Abstract
Affinity adsorption purification of hexahistidine-tagged (His-tagged) proteins using EDTA–chitosan-based adsorption was designed and carried out. Chitosan was elaborated with ethylenediaminetetraacetic acid (EDTA), and the resulting polymer was characterized by FTIR, TGA, and TEM. Different metals including Ni2+, Cu2+, and Zn2+ were immobilized with EDTA–chitosan, and their capability to the specific adsorption of His-tagged proteins were then investigated. The results showed that Ni2+–EDTA–chitosan and Zn2+–EDTA–chitosan had high affinity toward the His-tagged proteins, thus isolating them from protein mixture. The target fluorescent-labeled hexahistidine protein remained its fluorescent characteristic throughout the purification procedure when Zn2+–EDTA–chitosan was used as a sorbent, wherein the real-time monitor was performed to examine the immigration of fluorescent-labeled His-tagged protein. Comparatively, Zn2+–EDTA–chitosan showed more specific binding ability for the target protein, but with less binding capacity. It was further proved that this purification system could be recovered and reused at least for 5 times and could run on large scales. The presented M2+–EDTA–chitosan system, with the capability to specifically bind His-tagged proteins, make the purification of His-tagged proteins easy to handle, leaving out fussy preliminary treatment, and with the possibility of continuous processing and a reduction in operational cost in relation to the costs of conventional processes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Easily handling and effective purification of recombinant proteins have attracted much attention because of the increased requirement of these biomolecules in research and biopharmaceutical fields (Jain et al. 2010). Immobilized metal (ion) affinity chromatography (IMAC) established by Porath et al. (1975) is the foundation of most protein isolation procedures due to its high selectivity (Arnau et al. 2006). However, fussy pre-treatment such as the ultracentrifugation, collection, and resuspension of cell lysis, long separation times, difficulties in packing large columns, relatively high-pressure drops, as well as slow intra-bead diffusion of solutes substantially, make this traditional affinity-based purification method with packed-bed single column chromatography limiting in life sciences (Zou et al. 2001; Ghosh 2002). Thus, this bio-affinity feature continuously causes attention of biochemists and biologists to employ IMAC and enhance it to suit their particular applications (Ueda et al. 2003).
However, most of the developed IMAC systems have some restrictions in terms of complexity and time-consuming processes. As an ideal His-tagged protein purification system, the materials used should be biocompatible, biodegradable, eco-friendly, economic, while the purification procedure should be easy handle and high-throughput. Natural polymers, such as cellulose, agarose, dextran, and chitosan, have been frequently used as adsorption materials for protein separation (Zhang et al. 2011). However, the modification of natural polymers to be an affinity adsorbent, to our best knowledge, has not been studied and used in His-tagged protein purification.
Chitosan, an aminopolysaccharide comprising (1→4)-linked 2-amino-2-deoxy-β-d-glucose (GlcN) and 2-acetamido-2-deoxy-β-d-glucose (GlcNAc) is economically attractive because it is the second most abundant natural polymer, after cellulose, that can be considered appropriate material for the development of new medium for protein purification due to many characteristics, including high affinity to proteins, good biocompatibility, reduced nonspecific adsorption, improved resistance to chemical degradation, antibacterial properties, and the aptness in preparation of a variety of physical forms (Adriano et al. 2008; Bailey et al. 1999; Qiu et al. 2014). Chitosan has been used as a new support for enzyme (Klein et al. 2012), antibody (Yuan et al. 2007), and protein immobilization (Demolliens et al. 2008). It has also been used as an adsorbent, exhibiting relatively high sorption capacities and fast removal kinetics for heavy metals because of the reactivity of amine groups and stable chelation (Ali 2012; Reddy and Lee 2013). These intrinsic properties have been further enhanced by the elaboration of chelating agents such as ethylenediaminetetraacetic acid (EDTA) onto the chitosan scaffolds (Repo et al. 2010; Zhao et al. 2015; Shi et al. 2008). EDTA–chitosan fibers coordinated with nickel metal can mediate the assembly of histidine-tagged proteins (Shi et al. 2008). Inspired by these examples, we assumed that EDTA-attached chitosan would be more effective for His-tagged protein purification than chitosan used alone. Herein, EDTA–chitosan immobilized with different metals, including Ni2+, Cu2+, and Zn2+, were synthesized and were further investigated as adsorbents for His-tagged protein purification.
Materials and methods
Materials and instruments
Chitosan with 90 % degree of deacetylation and a centipoise viscosity of 150 mPa s (1 %, 20 °C) was purchased from Qingdao Honghai Biotechnology Company (Shandong, China). The N-terminal fused with enhanced green fluorescent protein (EGFP)–(His)6–NH2 (purity >95 %) and thioredoxin (Trx)–EGFP–(His)6–NH2 with a molecular weight of 39.5 kDa expressed in Escherichia coli (E. coli) cytoplasm were provided by Shanghai Sangon Biotech Co., Ltd, while Kana–(His)6–NH2 protein (43 kDa) expressed in E. coli cytoplasm was provided by Nanjing Detai Bio–technology Co., Ltd. BCA Protein Quantitation Kit was purchased from Shanghai Labaide Bio-Technology Co., Ltd. Chemicals were commercial reagent grade and used as received without further purification.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) analyses were carried out using 12 % gels with protein zones stained by standard Coomassie brilliant blue staining methods. The gel image was taken by Gel Doc XR system. Protein concentrations were determined with ELISA Infinite M200 at a wavelength of 562 nm based on bicinchoninic acid (BCA) kits, and the working range of the BCA method was 0.05–2 mg/mL. Fluorescence spectra were recorded on a Shimadzu RF–5301PC fluorescence spectrometer equipped with a 480-nm emission filter, while the images of fluorescent change during the purification procedures were taken under UV (254 nm) excitation. Ultraviolet–visible (UV-Vis) adsorption spectra of peptides or proteins were conducted with a Shimadzu UV–2550 Series instrument. Fourier transform infrared spectroscopy (FTIR) spectra were recorded on a Nicolet Nexus 870 spectrometer with KBr pellets. The thermogravimetric analysis (TGA) was performed using a USA TA thermal analyzer Model Q50, at a heating rate of 10 °C·min−1, from room temperature up to 600 °C under an argon atmosphere. Transmission electron microscopy (TEM) was taken by Hitachi HT7700 high-resolution transmission electron microscopy (HRTEM), which was performed using a double aberration corrected JEOL 2200FS (Japan) microscope equipped with a field emission gun operated at 100 kV. Surface morphology of samples was viewed and photographed with a scanning electron microscopy (SEM) (TM-1000, Hitachi, Japan).
