Abstract
Sucrose synthase (SuSy) catalyzes the reversible conversion of sucrose and a nucleoside diphosphate into fructose and nucleotide (NDP)-glucose. To date, only SuSy’s from plants and cyanobacteria, both photosynthetic organisms, have been characterized. Here, four prokaryotic SuSy enzymes from the nonphotosynthetic organisms Nitrosomonas Europaea (SuSyNe), Acidithiobacillus caldus (SuSyAc), Denitrovibrio acetiphilus (SusyDa), and Melioribacter roseus (SuSyMr) were recombinantly expressed in Escherichia coli and thoroughly characterized. The purified enzymes were found to display high-temperature optima (up to 80 °C), high activities (up to 125 U/mg), and high thermostability (up to 15 min at 60 °C). Furthermore, SuSyAc, SuSyNe, and SuSyDa showed a clear preference for ADP as nucleotide, as opposed to plant SuSy’s which prefer UDP. A structural and mutational analysis was performed to elucidate the difference in NDP preference between eukaryotic and prokaryotic SuSy’s. Finally, the physiological relevance of this enzyme specificity is discussed in the context of metabolic pathways and genomic organization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sucrose (Suc), the most abundant disaccharide in nature, is commonly found in plants and plays an important role in their development, growth, carbon storage, stress protection, and signal transduction (Winter and Huber 2000; Reid and Abratt 2005). Within the prokaryotic domain, mainly Cyanobacteria and some Proteobacteria are known to accumulate Suc, where it serves as a compatible solute to protect against osmotic stress (Reed 1986; Empadinhas and da Costa 2008) and is thought to stabilize protein and membrane structure (Reed 1986; Leslie et al. 1995). In plants and cyanobacteria, many efforts have already been made to unravel the enzymes behind their Suc metabolism, whereas for nonphotosynthetic organisms, this remains largely unexplored.
In plants and cyanobacteria, Suc synthesis is catalyzed by the sequential action of sucrose-phosphate synthase (SPS, EC 2.4.1.14) and sucrose-phosphate phosphatase (SPP, EC 3.1.3.24) (Porchia and Salerno 1996; Salerno and Curatti 2003; Cumino et al. 2010). In these organisms, SPS performs the first step of Suc synthesis, generating sucrose 6-phosphate (Suc6P) from fructose 6-phosphate (Fru6P) and an activated sugar donor, such as uridine diphosphate glucose (UDP-Glc). The phosphate group of S6P is then cleaved off by SPP to yield Suc. Suc can either be irreversibly hydrolyzed by an invertase or be further metabolized together with NDP yielding fructose and NDP-Glc (Vargas et al. 2003). The latter reaction, which is equally reversible in vitro, is catalyzed by sucrose synthase (SuSy, EC 2.4.1.13). Since its discovery in 1995 by Cardini et al., various SuSy’s from plants and cyanobacteria have been characterized (Delmer 1972; Tsai 1974; Morell and Copeland 1985; Porchia et al. 1999; Tanase and Yamaki 2000; Curatti et al. 2000; Baroja-Fernández et al. 2012; Kolman et al. 2012). These studies show that plant SuSy’s preferentially use UDP as nucleotide substrate, although ADP, CDP, GDP, and TDP can serve to a lesser extent as alternative acceptors. Conversely, the SuSy from the thermophilic cyanobacterium Thermosynechococcus elongatus (SuSyTe) showed a clear preference for ADP, as reflected by the 7-fold lower Km compared to UDP (Figueroa et al. 2013). The produced sugar nucleotides are directed toward cell wall or starch biosynthesis in plants, whereas they play an important role in the synthesis of glycogen and other (structural) polysaccharides in cyanobacteria (Haigler et al. 2001; Baroja-Fernández et al. 2003; Koch 2004; Curatti et al. 2008).
Besides its biological significance, SuSy has also proven to be a versatile biocatalyst for practical applications. In 1993, Elling and coworkers demonstrated the production of expensive nucleotide sugars (NDP-Glc) starting from the abundant and cheap substrate Suc. Moreover, SuSy can also be coupled with a glycosyltransferase (GT), which has resulted in a cost-effective method for the glycosylation of small molecules (Brinkmann et al. 2001; Masada et al. 2007; Son et al. 2009; Terasaka et al. 2012; Bungaruang et al. 2013; Gutmann et al. 2014). However, low activities and poor stability of the reported SuSy enzymes have impeded their commercial exploitation so far.
In this contribution, novel SuSy’s from different phyla (Proteobacteria, Deferribacteres, and Ignavibacteriae) are characterized for the first time and provide more insight into the evolution of these important sucrose-metabolizing enzymes. In addition, these SuSy’s display interesting properties that render them promising candidates for industrial applications.
Materials and methods
Materials
Unless otherwise stated, all chemicals were bought from Sigma-Aldrich, Merck, or Carbosynth and were of the highest purity.
Sequence analysis
All amino acid sequences annotated as sucrose synthase were retrieved from the UniProtKB database. Sequences that were either not unique, did not start with a methionine, were too long (>2000 amino acids), too short (<600 amino acids), or contained undefined amino acids, were removed. In total, 67 prokaryotic sequences were retained and aligned with Clustal Omega (default parameters) (Sievers et al. 2011). “MEGA 6.0” (Tamura et al. 2013) was used to create a maximum likelihood (ML) unrooted phylogenetic tree, based on the LG + G + I + F model, with 1000 bootstrap replications, five discrete gamma categories, a nearest-neighbor-interchange heuristic ML method and a strong branch swap filter.
