xia X, Quan W, Wang W, Yu X (2014) Achieving high strength vinegar fermentation via regulating cellular growth status and aeration strategy. Process Biochem 49:1063–1070. doi:10.1016/j.procbio.2014.03.018
Saichana N, Matsushita K, Adachi O, Frebort I, Frebortova J (2015) Acetic acid bacteria: a group of bacteria with versatile biotechnological applications. Biotechnol Adv 33:1260–1271. doi:10.1016/j.biotechadv.2014.12.001
Sakurai K, Arai H, Ishii M, Igarashi Y (2011) Transcriptome response to different carbon sources in Acetobacter aceti. Microbiology 157(3):899–910. doi:10.1099/mic.0.045906-0
Sakurai K, Arai H, Ishii M, Igarashi Y (2012) Changes in the gene expression profile of Acetobacter aceti during growth on ethanol. J Biosci Bioeng 113:343–348. doi:10.1016/j.jbiosc.2011.11.005
Sakurai K, Yamazaki S, Ishii M, Igarashi Y, Arai H (2013) Role of the glyoxylate pathway in acetic acid production by Acetobacter aceti. J Biosci Bioeng 115:32–36. doi:10.1016/j.jbiosc.2012.07.017
Shafiei R, Zarmehrkhorshid R, Mounir M, Thonart P, Delvigne F (2017) Influence of carbon sources on the viability and resuscitation of Acetobacter senegalensis during high-temperature gluconic acid fermentation. Bioprocess Biosyst Eng 40(5):769–780. doi:10.1007/s00449-017-1742-x
Shen Y, Liang J, Li H, Wang M (2015) Hydroxypropyl-β-cyclodextrin-mediated alterations in cell permeability, lipid and protein profiles of steroid-transforming Arthrobacter simplex. Appl Microbiol Biotechnol 99(1):387–397. doi:10.1007/s00253-014-6089-5
Steiner P, Sauer U (2001) Proteins induced during adaptation of Acetobacter aceti to high acetate concentrations. Appl Environ Microbiol 67:5474–5481. doi:10.1128/AEM.67.12.5474-5481.2001
Takemura H, Kondo K, Horinouchi S, Beppu T (1993) Induction by ethanol of alcohol dehydrogenase activity in Acetobacter pasteurianus. J Bacteriol 175(21):6857–6866. doi:10.1128/jb.175.21.6857-6866.1993
Tan SM, Lee SM, Dykes GA (2015) Acetic acid induces pH-independent cellular energy depletion in Salmonella enterica. Foodborne Pathog Dis 12:183–189. doi:10.1089/fpd.2014.1853
Teitzel GM, Parsek MR (2003) Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl Environ Microbiol 69:2313–2320. doi:10.1128/aem.69.4.2313-2320.2003
Trcek J, Toyama H, Czuba J, Misiewicz A, Matsushita K (2006) Correlation between acetic acid resistance and characteristics of PQQ-dependent ADH in acetic acid bacteria. Appl Microbiol Biotechnol 70:366–373. doi:10.1007/s00253-005-0073-z
Trcek J, Jernejc K, Matsushita K (2007) The highly tolerant acetic acid bacterium Gluconacetobacter europaeus adapts to the presence of acetic acid by changes in lipid composition, morphological properties and PQQ-dependent ADH expression. Extremophiles 11:627–635. doi:10.1007/s00792-007-0077-y
Wang B, Shao Y, Chen F (2015a) Overview on mechanisms of acetic acid resistance in acetic acid bacteria. World J Microbiol Biotechnol 31(2):255–263. doi:10.1007/s11274-015-1799-0
Wang B, Shao Y, Chen T, Chen W, Chen F (2015b) Global insights into acetic acid resistance mechanisms and genetic stability of Acetobacter pasteurianus strains by comparative genomics. Sci Rep 5:18330. doi:10.1038/srep18330
Wang Z, Zang N, Shi J, Feng W, Liu Y, Liang X (2015c) Comparative proteome of Acetobacter pasteurianus Ab3 during the high acidity rice vinegar fermentation. Appl Biochem Biotechnol 177:1573–1588. doi:10.1007/s12010-015-1838-1
Xia K, Zang N, Zhang J, Zhang H, Li Y, Liu Y, Liang X (2016) New insights into the mechanisms of acetic acid resistance in Acetobacter pasteurianus using iTRAQ-dependent quantitative proteomic analysis. Int J Food Microbiol 238:241–251. doi:10.1016/j.ijfoodmicro.2016.09.016
Xu Z, Bo F, Xia J, Sun Z, Li S, Feng X, Xu H (2015) Effects of oxygen-vectors on the synthesis of epsilon-poly-lysine and the metabolic characterization of Streptomyces albulus PD-1. Biochem Eng J 94:58–64. doi:10.1016/j.bej.2014.11.009
Acknowledgments
This research was supported by the National Natural Science Foundation of China (31201406), National High Technology Research and Development Program of China (2013AA102106), National Key R&D Program of China (2016YFD0400505), Tianjin Municipal Science and Technology Commission (16YFZCNC00650, 13JCQNJC10000), and the Program for Changjiang Scholars and the Innovative University Research Team (IRT15R49).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary materials
ESM 1
(PDF 401 kb).
Rights and permissions
About this article
Cite this article
Zheng, Y., Zhang, R., Yin, H. et al. Acetobacter pasteurianus metabolic change induced by initial acetic acid to adapt to acetic acid fermentation conditions. Appl Microbiol Biotechnol 101, 7007–7016 (2017). https://doi.org/10.1007/s00253-017-8453-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8453-8