Synthesis of EDTA–chitosan
EDTA bisanhydride was prepared according to the previous literatures (Montembault et al. 1996; Roosen and Binnemans 2014) with 90 % yield.
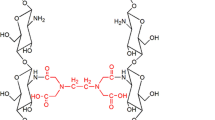
EDTA–chitosan was synthesized according to the previous report (Roosen and Binnemans 2014) with slight modifications (Scheme 1). Chitosan (5.0 g) was dissolved in a 10 % (v/v) aqueous acetic acid solution (100 mL), which was subsequently diluted in methanol (400 mL). EDTA bisanhydride (23.83 g, 93 mmol) suspended in methanol (100 mL) was then added dropwise to the above chitosan solution. After stirring at 40 °C for 24 h, the resulting mixture was filtered. After that, the filtrate was mixed with ethanol (200 mL), stirred for another 12 h, and the mixture was filtered again. The pH of filtrate was then adjusted to 11 with an aqueous sodium hydroxide (NaOH) solution (0.1 mol/L). After stirring for another 12 h, the resulting mixture was centrifuged for 10 min at 8000 rpm (3–30 K, Sigma). The precipitate was repeatedly washed with de-ionized water, centrifuged, until the pH value of supernatant was around 7. The resulting solid was washed with a 0.1 mol/L hydrochloric acid (HCl) solution and successively washed with de–ionized water, until the pH of the washing water was neutral. Finally, the precipitate was well dispersed in ethanol, filtered, dried in a vacuum oven at 40 °C overnight to give a white powder, EDTA–chitosan.
Adsorption of EDTA–chitosan with metal ions (Cu2+, Ni2+, and Zn2+)
The batch experiments of Cu(II), Zn(II), and Ni(II) sorption on adsorbents were carried out by mixing 5 mg of adsorbent with 1 mL of metal solution at concentrations of 0.4, 0.8, 1.2, 1.6, and 2.0 mg/mL respectively for 4 h to reach equilibrium. The mixture was then centrifuged at 6000 rpm for 3 min (3–30 K, Sigma). The resultant supernatant was discarded. The sorbent in the bottom layer was washed with 1 mL of NaOAc buffer (50 mmol/L, pH 4.0) for three times to remove nonspecifically and loosely bound metal ions. After that, the sorbent was digested with 2 mol/L HNO3 (2 mL) at 50 °C for 2 h to desorb the metal ion. After centrifuging, the supernatant was diluted with 2 % HNO3. The solution was then analyzed for the content of metal ion with inductive coupled plasma mass spectrometry (ICP–MS) (Agilent 7700X).
Metal immobilized with chitosan and EDTA–chitosan
After 5.0 mg EDTA–chitosan was incubated with 1 mL of 1.6 mg/mL aqueous copper chloride, nickel chloride, and zinc chloride solution respectively for 4 h at room temperature, the mixtures were then centrifuged at 6000 rpm for 3 min and discarded the supernatant. The residue was then washed with 1 mL acetic acid buffer (50 mmol/L, pH 4.0) for 3 times to completely wash away the non-binding metal ion. The M2+–chitosan (M═Cu, Ni, Zn) were similarly prepared as controls.
The affinity adsorption of purified EGFP–(His)6–NH2 with M2+–EDTA–chitosan and M2+–chitosan (M=Ni, Cu, Zn)
A mixture of M2+–EDTA–chitosan (5.0 mg) and 40 μL of EGFP–(His)6–NH2 (1.8 mg/mL in 20 mmol/L PBS buffer, pH 7.2, 500 mmol/L NaCl) were gently stirred in an ice bath for 40 min to reach adsorption equilibrium. After centrifugation at 6000 rpm for 3 min, the bottom layer residual was washed with 40 μL of PBS (20 mmol/L, pH 7.2, 500 mM NaCl) for three times to completely remove the unspecific adsorbed proteins. Subsequently, 40 μL of imidazole buffer (500 mmol/L, dissolved in 20 mmol/L PBS, pH 7.2, containing 500 mmol/L NaCl) was added to elute the specific adsorbed protein. This eluting process was repeated for twice. The concentration of His-tagged EGFP was determined by BCA method. M2+–chitosan (M═Ni, Cu, Zn) were used as control.
The binding behavior on purified EGFP–(His)6–NH2 with different quantity of M2+–EDTA–chitosan (M=Ni, Cu, Zn)
The solution batches of 40 μL of purified EGFP–(His)6–NH2 (1.8 mg/mL) were respectively incubated with different quantity of M2+–EDTA–chitosan (M═Ni, Cu, Zn) (1, 3, 5, 7, and 9 mg) with constant stirring for 40 min in an ice bath to reach adsorption equilibrium. After centrifuging at 6000 rpm for 3 min to remove the supernatant, the residue was washed with 40 μL of PBS (20 mmol/L, 1.0 mol/L NaCl, pH 7.2) for 3 times to remove the nonspecific proteins. The bound proteins were then eluted with 40 μL of imidazole buffer (500 mmol/L, dissolved in 20 mmol/L PBS, pH 7.2, containing 500 mmol/L NaCl) twice to desorb target protein completely. The elution was collected by centrifuging at 6000 rpm for 3 min, and the concentration of EGFP–(His)6–NH2 was determined by BCA method.
Adsorption behavior analyses of Trx–EGFP–(His)6–NH2 with M2+–EDTA–chitosan (M=Ni, Cu, Zn) under different pH value
A stock freeze-dried Trx–EGFP–(His)6–NH2 was dissolved in PBS (20 mmol/L, containing 500 mmol/L NaCl) with different pH value (5.2, 6.2, 7.2, 8.2, and 9.2) respectively to afford Trx-EGFP–(His)6–NH2 solutions with an ultimate concentration of 1.8 mg/mL. Then, 40 μL of those protein solutions were respectively incubated with 5 mg of M2+–EDTA–chitosan (M═Ni, Cu, Zn) adsorbent with constantly stirring for 40 min in an ice bath. The resultant mixtures were centrifuged at 6000 rpm for 3 min. After removal of the supernatant, the absorbent was washed with 40 μL of PBS (20 mmol/L, 500 mmol/L NaCl, with corresponding pH 5.2, 6.2, 7.2, 8.2, and 9.2). This process was repeated 3 times to wash out loosely bound proteins and unbound proteins. The target proteins were eluted with imidazole buffer solution (500 mmol/L dissolved in PBS, 500 mmol/L NaCl, with corresponding pH 5.2, 6.2, 7.2, 8.2, and 9.2). All of the portions including the upper layer in the affinity absorbent procedure were analyzed by SDS–PAGE, and the concentration of the specific binding Trx–EGFP–(His)6–NH2 eluted by imidazole buffer was determined with BCA method.