To calculate the percentage frequency of amino acids occurring at positions within 4 Å of the nucleobase ring of UDP (based on the crystal structure of SuSyAt1, PDB ID: 3S27), a multiple sequence alignment of 110 plant sequences and 67 prokaryotic sequences was used (Table S4).
To determine the gene organization of sucrose-metabolizing genes in prokaryotic organisms, the Prokaryotic Operon DataBase (ProOpDB) and the Database of prokaryotic Operons (DOOR) were used (Taboada et al. 2012; Mao et al. 2014).
Cloning and site-directed mutagenesis
The putative SuSy sequences from Acidithiobacillus caldus ATCC 51756 (SuSyAc, UniProt ID: A0A059ZV61), Denitrovibrio acetiphilus DSM 12809 (SuSyDa, UniProt ID: D4H6M0), Melioribacter roseus JCM 17771 (SuSyMr, UniProt ID: I7A3T6), and Thermosipho melanesiensis DSM 12029 (spsTm, UniProt ID: A6LKE9) were codon optimized for Escherichia coli, provided with a C-terminal His6-tag and chemically synthesized by GenScript (Piscataway, NJ, USA) (Fig. S1). The SuSy encoding sequence of Nitrosomonas europaea ATCC 19718 (SuSyNe, RefSeq: NP_841269.1, UniProt ID: Q820M5) was amplified from genomic DNA that was extracted from the organism and kindly provided by Prof. Nico Boon (Ghent University). The SuSy encoding sequences were cloned into the constitutive expression vector pCXP34h (Aerts et al. 2011) by means of a Gibson assembly procedure (Gibson et al. 2009). Primers used to amplify the genes and backbone are summarized in Table S1. In case of SuSyAc, SuSyDa, SuSyMr, and the pCXP34h backbone, the reaction mixture was composed of PrimeSTAR premix (Westburg), 2.5 μM forward and reverse primer and ~3 ng/μL template, in a total volume of 50 μL. The following program was used: initial denaturation of 5 min at 98 °C and 30 cycles of denaturation at 98 °C for 10 s, annealing at 55 °C for 5 s and elongation at 72 °C for 1 min/kb. For SuSyNe, the reaction mixture was composed of gDNA or pCXP34h plasmid (with C-terminal His6-tag), 5× Q5 reaction buffer, Q5 High-fidelity DNA polymerase (0.02 U/μL), dNTP mix (0.2 mM), forward primer (0.5 μM), and reverse primer (0.5 μM). The following program was used: initial denaturation of 30 s at 98 °C and 30 cycles of denaturation at 98 °C for 10 s, annealing at 65 °C for 20 s, and elongation at 72 °C for 30 s/kb, followed by a final elongation of 2 min at 72 °C. Next, PCR products were treated with DpnI (Westburg) to remove template DNA and were subsequently purified using the Qiagen purification kit, checked on a 1 % agarose gel, and the DNA concentration was measured with a Nanodrop ND-1000 (Thermo Scientific) at 260 nm. To ligate the SuSy encoding sequences and the pCXP34h backbone, a Gibson assembly mix (20 μL) containing 100 ng backbone and an equimolar amount of gene product was incubated for 1 h at 50 °C. Finally, the resulting expression plasmids were transformed in E. coli BL21 (DE3). All constructs were subjected to nucleotide sequencing (AGOWA sequence service, Berlin) to confirm that the ligation was correct and to exclude the presence of undesirable mutations.
Site-directed mutations were introduced with a modified two-stage megaprimer-based whole plasmid PCR method (Sanchis et al. 2008), using the primers described in Table S2. The PCR mix contained 5× Q5 reaction buffer, 0.02 U/μL Q5 High-Fidelity DNA Polymerase (Bioke), 0.2 mM dNTP mix, 0.002–0.02 ng/μL template plasmid DNA (pCXP34h SuSyAc), 0.5 μM forward, and reverse primer in a total volume of 50 μL. The amplification program started with an initial denaturation (30 s at 98 °C), followed by 5 cycles of denaturation for 10 s at 98 °C, annealing for 20 s at 66 °C, and extension for 30 s/kb (size megaprimer) at 72 °C. The second stage consisted of 25 cycles of 10 s at 98 °C and extension for 1 min/kb (size whole plasmid) at 72 °C, and one final extension of 2 min at 72 °C. After digestion of the template DNA by DpnI (Westburg), mutagenized plasmids were transformed in E. coli BL21 (DE3) (Novagen). All constructs were subjected to nucleotide sequencing (AGOWA sequence service, Berlin).
Enzyme production and purification
For enzyme production, transformed E. coli was first inoculated in 5-mL LB medium containing 10 g/L trypton, 10 g/L NaCl, 5 g/L yeast extract, and 100 μg/mL ampicillin and incubated overnight at 37 °C, with continuous shaking at 250 rpm. Next, 1 % (v/v) of the overnight culture was inoculated in shake flasks with fermentation medium (250 mL LB containing 100 μg/mL ampicillin) and incubated with continuous shaking at 200 rpm for at least 6 h at 37 °C, until an OD of about 4 was reached. The produced biomass was harvested by centrifugation for 15 min at 8000 rpm at 4 °C, and the obtained cell pellets were stored at −20 °C. Cell pellet from 250-mL culture was then redissolved in 10-mL lysis buffer (50 mM NaPB pH 7.4 and 500 mM NaCl (PBS), 40 mM imidazole, and 100 μM PMSF in ethanol) supplemented with lysozyme in a final concentration of 1 mg/mL. This cell suspension was incubated on ice for 10 min and sonicated 3 times for 2.5 min (Branson sonifier 250, level 3, 50 % duty cycle). After sonication, cell debris was removed by centrifugation at 9000 rpm for 45 min. The resulting supernatant, containing the soluble fraction of the protein, was collected. The His6-tagged proteins were purified by Ni-NTA chromatography as described by the supplier (Qiagen) in the protocol purification of His-tagged proteins using a gravity-flow column. First, equilibration was performed using a buffer composed of 50 mM NaPB pH 7.4, 500 mM NaCl (PBS), and 40 mM imidazole. Then, the protein solution was applied to the column and washed with a buffer containing 50 mM PBS and 60 mM (SuSyDa and SuSyTm) or 100 mM imidazole (SuSyNe, SuSyMr, SuSyAc, and mutants thereof). Afterward, elution occurred with a buffer composed of 50 mM PBS and 250 mM imidazole. Finally, the buffer was exchanged to 100 mM MOPS pH 7.0 in a 30 K Amicon Ultra centrifugal filter. The protein concentration was measured with a Nanodrop ND-1000 (Thermo Scientific).
Enzyme assays
The bicinchoninic acid (BCA) method detects reducing sugars (Waffenschmidt and Jaenicke 1987), such as fructose and glucose, and has already been used before to measure the activity of enzymes, such as cellulases (Chen et al. 2004) and phosphorylases (Cerdobbel et al. 2011). As SuSy releases fructose during the cleavage of Suc, the BCA assay is also able to measure SuSy activity accurately. The color reagent is prepared by combining 23 parts of a solution containing 1.5 g/L 4,4′-dicarboxy-2,2′-biquinoline dipotassium salt and 62.3 g/L anhydrous Na2CO3, 1 part of a solution composed of 23 g/L aspartic acid, 33 g/L anhydrous Na2CO3, and 7.3 g/L CuSO4 and 6 parts ethanol. Sample (25 μL) is added to 150 μL of assay solution. Afterward, the microtiter plate is covered by a plastic foil and incubated for 30 min at 70 °C. After cooling to room temperature, the absorbance is measured at 560 nm. One unit of SuSy activity is defined as the amount of enzyme that released 1 μmol of fructose min−1 under the specified conditions. Kinetic parameters (apparent Km and Vmax values) were calculated by nonlinear regression of the Michaelis-Menten equation using Sigma Plot 11.0. Alternatively, substrate inhibition was fitted according to the equation (Vmax*S) / (S + Km + (S2/Ki)) with Vmax = maximal reaction velocity (U/mg); S = substrate concentration (mM); Ki = inhibition dissociation constant; Km = Michaelis-Menten constant (Copeland 2000).
Influence of divalent cations, thermostability, temperature, and pH dependence of SuSy activity
A universal Britton-Robinson (BR) buffer system, consisting of 25 mM H3BO3, H3PO4, and CH3COOH was used to determine pH profiles of SuSyAc, SuSyDa, SuSyMr, and SuSyNe in the Suc cleavage direction. One part of 50 mM BR buffer was mixed with 1 part of substrate mix (Suc and ADP in milliQ) and titrated to the desired pH with NaOH. Concentrations of Suc and ADP in the final reaction mixture were 200 and 5 mM, respectively.
Temperature profiles were made by determining the activity of SuSy’s in the direction of Suc cleavage from 30 to 90 °C. Thermal stability of these SuSy’s was evaluated by incubating the enzyme (~0.17 mg/mL), without the presence of any substrate, for 15 min at 60 °C in 100 mM MOPS pH 7.0. After incubation, residual activity in the Suc cleavage direction was determined (200 mM Suc, 5 mM ADP, 100 mM MOPS pH 7.0).
Influence of MgCl2 on SuSy was determined by measuring the activity at 60 °C in the presence of 100 mM MOPS pH 7.0, 200 mM Suc, 5 mM ADP, and concentrations of MgCl2 ranging from 0 to 10 mM.
Nucleotide sequence accession numbers
The DNA sequences of the codon-optimized genes have been submitted to GenBank (ID 1782677) under accession numbers KP284426 (SuSyAc), KP284427 (SuSyDa), KP284428 (SuSyMr), and KP284429 (spsTm). The sequences are also provided in Fig. S1.
Results
Phylogenetic analysis and expression of prokaryotic SuSy sequences
To date, only SuSy enzymes from photosynthetic organisms like plants and cyanobacteria have been characterized. However, several studies on genome annotation have revealed the occurrence of predicted SuSy sequences in (non-)photosynthetic proteobacterial organisms (Lunn 2002; Subbaiah et al. 2006; Jayashree et al. 2008), although this was never confirmed experimentally. Furthermore, the increasing amount of genomic data that has become available in the past few years calls for a revision of the taxonomic distribution of putative prokaryotic SuSy enzymes. Hence, a phylogenetic tree was constructed with all available prokaryotic sequences (~68) from the UniProtKB database that were annotated as SuSy (Fig. 1).