Adsorption behavior analyses of Trx–EGFP–(His)6–NH2 under different ionic strength
The freeze-dried cell lysate Trx–EGFP–(His)6–NH2 powder was dissolved in PBS (pH 7.2, 20 mmol/L) solution with different ion strength (containing 0, 500, and 1000 mmol/L NaCl, respectively) to give a final concentration of 1.8 mg/mL. Then, 40 μL of Trx–EGFP–(His)6–NH2 with different NaCl concentrations was separately incubating with 5 mg M2+–EDTA–chitosan (M═Ni, Cu, Zn) for 40 min and then centrifuged at 6000 rpm for 3 min, and 40 μL of PBS (20 mmol/L, pH 7.2, with corresponding 0, 500, 1000 mmol/L NaCl) was added to wash out loosely bound proteins and unbound proteins. After that, 40 μL of imidazole buffer (500 mmol/L, pH 7.2, with corresponding 0, 500, 1000 mmol/L NaCl) was used to desorb the targeted protein. All of the portions including the supernatant in the affinity adsorbent procedure were analyzed by SDS–PAGE, and the concentration of the specific binding Trx–EGFP–(His)6–NH2 eluted by imidazole buffer was determined with BCA method.
Purification of Trx–EGFP–(His)6–NH2 from crude E. coli cell lysate with M2+–EDTA–chitosan (M=Ni, Zn)
The solution batches of 40 μL of 1.8 mg/mL crude proteins containing Trx–EGFP–(His)6–NH2 in 20 mmol/L PBS (pH 7.2, 500 mmol/L NaCl) were mixed with 5 mg M2+–EDTA–chitosan (M═Ni, Zn). The mixtures were incubated in an ice bath for 40 min with gently stirring. After centrifugation at 6000 rpm for 3 min, the sorbent was washed with 40 μL PBS (pH 7.2, 500 mmol/L NaCl) for 3 times to remove the nonspecific proteins. After that, 40 μL of imidazole buffer (500 mmol/L, pH 7.2, 500 mmol/L NaCl) was used to desorb the targeted protein. After this process was repeated twice, all the portions including the supernatant in the affinity adsorbent procedure were analyzed by SDS–PAGE, and the concentration of the specific binding Trx–EGFP–(His)6–NH2 eluted by imidazole buffer was determined with BCA method.
Purification of Kana–(His)6–NH2 from crude E. coli cell lysate with M2+–EDTA–chitosan (M=Ni, Zn)
After 80 μL of crude protein solution containing Kana–(His)6–NH2 (6 mg/mL, dissolved in PBS, pH 6.2, 500 mmol/L NaCl) was mixed with 10 mg M2+–EDTA–chitosan, the mixture was gently stirred in an ice bath for 40 min. The mixture was then centrifuged at 6000 rpm for 3 min, and the sorbent was washed with 40 μL PBS (20 mmol/L, pH 6.2, 500 mmol/L NaCl) 3 times to remove the nonspecific proteins. After that, 40 μL of imidazole buffer (200 mmol/L, pH 6.2, 500 mmol/L NaCl) was used to desorb target proteins. All the portions including the supernatant of affinity adsorbent procedures were analyzed by SDS–PAGE, and the concentration of the specific binding Kana–(His)6–NH2 eluted by imidazole buffer was determined with BCA method.
Reuse of M2+–EDTA–chitosan (M=Ni, Cu, Zn)
A mixture of 5.0 mg M2+–EDTA–chitosan and 40–μL EGFP–(His)6–NH2 (1.8 mg/mL, dissolved in 20 mmol/L PBS, pH 7.2) containing 500 mmol/L NaCl was gently stirred in ice bath for 40 min to reach adsorption equilibrium. The mixture was then centrifuged at 6000 rpm for 3 min. After removing the supernatant, the medium (EDTA–chitosan) was successively mixed with PBS (20 mmol/L PBS, pH 7.2, 3 × 40 μL) to remove loosely bound proteins and unbound proteins. After that, imidazole buffer (500 mmol/L imidazole, pH 7.2, 500 mmol/L NaCl, 3 × 40 μL) was added to extract the specific binding protein. The sorbent was then washed with PBS (20 mmol/L, pH 7.2, 500 mmol/L NaCl) for 5 times to remove the residual imidazole. This batch adsorption purification was repeated 5 operation times. The concentration of adsorbed proteins was determined with BCA method.
Results
Characterization of EDTA–chitosan
FTIR analysis
As shown in the FTIR spectra of chitosan and EDTA–chitosan (Fig. 1), chitosan (a) has the characteristic C═O stretching (amide I) bands at 1654 cm−1, N–H angular deformation band of amino group at 1584 cm−1, –CH2 bending vibration at 1419 cm−1, C–H (in plane) bending at 1364 cm−1, and amide III band at 1314 cm−1, as well as the band at 1080 cm−1 corresponding to the C–O stretching (primary alcoholic groups). The C–N stretching band is observed at 1157 cm−1. The broad band at 3411 cm−1 corresponds to –OH and –NH stretching absorption, whereas the aliphatic C–H symmetric and asymmetric stretching band is observed at 2875 cm−1.
The FTIR spectrum of EDTA–chitosan shows new peaks at 1729 and 1637 cm−1 associated with the carbonyl groups vibration of −COOH, and −CONH−, respectively (Fig. 1b), indicating that EDTA was introduced to the backbone of chitosan. Furthermore, a broad band around 3410 cm−1 attributed to the vibration stretch of O–H and N–H. The peaks at 2955 and 2878 cm−1 correspond to the CH2 symmetric stretching, which is accordance with the previous literatures (Repo et al. 2010; Zhao et al. 2015; Shi et al. 2008). Comparing to the unmodified chitosan, the adsorption peak of C–N stretch (primary amine) of EDTA–chitosan is at 1067 cm−1 with a red shift and enhanced intensity.
TGA analysis
As shown in Fig. 2, two samples were thermally stable. All thermal decomposition profiles exhibited two main stages, with one starting at around 100 °C and another at 200 °C (EDTA–chitosan, dash line), and 280 °C (chitosan, solid line). At high temperature, the mass lose of EDTA–chitosan was higher than that of chitosan, indicating that the introduction EDTA group onto the chitosan backbone made it easy to degrade by comparison with that of chitosan. This result is in accordance with the literature that alien groups could influence the thermal stability of chitosan (Tirtom et al. 2012).