Most of these prokaryotic organisms belong to the Cyanobacteria and Proteobacteria, which is in good agreement with other reports (Lunn 2002; Subbaiah et al. 2006; Jayashree et al. 2008). Remarkably, our phylogenetic analysis revealed that also organisms belonging to other phyla contain predicted SuSy’s. Indeed, Denitrovibrio acetiphilus, Desulfurispirillum indicum, Dethiobacter alkaliphilus, Melioribacter roseus belong, respectively, to the phyla Deferribacteres, Chrysiogenetes, Firmicutes, and Ignavibacteriae. Subsequently, two putative proteobacterial SuSy’s and two from the rather unusual phyla Deferribacteres and Ignavibacteriae were selected for characterization (Table 1).
The sequences, provided with a C-terminal His6-tag, were expressed in E. coli BL21 (DE3) and purified by Ni-NTA metal affinity chromatography to apparent homogeneity (>95 %) under optimized purification conditions (Fig. 2). All enzymes were mainly present in the soluble fraction, but expression of SuSyMr was very poor (~75 μg enzyme, starting from 250-mL culture medium). Their electrophoretic behavior corresponds well with their predicted molecular mass of about 92 kDa.
SDS-PAGE analysis of the recombinantly expressed prokaryotic SuSy’s from A. caldus (SuSyAc), N. europaea (SuSyNe), M. roseus (SuSyMr), and D. acetiphilus (SuSyDa). Lanes 1–4 purified enzymes; lanes 5–8 crude cell extract (soluble fraction); 1 and 5 = SuSyAc; 2 and 6 = SuSyNe; 3 and 8 = SuSyMr; 4 and 8 = SuSyDa
pH optimum, temperature profile, thermostability, and effect of divalent cations
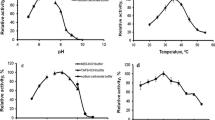
To determine the optimal conditions of the enzymes in the Suc cleavage direction, the effect of temperature, pH, and MgCl2 on the activity was studied. Results are summarized in Table 2.
The pH optima of SuSyAc, SuSyDa, SuSyMr, and SuSyNe were around 5.5, 6, 7, and 5, respectively. All SuSy’s displayed at least 40 % of their maximal activity within a range of pH 5.5–7.5 (Fig. S2). Temperature profiles were determined in the presence of 200 mM Suc, 5 mM ADP, and 2 mM MgCl2 at pH 7 (Fig. S3). The highest activities were obtained at 60, 65, 80, and 75 °C for SuSyAc, SuSyDa, SuSyMr, and SuSyNe, respectively. These are remarkably high-temperature optima, especially since the source organisms of these SuSy’s only have optimal growth temperatures between 20 and 55 °C (Table 1). The cyanobacterial SuSyTe also displays an optimum of 70 °C, whereas plants SuSy’s have optima between 40 and 55 °C (Sebková et al. 1995; Elling and Kula 1995; Klotz et al. 2003; Figueroa et al. 2013).
In addition, the thermostability of the selected SuSy’s was assessed by determining the residual activity after incubating the enzymes for 15 min at 60 °C. It should be noted that the enzymes were incubated without the presence of any substrates, since Suc is known to act as a stabilizing agent (Lee and Timasheff 1981; Leslie et al. 1995). Unlike the others, SuSyDa was completely inactivated within 15 min. The most thermostable SuSy appeared to be SuSyAc with a residual activity of 96 %.
Mg2+ or other cations have been frequently reported to either positively (Morell and Copeland 1985) or negatively (Tsai 1974; Huang and Wang 1998) influence the activity of SuSy in the Suc cleavage direction. To scrutinize the effect of cations on the different SuSy enzymes, the activity was determined in the presence of 200 mM Suc, 5 mM ADP, and varying concentrations of MgCl2 (Fig. S4). For SuSyDa, SuSyMr, and SuSyNe, a decrease in activity was observed for increasing concentrations of MgCl2. In contrast, MgCl2 slightly stimulates the activity of SuSyAc.
Kinetic properties and substrate specificity
To investigate the nucleotide preference of SuSyNe, apparent kinetic parameters were determined for Suc, ADP, UDP, GDP, and CDP at 60 °C and pH 7.0 in the Suc breakdown direction (Table 3).
Substrate inhibition occurred in the presence of GDP at concentrations above 10 mM (Ki ≈ 50 mM) whereas typical Michaelis-Menten kinetics were observed for the other substrates. A significant difference was observed between the affinity for Suc in the presence of either ADP or UDP. Apparently, the Km for Suc is about 8 times lower with ADP as cosubstrate instead of UDP. Conversely, Km values for ADP and UDP are in the same range. This indicates that, in vivo, this enzyme will probably metabolize sucrose mainly using ADP. For plant SuSy’s, reported Km values for Suc are also dependent on the used cosubstrate, but for these enzymes, the affinity for Suc was highest with UDP (Delmer 1972; Baroja-Fernández et al. 2003, 2012).
Based on the affinities for the different nucleotides, SuSyAc also showed a clear preference for ADP. The enzyme displayed Km values of 0.17, 7.8, 8.5, and 16.9 mM for ADP, UDP, GDP, and CDP, respectively (Table S3). The Km for ADP is thus at least 45 times lower compared to the other nucleotides. In fact, it is the first time that such high Km values for these nucleotides are reported for a SuSy enzyme.