SEM
The scanning electron microscopy of EDTA–chitosan and Ni–EDTA–chitosan is shown in Fig. 3a, b. The surface changes in the SEM micrographs indicated the structural changes in the adsorbents before and after nickel treatment. The general morphology of EDTA–chitosan could be characterized as rough and folded, while the surfaces of the Ni loaded EDTA–chitosan were noted to have much asperity.
TEM
The morphologies of EDTA–chitosan and Ni–EDTA–chitosan were investigated by TEM (Fig. 3). Uniformed offwhite spots with sizes of 100 nm were observed in the TEM graph of EDTA–chitosan (Fig. 3c), while the TEM graph of Ni2+–EDTA–chitosan (Fig. 3d) exhibited gray to black spherical clusters with sizes of 30–50 nm except for the offwhite spots standing for the coexistence of pristine chitosan.
The binding behavior of EDTA–chitosan and chitosan with metal ions Cu2+, Ni2+, and Zn2+
Initially, the adsorption capacity of EDTA–chitosan (with fixed amount of 5 mg) with a chemical concentration gradient of the first row transition metal ions (Cu2+, Ni2+, and Zn2+) was investigated. As shown in Fig. 4, the adsorption capacity was dose-dependent before 1.6 mg/mL. Subsequently, it trended to be saturated. The maximum binding capacity of EDTA–chitosan on Cu2+, Ni2+, and Zn2+ reached 86.32 mg/per gram of EDTA–chitosan, 74.77 mg/per gram of EDTA–chitosan, and 85.01 mg/per gram of EDTA–chitosan, respectively (Fig. 4). While the maximum binding capacity of chitosan on Cu2+, Ni2+, and Zn2+ was 38.71 mg/per gram of chitosan, 32.81 mg/per gram of chitosan, and 35.25 mg/per gram of chitosan, respectively (Fig. S2).
The binding behavior of M2+–EDTA–chitosan and M2+–chitosan with EGFP–(His)6–NH2 (M=Ni, Cu, Zn)
From the data presented in Fig. 5, it was obviously resulted that the binding capacities of all of M2+–EDTA–chitosan (M═Ni, Cu, Zn) were much higher than those of the corresponding M2+–chitosan (M═Ni, Cu, Zn), albeit the binding capacities of M2+–EDTA–chitosan with different metals showed slight differences. Among three metals tested, Cu2+–EDTA–chitosan exhibited the highest binding capacity (12.48 mg/g, 86.7 %), followed by Zn2+–EDTA–chitosan (11.52 mg/g, 80.0 %), and Ni2+–EDTA–chitosan (10.48 mg/g, 72.8 %). Whereas the binding capacities of Cu2+–chitosan, Zn2+–chitosan, and Ni2+–chitosan were almost negligible (Cu2+–chitosan: 1.2 mg/g, 8.3 %; Zn2+–chitosan: 0.96 mg/g, 6.7 %; Ni2+–chitosan: 0.80 mg/g, 5.6 %).
SDS–PAGE analysis of binding behavior of purified EGFP–(His)6–NH2 with M2+–EDTA–chitosan and M2+–chitosan (M═Ni, Cu, Zn). Lane 1, Pure EGFP; lane 2, the supernatant after adsorption by Cu2+–EDTA–chitosan; lane 3, PBS elution after adsorption by Cu2+–EDTA–chitosan; lane 4, the elution of imidazole buffer solution; lane 5, the supernatant after adsorption by Cu2+–chitosan; lane 6, PBS elution after adsorption by Cu2+–EDTA–chitosan; lane 7, the elution of imidazole buffer solution; lane 8, the supernatant after adsorption by Ni2+–EDTA–chitosan; lane 9, PBS elution after adsorption by Ni2+–EDTA–chitosan; lane 10, the elution of imidazole buffer solution; lane 11, the supernatant after adsorption by Ni2+–chitosan; lane 12, PBS elution after adsorption by Ni2+–chitosan; lane 13, the elution of imidazole buffer solution; lane 14, the supernatant after adsorption by Zn2+–EDTA–chitosan; lane 15, PBS elution after adsorption by Zn2+–EDTA–chitosan; lane 16, the elution of imidazole buffer solution; lane 17, the supernatant after adsorption by Zn2+–chitosan; lane 18, PBS elution after adsorption by Zn2+–chitosan; lane 19, the elution of imidazole buffer solution; lane 20, protein molecular weight markers (unit, kDa)
As shown in Fig. 6, the distribution of EGFP–(His)6–NH2 between two layers was visually observed by the bright green fluorescence. The fluorescence diminished in the presence of Ni2+–EDTA–chitosan and Cu2+–EDTA–chitosan (Fig. 6b, c). The fluorescence was re-observed in the aqueous phase (upper layer) when imidazole buffer (500 mmol/L dissolved into 20 mmol/L PBS, pH 7.2, 500 mmol/L NaCl) was added to the mixture to elute the targeted protein. In the case of Zn2+–EDTA–chitosan, the green fluorescence remained throughout the purification procedure.
The binding behavior of M2+–EDTA–chitosan with EGFP–(His)6–NH2
To determine the binding stoichiometry of these adsorbents, a fixed concentration of EGFP–(His)6–NH2 was adsorbed by a gradient quantity of M2+–EDTA–chitosan. As shown in Fig. 7, the binding capacities were dose-dependent. As 1 mg of M2+–EDTA–chitosan was used, only a part of protein was bound to three adsorbents (Cu2+–EDTA–chitosan, 9.28 × 10−3 mg; 64.4 %; Zn2+–EDTA–chitosan, 7.12 × 10−3 mg, 49.4 %; Ni2+–EDTA–chitosan, 4.88 × 10−3 mg, 33.9 %). When the quantity of M2+–EDTA–chitosan was up to 5 mg, the binding capacities reached up to 62.4 × 10−3 mg, 86.7 % (Cu2+–EDTA–chitosan); 57.60 × 10−3 mg, 80.0 % (Zn2+–EDTA–chitosan); 52.40 × 10−3 mg, 72.8 % (Ni2+–EDTA–chitosan), respectively.
Adsorption behavior analyses of Trx–EGFP–(His)6–NH2 under different ionic strengths
It has been reported that ion strength was one of the important influence factors in recombinant proteins separation and purification, so adsorption isotherm analyses of crude Trx–EGFP–(His)6–NH2 which was expressed in E. coli was investigated under different ionic strengths. As shown in Fig. 8, it was obviously found that EDTA–chitosan immobilized with different metals showed different behavior. Cu2+–EDTA–chitosan exhibited a strong affinity on proteins with 75 % purity (Fig. 8c, lane 6) when the concentration of NaCl was 500 mmol/L. While Ni2+–EDTA–chitosan and Zn2+–EDTA–chitosan had lower absorption capacities with high purities (nearly 95 %) at 500 mmol/L of NaCl concentration (Fig. 8a, lane 6, Ni2+–EDTA–chitosan; and Fig. 8b, lane 15, Zn2+–EDTA–chitosan).
The influence of ion strength and SDS–PAGE analysis of the influence on ion strength (a Ni2+–EDTA–chitosan, b Zn2+–EDTA–chitosan, c Cu2+–EDTA–chitosan extraction respectively), a: Lane 1, the supernatant after adsorption by Ni2+–EDTA–chitosan; lane 2, PBS (0 M NaCl) elution after adsorption by Ni2+–EDTA–chitosan; lane 3, the elution of imidazole buffer (0 M NaCl) solution; lane 4, the supernatant after adsorption by Ni2+–EDTA–chitosan; lane 5, PBS (500 mM NaCl) elution after adsorption by Ni2+–EDTA–chitosan; lane 6, the elution of imidazole buffer (500 mM NaCl) solution; lane 7, the supernatant after adsorption by Ni2+–EDTA–chitosan; lane 8, PBS (1000 mM NaCl) elution after adsorption by Ni2+–EDTA–chitosan; lane 9, the elution of imidazole buffer (1000 mM NaCl) solution; b: lane 10, the supernatant after adsorption by Zn2+–EDTA–chitosan; lane 11, PBS (0 mM NaCl) elution after adsorption by Ni2+–EDTA–chitosan; lane 12, the elution of imidazole buffer (0 M NaCl) solution; lane 13, the supernatant after adsorption by Zn2+–EDTA–chitosan; lane 14, PBS (500 mM NaCl) elution after adsorption by Ni2+–EDTA–chitosan; lane 15, the elution of imidazole buffer (500 mM NaCl) solution; lane 16, the supernatant after adsorption by Zn2+–EDTA–chitosan; lane 17, PBS (1000 mM NaCl) elution after adsorption by Ni2+–EDTA–chitosan; lane 18, the elution of imidazole buffer (1000 mM NaCl) solution; lane 19, cell lysate feedstock; c: lane 1, the supernatant after adsorption by Cu2+–EDTA–chitosan; lane 2, PBS 0 mM NaCl) elution after adsorption by Cu2+–EDTA–chitosan; lane 3, the elution of imidazole buffer (0 M NaCl) solution; lane 4, the supernatant after adsorption by Cu2+–EDTA–chitosan; lane 5, PBS (500 mM NaCl) elution after adsorption by Cu2+–EDTA–chitosan; lane 6, the elution of imidazole buffer (500 mM NaCl) solution; lane 7, the supernatant after Cu2+–EDTA–chitosan adsorbed; lane 8, PBS (1000 mM NaCl) elution after adsorption by Cu2+–EDTA–chitosan; lane 9, the elution of imidazole buffer (1000 mM NaCl) solution; lane 10, cell lysate feedstock
The reuse of M2+–EDTA–chitosan (M=Ni, Cu, Zn)
With the ever increasing requirements to explore eco-friendly and cost-effective protocols for the purification of His-tagged proteins, the feasible reuse of EDTA–chitosan-based adsorbents was investigated using pure EGFP–(His)6–NH2 as model protein. From the results presented in Table 1, the absorption capacities of the fresh Ni2+–EDTA–chitosan, Cu2+–EDTA–chitosan, and Zn2+–EDTA–chitosan were found to be 10.48, 12.40, and 11.44 mg/g, respectively. At the second recycle, the absorption capacities and recycle rates (based on the absorption capacities in the first time) were 10.32 mg/g (98.5 %), 12.08 mg/g (97.4 %), and 11.20 mg/g (97.9 %), respectively. The recycle rates (based on the absorption capacities in the first time) still reached 84.7, 78.1, and 79.0 %, respectively, corresponding to Ni2+–EDTA–chitosan, Cu2+–EDTA–chitosan, Zn2+–EDTA–chitosan at the fifth recycle.
Purification of Trx–EGFP–(His)6–NH2 from crude E. coli cell lysate
With the optimized extraction conditions at hand, a further extension of the use of this one-pot adsorption method to the purification of crude lysate containing Trx–EGFP–(His)6–NH2 expressed in E. coli was investigated. According to the data assessed from SDS–PAGE, efficient adsorption purification of Trx–EGFP–(His)6–NH2 was achieved at around 0.43 mg/mL (Fig. 9, lane 4, Ni2+–EDTA–chitosan) and 0.46 mg/mL (Fig. 9, lane 7, Zn2+–EDTA–chitosan) from 1.8 mg/mL E. coli cell lysate.
SDS–PAGE analysis of adsorption purification of Trx–EGFP–(His)6–NH2 in E. coli cell lysate with M2+–EDTA–chitosan (M═Ni2+, Zn2+). Lane 1, cell lysate feedstock; lane 2, the supernatant after adsorption by Ni2+–EDTA–chitosan; lane 3, PBS elution after adsorption by Ni2+–EDTA–chitosan; lane 4, the elution of imidazole buffer solution; lane 5, the supernatant after adsorption by Zn2+–EDTA–chitosan; lane 6, PBS elution after adsorption by Zn2+–EDTA–chitosan; lane 7, the elution of imidazole buffer solution; lane 8, protein molecular weight markers (unit, kDa). The band at 39.5 kDa was assigned to Trx–EGFP–(His)6–NH2. Electrophoresis was performed under reduced conditions on 12 % polyacrylamide gel
Purification of Kana–(His)6–NH2 from crude E. coli lysate
To further evaluate the generality of the present adsorption system, another His-tagged protein, Kana–(His)6–NH2 expressed in E. coli lysate was purified using this adsorption method. As shown in Fig. 10, around 90 % purity of the target protein was obtained from the crude E. coli lysate by using the absorption Ni2+–EDTA–chitosan (lane 3) or Zn2+–EDTA–chitosan (lane 6). The concentration of extracted protein was 0.70 mg/mL with Ni2+–EDTA–chitosan and 0.89 mg/mL with Zn2+–EDTA–chitosan, respectively.