Finally, the predilection for ADP could also be extended toward SuSyDa. Indeed, the specific activity of this enzyme with ADP (125 U/mg) was about 20-fold higher than that with the other nucleotides. With a few exceptions (Baroja-Fernández et al. 2003, 2012), typical values for specific activity of SuSy enzymes are between 1 and 14 U/mg (Figueroa et al. 2013), and thus a 10- to 100-fold lower than that observed for SuSyDa. Conversely, specific activities for SuSyMr were about 2–4 U/mg for both ADP and UDP. Detailed kinetic characterization of SuSyMr is not provided because of the poor expression and low activities compared to the other SuSy’s.
Structure-function relationship
It is known from literature that plant SuSy’s generally prefer UDP, although they can also use other nucleotides to a certain extent (Delmer 1972; Tsai 1974; Morell and Copeland 1985; Ross and Davies 1992; Porchia et al. 1999; Tanase and Yamaki 2000; Baroja-Fernández et al. 2012). As shown in the previous sections, SuSyAc, SuSyNe, and SuSyDa displayed a clear preference for ADP. To elucidate the structural determinants responsible for this difference in nucleotide preference, residues surrounding the nucleotide substrate were compared between prokaryotic and eukaryotic SuSy sequences. To identify these residues, the crystal structure of SuSyAt1 (PDB ID: 3S27) in complex with UDP was used as this is the only SuSy structure available to date. Table S4 lists all the residues of SuSyAt1 within 4 Å of the nucleobase ring of UDP and the relative abundance of amino acids at the corresponding positions for both plant and bacterial SuSy’s.
Interestingly, the amino acid distribution at the positions surrounding the base moiety differed remarkably between both domains. Generally, the positions are much more conserved in plants compared to the prokaryotic SuSy’s. Seven positions (296, 270, 578, 579, 580, 609, 647, and 649 in SuSyAt1) are comparable between bacterial and plant SuSy’s and are characterized mainly by hydrophobic amino acid side chains. In contrast, the glutamine at position 648 and the asparagine at position 654 are 100 % conserved in plants, while at the corresponding positions in bacteria, a whole range of other amino acids occur but rarely a glutamine (at 654) and never an asparagine (at 648) (Table S4 and Fig. 3). Remarkably, these two positions are the only ones that make hydrogen bonds with the uracyl ring of UDP in the structure of SuSyAt1. This led to the hypothesis that these residues are responsible for the difference in nucleotide preference between plants and bacteria. Another potentially interesting residue is found at position 653, where arginine is almost 100 % conserved in plants while in bacteria, the predominant amino acids are hydrophobic or negatively charged. Furthermore, R653 makes a hydrogen bond with Q648 and could thus indirectly contribute to the NDP preference.
Comparison and visualization of the residues in plant and bacterial SuSy’s near the nucleotide binding pocket (a). Sequence alignment of amino acids in plant and bacterial SuSy’s close to residues Gln648 and Asn654 of SuSyAt1. These two residues make H-bonds with the uracil ring of UDP and are indicated by boxes of solid lines. Numbers above the alignment indicate the amino acid position in SuSyAt1 (b). Bacterial amino acid distributions at positions corresponding to Gln648 and Asn654 of SuSyAt1 (c). Molecular visualization of the residues Gln648, Asn654, and Arg653 (which were selected for mutagenesis), the substrate UDP, and their interactions. H-bridges are represented by dashed yellow lines
To test the importance of these three residues, the amino acids of SuSyAc at these positions were replaced by those occurring in plant SuSy’s. To that end, one single mutant (A642N), one double mutant (L636Q-A642N), and one triple mutant (L636Q-V641R-A642N) were created. Unfortunately, the Km value for UDP could not be lowered by none of these mutations (Fig. S5). These results could thus not confirm the hypothesis that the evaluated positions are responsible for the difference in nucleotide preference between bacteria and plants.
Discussion
Phylogenetic analysis and genomic organization
To explore the arsenal of SuSy’s that nature has to offer us, a phylogenetic tree was constructed of all annotated prokaryotic SuSy’s from the UniProtKB Web site. Due to the increasing availability of genomic data, this phylogenetic analysis and subsequent activity measurements revealed that not only photosynthetic organisms such as plants and cyanobacteria harbor active SuSy’s but also other phyla such as the Proteobacteria, Deferribacteres, Chrysiogenetes, Ignavibacteriae, and Firmicutes. Interestingly, a previous report also described the presence of a putative SuSy in the genome of Thermosipho melanesiensis, which belongs to the Thermotogae (Jayashree et al. 2008). Therefore, we also cloned, expressed, and purified that enzyme but found that it is a sucrose-phosphate synthase instead of a SuSy (data not shown). This sequence is thus wrongly annotated in the UniprotKB database.
In contrast, the sequences from the nonphotosynthetic prokaryotes M. roseus, D. acetiphilus, N. europaea, and A. caldus were found to be true SuSy enzymes and were fully characterized. To check whether these organisms also possessed other sucrose-synthesizing enzymes, their genomes were screened for the occurrence of putative SPS and SPP encoding genes (Fig. 4). Interestingly, in all cases, SuSy was clustered in an operon together with a putative fructokinase and SPS/SPP bimodular enzyme. Although genetic clustering of SPS and SPP was already observed for the cyanobacterium Synechococcus sp. PCC 7002 (Cumino et al. 2010), the contiguous location of both SPS, SPP, and SuSy has not been reported so far. Bifunctional enzymes, with both SPS and SPP activity, have been described for both proteobacterial and cyanobacterial organisms (Martínez-Noël et al. 2013; But et al. 2013). In case of the putative SPS/SPP encoding sequences of M. roseus, D. acetiphilus, N. europaea, and A. caldus, all HAD-phosphatase residues required for SPP activity were present and other homologous SPP sequences were not found, indicating that they are probably functional bimodular enzymes (Fig. 4 and Table S5). However, this still remains to be confirmed experimentally.
Genomic organization of sucrose-metabolizing genes in nonphotosynthetic and photosynthetic prokaryotes. Position in the genome is indicated above the arrows. Blue box: seemingly futile cycle of Suc metabolism. Gene abbreviations: sps = sucrose-phosphate synthase, spp = sucrose-phosphate phosphatase, susy = sucrose synthase, sp = sucrose phosphorylase, frk = fructokinase, pfkb = PfkB family of carbohydrate kinase, amsA = amylosucrase
The occurrence of both sucrose-synthesizing enzymes and sucrose-degrading enzymes in the same operon raises metabolic questions about the function of these enzymes in nonphotosynthetic organisms. The seemingly futile cycle of Suc metabolism, resulting from these coexpressed enzymes, could be an ingenious mechanism to fine-tune the supply of Suc and nucleotide sugars, depending on the cell’s demand under certain environmental conditions. Indeed, it has been suggested before that Suc cycles in plants, characterized by a permanent process of formation and degradation, could allow organisms to respond with a high degree of sensitivity to factors influencing sugar accumulation, osmotic potential, respiration, and sugar signaling (Roby et al. 2002; Cumino et al. 2007). However, additional studies are needed to unravel the specific role and regulation of sucrose-metabolizing enzymes in nonphotosynthetic bacteria.
Substrate preference
The kinetic parameters, determined for SuSyAc, SuSyDa, and SuSyNe, imply a preference for ADP. For SuSyTe, a similar observation was made which indicates that this is probably a common feature for prokaryotic SuSy’s, in contrast to plant SuSy’s which generally prefer UDP. It was already suggested before that this preference for adenine nucleotides links Suc metabolism directly to glycogen metabolism (Cumino et al. 2007; Curatti et al. 2008). Production of glycogen is catalyzed by glycogen synthase which uses ADP-glucose (ADP-Glc) as glucosyl donor to elongate an α-1,4-glucosidic chain. ADP-Glc is mainly generated from glucose 1-phosphate by ADP-Glc pyrophosphorylase (AGPase, EC 2.7.7.27). However, it has been demonstrated that a concomitant supply of ADP-Glc for glycogen biosynthesis should also be attributed to the Suc cleavage action of SuSy (Cumino et al. 2007; Curatti et al. 2008). The clear preference for ADP, observed for the SuSy’s from nonphotosynthetic species, could thus indicate a similar function in regulating the C-flux between Suc and glycogen.
Differences in nucleotide preference are also observed for SPS enzymes. Indeed, plant SPSs are highly specific for UDP-Glc, whereas bacterial SPSs can also use other NDP-Glc substrates (Porchia and Salerno 1996; Lunn et al. 1999). Chua and coworkers (2008) already suggested three nucleotide binding residues of Halothermothrix orenii SPS, conserved in plants but highly variable among bacteria, which could be responsible for the different binding modes in plant and bacterial SPSs. However, this hypothesis was never verified experimentally. In this article, a similar hypothesis was tested by mutating three putative substrate preference determining residues in the bacterial SuSyAc to the corresponding amino acids occurring in plant SuSy’s. Unfortunately, no switch from ADP to UDP preference could be obtained, indicating that this phenomena is much more complex than initially thought.
Industrial applications
One of the major hurdles of large-scale glycosylation processes is the high price of nucleotide sugars (UDP-glucose ≈ 150 €/g). In this respect, SuSy enzymes are interesting biocatalysts for the production of these activated sugars starting from the abundant and cheap substrate sucrose and for the cost-effective glycosylation of small molecules by coupling to a GT (Zervosen et al. 1998; Masada et al. 2007; Son et al. 2009; Terasaka et al. 2012; Bungaruang et al. 2013; Gutmann et al. 2014). Key requirements for successful application in industry predominantly consist of highly active, (thermo) stable enzymes, and a high number of regeneration cycles for UDP (De Bruyn et al. 2015). To this end, SuSyAc could be a novel promising alternative to the plant SuSy’s currently used. Indeed, the enzyme displays high maximal activities on both UDP and ADP (at least four times higher than most of the plant enzymes and the thermophilic SuSyTe) but most of all because of its high stability. After 15 min, the enzyme still displays 96 % of its activity while, e.g., SuSyTe only has 30 % activity left after 10 min of incubation at 60 °C and plant SuSy’s are often even less stable at that temperature. Such an elevated temperature is of interest if microbial contamination needs to be avoided during the reaction process. Furthermore, compared to plants, expression yields are high. At least 2 mg of SuSyAc could be recovered after purification, starting from 250-mL culture medium. All these characteristics determine the final efficiency and cost-effectiveness of any SuSy-coupled glycosylation reaction in future industrial applications.