SDS–PAGE analysis of adsorption purification of Kana–(His)6–NH2 in E. coli cell lysate, lane 1, the supernatant after adsorption by Ni2+–EDTA–chitosan; lane 2, PBS elution after adsorption by Ni2+–EDTA–chitosan; lane 3, the elution of imidazole buffer solution; lane 4, the supernatant after adsorption by Zn2+–EDTA–chitosan; lane 5, PBS elution after adsorption by Zn2+–EDTA–chitosan; lane 6, the elution of imidazole buffer solution; lane 7, cell lysate feedstock; lane 8, protein molecular weight markers (unit, kDa). The band at 43 kDa was assigned to Kana–EGFP–(His)6–NH2. Electrophoresis was performed under reduced conditions on 12 % polyacrylamide gel
Discussion
Polyhistidine is the most common affinity tag for protein purification (Groll et al. 2005). Inspired of that the natural polymers, such as cellulose, agarose, and dextran, have been frequently used as adsorbent materials for protein separation, the inherent nature of chitosan was explored to be used as affinity adsorbents for the purification of His-tagged proteins. One of the most important goals of this work is the application of chitosan derivatives as a class of affinity adsorbents for selective binding of His-tagged proteins with a high capacity. Chitosan exists widely in nature as a kind of biocompatible polysaccharide. The well-known beneficial properties of chitosan, such as biocompatibility, degradability, bioadhesion, and antimicrobial activity, can thus be complemented with a procedure allowing flexible and oriented protein conjugation to chitosan via coiled–coil interactions, providing multiple additional functionalities in applications such as gene delivery and regenerative medicine (Demolliens et al. 2008). In the present studies, EDTA–chitosan was easily obtained via acylation with amino functional groups of chitosan and EDTA bis-anhydride, forming very strong chelates with metals. However, the resulting polymer is not water-soluble and fat-soluble because of the crosslinking of chitosan through the interaction of its amino groups with EDTA. The structure of resulting polymer was characterized by FTIR, and the main characteristic peaks of FTIR were in agreement with the previous literature (Zhao et al. 2015).
As shown in Fig. 2, a slight difference in the TGA curve slope also illustrated slight change of the prepared EDTA–chitosan compared with the pristine chitosan. According to the previous literature, for all samples, the weight loss at temperature below 200 °C could be attributed to the release of moisture on the surface of samples (Wang et al. 2010). From the data presented in Fig. 2, EDTA–chitosan adsorbed physically a little bit more water than chitosan, and then it rapidly lost part of its masses when the temperature is higher than 200 °C due to the decomposition of the organic polymers or nitrogen-containing functional groups. This is similar to the polymers decomposition period of magnetic EDTA–and/or DTPA–cross-linked chitosan adsorbents reported by Zhao et al. (2015). Upon the different microsphere surfaces and morphologies of the EDTA–chitosan and Ni2+–EDTA–chitosan samples observed by SEM (Fig. 3a, b) and TEM (Fig. 3c, d) exhibited that nickel ions were loaded into EDTA–chitosan.
The efficiency of IMAC-based affinity adsorption of His-tagged proteins is correlated with the binding capacity and specific binding ability of the adsorbents. The improved metal ion-binding capacity of EDTA–chitosan because of the existence of carboxyl groups of alien EDTA has been demonstrated (Repo et al. 2010; Zhao et al. 2015). Herein, the first row transition metal ions (Cu2+, Ni2+, and Zn2+) were chosen to coordinate with chitosan and EDTA–chitosan because they were predominantly involved in IMAC procedures due to the easily polarized nature of their d–electron shells. From the data presented in Fig. 4, Cu2+ exhibited higher binding capacity loaded with EDTA–chitosan than that of Ni2+ and Zn2+ that was accordance with our previous report that the binding capacity of copper ion was higher than nickel and zinc ions (Xu et al. 2014; Ren et al. 2015).
Therefore, the synthesized EDTA–chitosan with excellent metal-binding properties was subsequently immobilized with different metals to be used as affinity adsorbent for His-tagged protein purification. The adsorption experiments indicated that the protein-binding capabilities of M2+–EDTA–chitosan were higher than those of M2+–chitosan (Fig. 5). Among these adsorbents, Cu2+–EDTA–chitosan exhibited the best protein-binding capability, which was correlated with the metal-binding behavior of EDTA–chitosan (Fig. 4). This result is also in agreement with the previous reports that metals played an important role in protein absorption because histidine residues in the proteins could coordinate with metal ions, serving as specific affinity handles in the purification processes (Porath et al. 1975; Gutierrez et al. 2007; Chaga 2001).
From the data obtained from the binding behavior of the M2+–EDTA–chitosan with EGFP–(His)6–NH2, it was obviously demonstrated that the protein-binding capacities of different metals were dose-dependent, and then achieved maximal capacity when 5 mg of the adsorbent was used (Fig. 7). The sequence of adsorption capacity was Cu2+–EDTA–chitosan > Zn2+–EDTA–chitosan > Ni2+–EDTA–chitosan. The amino group of chitosan is the principal group for binding metal ions. It is widely accepted that a copper ion coordinates with four amino groups in a square–planar geometry, by which the copper ion is immobilized on chitosan (Kofuji et al. 2005; Li et al. 2007), and the adsorption capacity of the bound metal ion depends on the degree of deacetylation (Tikhonov et al. 1996). The carbonyl groups that were introduced onto the backbone of chitosan caused good adsorption capacities of M2+–EDTA–chitosan, especially Cu2+–EDTA–chitosan.
Ionic strength is one of the most important factors that affect the equilibrium characteristics of the adsorption of proteins to affinity adsorbents, and the effect of ionic strength on the protein adsorption onto affinity adsorbents is a very complex phenomenon. It is supposed that (1) the salt counter ions compete against the protein ions for binding sites; (2) the salt co-ions shield the protein ions and the charged binding sites from each other; (3) the change of ionic strength changes the folding and configuration of protein molecules, resulting in variation of the hydrophobic interaction between the protein and the adsorbents; and (4) the increase of ionic strength may cause the network of the adsorbents to shrink, reducing the porosity of the adsorbents and hence the availability of binding sites (Stigter and Dill 1990; Khare and Peppas 1995; Lan et al. 2001; Xu et al. 2014).
Hearn and co-workers had also reported that metal ion-chelating system exhibited selectivity characteristic of coordination interactions upon an increase of ionic strength (Zachariou and Hearn 1996), while other literatures demonstrated that the increase of ionic strength could reduce the binding strength and lower the binding capacity upon the study of adsorption of various proteins/adsorbents (Janzen et al. 1990; Lan et al. 2001).