Taken all together, we succeeded in expanding the pool of (industrially relevant) SuSy’s, which will be of crucial importance to sustain and improve the quality of biocatalytic processes and also gives the opportunity to conduct further research on the evolution and function of sucrose-metabolizing enzymes in nonphotosynthetic organisms.
References
Aerts D, Verhaeghe T, De Mey M, Desmet T, Soetaert W (2011) A constitutive expression system for high-throughput screening. Eng Life Sci 11:10–19. doi:10.1002/elsc.201000065
Baroja-Fernández E, Muñoz FJ, Saikusa T, Rodríguez-López M, Akazawa T, Pozueta-Romero J (2003) Sucrose synthase catalyzes the de novo production of ADPglucose linked to starch biosynthesis in heterotrophic tissues of plants. Plant Cell Physiol 44:500–509
Baroja-Fernández E, Muñoz FJ, Li J, Bahaji A, Almagro G, Montero M, Etxeberria E, Hidalgo M, Sesma MT, Pozueta-Romero J (2012) Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc Natl Acad Sci U S A 109:321–326. doi:10.1073/pnas.1117099109
Brinkmann N, Malissard M, Ramuz M, Römer U, Schumacher T, Berger EG, Elling L, Wandrey C, Liese A (2001) Chemo-enzymatic synthesis of the Galili epitope Gal(alpha)(1→3)Galbeta(1→4)GlcNAc on a homogeneously soluble PEG polymer by a multi-enzyme system. Bioorg Med Chem Lett 11:2503–2506
Bungaruang L, Gutmann A, Nidetzky B (2013) Leloir Glycosyltransferases and Natural Product Glycosylation: Biocatalytic Synthesis of the C-Glucoside Nothofagin, a Major Antioxidant of Redbush Herbal Tea. Adv Synth Catal 355:2757–2763. doi:10.1002/adsc.201300251
But SY, Khmelenina VN, Reshetnikov AS, Trotsenko YA (2013) Bifunctional sucrose phosphate synthase/phosphatase is involved in the sucrose biosynthesis by Methylobacillus flagellatus KT. FEMS Microbiol Lett 347:43–51. doi:10.1111/1574-6968.12219
Cardini CE, Leloir LF, Chiriboga J (1955) The biosynthesis of sucrose. J Biol Chem 214:149–155
Cerdobbel A, De Winter K, Aerts D, Kuipers R, Joosten H-J, Soetaert W, Desmet T (2011) Increasing the thermostability of sucrose phosphorylase by a combination of sequence- and structure-based mutagenesis. Protein Eng Des Sel 24:829–834. doi:10.1093/protein/gzr042
Chen P-J, Wei T-C, Chang Y-T, Lin L-P (2004) Purification and characterization of carboxymethyl cellulase from Sinorhizobium fredii. Bot Bull Acad Sin 45:111–118
Chua TK, Bujnicki JM, Tan T-C, Huynh F, Patel BK, Sivaraman J (2008) The structure of sucrose phosphate synthase from Halothermothrix orenii reveals its mechanism of action and binding mode. Plant Cell 20:1059–1072. doi:10.1105/tpc.107.051193
Copeland R (2000) Enzymes. A practical introduction to structure, mechanism and data analysis. Wiley-VCH, New York
Cumino AC, Marcozzi C, Barreiro R, Salerno GL (2007) Carbon cycling in Anabaena sp. PCC 7120. Sucrose synthesis in the heterocysts and possible role in nitrogen fixation. Plant Physiol 143:1385–1397. doi:10.1104/pp. 106.091736
Cumino AC, Perez-Cenci M, Giarrocco LE, Salerno GL (2010) The proteins involved in sucrose synthesis in the marine cyanobacterium Synechococcus sp. PCC 7002 are encoded by two genes transcribed from a gene cluster. FEBS Lett 584:4655–4660. doi:10.1016/j.febslet.2010.10.040
Curatti L, Porchia AC, Herrera-Estrella L, Salerno GL (2000) A prokaryotic sucrose synthase gene (susA) isolated from a filamentous nitrogen-fixing cyanobacterium encodes a protein similar to those of plants. Planta 211:729–735
Curatti L, Giarrocco LE, Cumino AC, Salerno GL (2008) Sucrose synthase is involved in the conversion of sucrose to polysaccharides in filamentous nitrogen-fixing cyanobacteria. Planta 228:617–625. doi:10.1007/s00425-008-0764-7
De Bruyn F, Maertens J, Beauprez J, Soetaert W, De Mey M (2015) Biotechnological advances in UDP-sugar based glycosylation of small molecules. Biotechnol Adv. doi:10.1016/j.biotechadv.2015.02.005
Delmer DP (1972) The Purification and Properties of Sucrose Synthetase from Etiolated Phaseolus aureus Seedlings. J Biol Chem 247:3822–3828
Elling L, Kula M-R (1995) Characterization of sucrose synthase from rice grains for the enzymatic synthesis of UDP and TDP glucose. Enzym Microb Technol 17:929–934. doi:10.1016/0141-0229(94)00017-L
Elling L, Grothus M, Kula MR (1993) Investigation of sucrose synthase from rice for the synthesis of various nucleotide sugars and saccharides. Glycobiology 3:349–355