In our study, the effect of different ionic strength of the equilibration buffer (20 mM sodium phosphate, pH 7.2) was investigated in crude Trx–EGFP–(His)6–NH2 cell lysate absorptions with different M2+–EDTA–chitosan. The results showed that both absorption capacity and specific binding were influenced by adding different concentrations of sodium chloride (Fig. 8). By comparison with Ni2+–EDTA–chitosan and Zn2+–EDTA–chitosan, Cu2+–EDTA–chitosan could bind more protein from cell lysate feedstock but with lower purity of target protein (75 %) at 500 mmol/L concentration of NaCl. Although Ni2+–EDTA–chitosan and Zn2+–EDTA–chitosan exhibited higher binding capacities at 1000 mmol/L (4.08 mg/g, and 4.32 mg/mL, respectively), the purity of the targeted protein was lower than that of the protein obtained at 500 mmol/L concentration of NaCl (95 %, and 95 %). Cu2+-immobilized adsorbent possessed well binding capability, but showed lower specific binding. It is supposed that the addition of a salt in His-tagged proteins adsorption is mainly to neutralize charged groups on the support surface making the histidine–chelate more selective. At low ionic strength (0 mM of NaCl), the adsorption will be mixed (histidine–chelate plus ionic interaction); at medium ionic strength (500 mM of NaCl), the adsorption will be selective (only histidine–chelate), while at high ionic strength (1000 mM of NaCl) the adsorption will be mixed (histidine–chelate plus hydrophobic interaction). This result is accordance with our previous reports (Xu et al. 2014; Ren et al. 2015).
pH is the other factor influencing the protein binding ability due to the protonation of histidine. Normally, the effect of pH on adsorption located at the range of 6.0 to 8.0 (Lee et al. 2007). In our study, the efficiency of adsorption was achieved when pH was 7.2 (Fig. S1), which was accordance with the previous report (Lee et al. 2007).
Upon the fluorescent change of EGFP–(His)6–NH2 during the adsorption processes, Cu2+–EDTA–chitosan and Ni2+–EDTA–chitosan could quench the fluorescence, while Zn2+–EDTA–chitosan remained the fluorescence during the purification processes, even when the EGFP–(His)6–NH2 was bound to Zn2+–EDTA–chitosan (Fig. 6). Thereby, real-time monitoring could be achieved with the immigration of fluorescence-labeled hexahistidine. Comparatively, Zn2+–EDTA–chitosan showed more specific binding to the target protein with less affinity binding capacity (Fig. 5, lanes 16 (Zn2+–EDTA–chitosan) and 19 (Zn2+–chitosan) vs lanes 16 (Zn2+–EDTA–chitosan) and 7 (Cu2+–EDTA–chitosan)) examined with the fluorescent characteristic. These results indicated that selective binding of proteins could be profoundly affected by not only the chemical nature of the chelating ligand but also the metal ions (Chaouk and Hearn 1999).
Except for the easy availability and high efficiency, the environmental and economic factors are also essential to an ideal adsorption system. Therefore, we further developed a practical reuse method for the purification system based on M2+–EDTA–chitosan absorption. From the results presented in Table 1, the M2+–EDTA–chitosan (M═Ni, Cu, Zn) adsorbents were recycled to adsorb His-tagged protein. At the 5th recycle number, M2+–EDTA–chitosan adsorbent still bounds to about 80 % proteins, which suggested that the interaction between the M2+–EDTA–chitosan and protein was dominated by a physical process instead of a chemical reaction and therefore making M2+–EDTA–chitosan to be easily reused. Additionally, carbonyl groups of EDTA could well bind with metal ions, so less loose of metal was observed when the adsorbents were washed with PBS buffer and imidazole solution, also avoiding the denaturation which was caused by the release of metal ions from the adsorbents.
Furthermore, two His-tagged proteins, Trx–EGFP–(His)6–NH2 and Kana–(His)6–NH2 expressed in E. coli, were purified using this affinity adsorption system. It is noted that different purification condition was used in the purification procedure of Kana–(His)6–NH2 because the purity and efficiency were not good as the results of Trx–EGFP–(His)6–NH2 purification. As shown in Fig. S3, the target protein (lane 3 and lane 6) eluted by imidazole solution was impurity, while most of target proteins obtained in PBS solution (lane 2 and lane 4). Thus, to the purification procedure of Kana–(His)6–NH2, the experimental conditions were re-optimized. Fortunately, all experiment results showed that they could be quickly and easily purified from crude E. coli cell lysate without pre-treatment with the purity of up to 95 and 90 % (Figs. 9 and 10).
In summary, The M2+–EDTA–chitosan could be served as the adsorbents of His-tagged protein because it is readily available, and the adsorption procedure is easy to handle and in high purity without fussy pre-treatment. As shown in these studies, EDTA–chitosan was found to be especially promising in the protein purification field. The presented M2+–EDTA–chitosan-based purification system is eminently capable of performing specific binding with His-tagged proteins, in which the affinity protocol is compatible and practical.
References
Adriano WS, Mendonca DB, Rodrigues DS, Mammarella EJ, Giordano RLC (2008) Improving the properties of chitosan as support for the covalent multipoint immobilization of chymotrypsin. Biomacromolecules 9(8):2170–2179
Ali I (2012) New generation adsorbents for water treatment. Chem Rev 112:5073–5091
Arnau J, Lauritzen C, Petersen GE, Pedersen J (2006) Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. J Protein Expres Purifi 48:1–13
Bailey SE, Olin TJ, Bricka RM, Adrian DD (1999) A review of potentially low-cost sorbents for heavy metals. Water Res 33(11):2469–2479
Chaga GS (2001) Twenty–five years of immobilized metal ion affinity chromatography: past, present and future. J Biochem Biophys Methods 49(1):313–334
Chaouk H, Hearn MTW (1999) Examination of the protein binding behaviour of immobilised copper (II)–2, 6–diaminomethylpyridine and its application in the immobilised metal ion affinity chromatographic separation of several human serum proteins. J Biochem Biophys Methods 39:161–177
Demolliens A, Boucher C, Durocher Y, Jolicoeur M, Buschmann MD, Decrescenzo G (2008) Tyrosinase–catalyzed synthesis of a universal coil–chitosan bioconjugate for protein immobilization. Bioconjugate Chem 19:1849–1854
Ghosh R (2002) Protein separation using membrane chromatography: opportunities and challenges. J Chromatogr A 952:13–27
Gutierrez R, Martín del Valle E, Galan M (2007) Immobilized metal ion affinity chromatography: status and trends. Sep Purif Rev 36(1):71–111
Groll J, Haubensak W, Ameringer T, Moeller M (2005) Ultrathin coatings from isocyanate terminated star PEG prepolymers: patterning of proteins on the layers. Langmuir 21:3076–3083
Jain P, Vyas MK, Geiger JH, Baker GL, Bruening ML (2010) Protein purification with polymeric affinity membranes containing functionalized poly (acid) brushes. Biomacromolecules 11:1019–1026
Lee JJ, Bruley DF, Kang KA (2007) Effect of pH and imidazole on protein C purification from Cohn fraction IV–1 by IMAC. Adv Eexp Med Biol 599:61–66
Janzen R, Unger K, Müller W, Hearn M (1990) Adsorption of proteins on porous and non–porous poly(ethyleneimine) and tentacle–type anion exchangers. J Chromatogr A 522:77–93
Khare AR, Peppas NA (1995) Swelling/deswelling of anionic copolymer gels. Biomaterials 16:559–567
Klein MP, Nunes MR, Rodrigues RC, Benvenutti EV, Costa TMH, Hertz PF, Ninow JL (2012) Effect of the support size on the properties of β-galactosidase immobilized on chitosan: advantages and disadvantages of macro and nanoparticles. Biomacromolecules 13:2456–2464
Kofuji K, Murata Y, Kawashima S (2005) Sustained insulin release with biodegradation of chitosan gel beads prepared by copper ions. Int J Pharm 303:95–103
Lan Q, Bassi AS, Zhu JX, Margaritis A (2001) A modified Langmuir model for the prediction of the effects of ionic strength on the equilibrium characteristics of protein adsorption onto ion exchange/affinity adsorbents. Chem Eng J 81(1–3):179–186
Li F, Du P, Chen W, Zhang SS (2007) Preparation of silica-supported porous sorbent for heavy metal ions removal in wastewater treatment by organic–inorganic hybridization combined with sucrose and polyethylene glycol imprinting. Anal Chim Acta 585:211–218
Montembault V, Soutif JC, Brosse JC (1996) Synthesis of chelating molecules as agents for magnetic resonance imaging, polycondensation of diethylenetriaminepentaacetic acid bisanhydride with diols and diamines. React Func Polym 29:29–39
Porath J, Carlsson J, Olsson I, Belfrage G (1975) Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 258:598–599
Qiu M, Wu C, Ren GR, Liang XL, Wang XY, Huang JY (2014) Effect of chitosan and its derivatives as antifungal and preservative agents on postharvest green asparagus. Food Chem 155:105–11
Reddy DHK, Lee SM (2013) Synthesis and characterization of a chitosan ligand for the removal of copper from aqueous media. J Appl Polym Sci 130:4542–4550
Ren GW, Gong XW, Wang BL, Chen YY, Huang JY (2015) Affinity ionic liquids for the rapid liquid–liquid extraction purification of hexahistidine tagged proteins. Sep Purif Technol 146:114–120
Repo E, Warchol JK, Kurniawan TA, Sillanpaa MET (2010) Adsorption of Co(II) and Ni(II) by EDTA– and/or DTPA–modified chitosan: kinetic and equilibrium modeling. Chem Eng J 161:73–82
Roosen J, Binnemans K (2014) Adsorption and chromatographic separation of rare earths with EDTA and DTPA–functionalized chitosan biopolymers. J Mater Chem A 2:1530–1540
Shi XW, Wu HC, Liu Y, Tsao CY, Wang K, Kobatake E, Bentley WE, Payne GF (2008) Chitosan fibers: versatile platform for nickel–mediated protein assembly. Biomacromolecules 9:1417–1423
Stigter D, Dill A (1990) Charge effects on folded and unfolded proteins. Biochemistry 29:1262–1271
Tikhonov VE, Radigina LA, Yamskov YA (1996) Metal–chelating chitin derivatives via reaction of chitosan with nitrilotriacetic acid. Carbohyd Res 290:33–41
Tirtom VN, Dinçer A, Becerik S, Aydemir T, Çelik A (2012) Comparative adsorption of Ni(II) and Cd(II) ions on epichlorohydrin crosslinked chitosan–clay composite beads in aqueous solution. Chem Eng J 197:379–386
Ueda EKM, Gout PW, Morganti L (2003) Current and prospective applications of metal ion–protein binding. J Chromatogr A 988:1–23
Wang J, Zheng S, Shao Y, Liu J, Xu Z, Zhu D (2010) Aminofunctionalized Fe3O4@SiO2 core−shell magnetic nanomaterial as a novel adsorbent for aqueous heavy metals removal. J Colloid Interface Sci 349:293–299
Xu WY, Cao HZ, Ren GW, Xie HJ, Huang JY, Li SJ (2014) An AIL/IL–based liquid/liquid extraction system for the purification of his–tagged proteins. Appl Microbiol Biot 98:5665–5675
Yuan WY, Dong H, Li CM, Cui XQ, Yu L, Lu ZS, Zhou Q (2007) pH–controlled construction of chitosan/alginate multilayer film: characterization and application for antibody immobilization. Langmuir 23:13046–13052
Zachariou M, Hearn MT (1996) Application of immobilized metal ion chelate complexes as pseudocation exchange adsorbents for protein separation. Biochemistry 35(1):202–211
Zhang WC, Sun CY, Zhao Y, Lu XM (2011) One–pot synthesis and characterization of cross–linked quaternized chitosan microspheres as protein adsorbent. Int J Biol Macromol 49:688–692
Zhao FP, Sillanpa EPM, Meng Y, Yin DL, Tang WZ (2015) Green synthesis of magnetic EDTA–and/or DTPA–Cross–Linked chitosan adsorbents for highly efficient removal of metals. Ind Eng Chem Res 54:1271–1281
Zou H, Luo Q, Zhou D (2001) Affinity membrane chromatography for the analysis and purification of proteins. J Biochem Bioph Meth 49:199–240
Funding
This study was funded by the Natural Science Foundation of Zhejiang Province of China (Y3110204), the Technology Planning Project of Zhejiang Province of China (2011C12031), the National Science Foundation of China (21102129), and the Natural Key Technology R & D Program of China during the 12th Five-Year Plan Period (2012BAD29B06).
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 183 kb)
Rights and permissions
About this article
Cite this article
Hua, W., Lou, Y., Xu, W. et al. Batch affinity adsorption of His-tagged proteins with EDTA-based chitosan. Appl Microbiol Biotechnol 100, 879–891 (2016). https://doi.org/10.1007/s00253-015-7137-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7137-5