Empadinhas N, da Costa MS (2008) Osmoadaptation mechanisms in prokaryotes: distribution of compatible solutes. Int Microbiol 11:151–161
Figueroa CM, Asención Diez MD, Kuhn ML, McEwen S, Salerno GL, Iglesias AA, Ballicora MA (2013) The unique nucleotide specificity of the sucrose synthase from Thermosynechococcus elongatus. FEBS Lett 587:165–169. doi:10.1016/j.febslet.2012.11.011
Gibson DG, Young L, Chuang R, Venter JC, Iii CAH, Smith HO, America N (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. 6:12–16. doi:10.1038/NMETH.1318
Gutmann A, Bungaruang L, Weber H, Leypold M, Breinbauer R, Nidetzky B (2014) Towards the synthesis of glycosylated dihydrochalcone natural products using glycosyltransferase-catalysed cascade reactions. Green Chem 16:4417–4425. doi:10.1039/C4GC00960F
Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47:29–51
Huang DY, Wang AY (1998) Purification and characterization of sucrose synthase isozymes from etiolated rice seedlings. Biochem Mol Biol Int 46:107–113
Jayashree B, Pradeep R, Kumar A, Gopal B (2008) Correlation between the Sucrose Synthase Protein Subfamilies, Variations in Structure and Expression in Stress-derived Expressed Sequence Tag Datasets. J Proteomics Bioinform 01:408–423. doi:10.4172/jpb.1000050
Klotz KL, Finger FL, Shelver WL (2003) Characterization of two sucrose synthase isoforms in sugarbeet root. Plant Physiol Biochem 41:107–115. doi:10.1016/S0981-9428(02)00024-4
Koch K (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7:235–246. doi:10.1016/j.pbi.2004.03.014
Kolman MA, Torres LL, Martin ML, Salerno GL (2012) Sucrose synthase in unicellular cyanobacteria and its relationship with salt and hypoxic stress. Planta 235:955–964. doi:10.1007/s00425-011-1542-5
Lee JC, Timasheff SN (1981) The stabilization of proteins by sucrose. J Biol Chem 256:7193–7201
Leslie SB, Israeli E, Lighthart B, Crowe JH, Crowe LM (1995) Trehalose and sucrose protect both membranes and proteins in intact bacteria during drying. Appl Environ Microbiol 61:3592–3597
Lunn JE (2002) Evolution of sucrose synthesis. Plant Physiol 128:1490–1500. doi:10.1104/pp. 010898
Lunn JE, Price GD, Furbank RT (1999) Cloning and expression of a prokaryotic sucrose-phosphate synthase gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 40:297–305
Mao X, Ma Q, Zhou C, Chen X, Zhang H, Yang J, Mao F, Lai W, Xu Y (2014) DOOR 2.0: presenting operons and their functions through dynamic and integrated views. Nucleic Acids Res 42:D654–D659. doi:10.1093/nar/gkt1048
Martínez-Noël GM, Cumino AC, Kolman Mde L, Salerno GL (2013) First evidence of sucrose biosynthesis by single cyanobacterial bimodular proteins. FEBS Lett 587:1669–1674. doi:10.1016/j.febslet.2013.04.012
Masada S, Kawase Y, Nagatoshi M, Oguchi Y, Terasaka K, Mizukami H (2007) An efficient chemoenzymatic production of small molecule glucosides with in situ UDP-glucose recycling. FEBS Lett 581:2562–2566. doi:10.1016/j.febslet.2007.04.074
Morell M, Copeland L (1985) Sucrose synthase of soybean nodules. Plant Physiol 78:149–154
Porchia AC, Salerno GL (1996) Sucrose biosynthesis in a prokaryotic organism: Presence of two sucrose-phosphate synthases in Anabaena with remarkable differences compared with the plant enzymes. Proc Natl Acad Sci U S A 93:13600–13604
Porchia AC, Curatti L, Salerno GL (1999) Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably different from the plant enzymes with respect to substrate affinity and amino-terminal sequence. Planta 210:34–40
Reed R (1986) Organic solute accumulation in osmotically stressed cyanobacteria. FEMS Microbiol Lett 39:51–56. doi:10.1016/0378-1097(86)90060-1
Reid SJ, Abratt VR (2005) Sucrose utilisation in bacteria: genetic organisation and regulation. Appl Microbiol Biotechnol 67:312–321. doi:10.1007/s00253-004-1885-y
Roby C, Cortès S, Gromova M, Le Bail J-L, Roberts JKM (2002) Sucrose cycling in heterotrophic plant cell metabolism: first step towards an experimental model. Mol Biol Rep 29:145–149
Ross HA, Davies HV (1992) Purification and Characterization of Sucrose Synthase from the Cotyledons of Vicia faba L. Plant Physiol 100:1008–1013
Salerno GL, Curatti L (2003) Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci 8:63–69. doi:10.1016/S1360-1385(02)00029-8
Sanchis J, Fernández L, Carballeira JD, Drone J, Gumulya Y, Höbenreich H, Kahakeaw D, Kille S, Lohmer R, Peyralans JJ-P, Podtetenieff J, Prasad S, Soni P, Taglieber A, Wu S, Zilly FE, Reetz MT (2008) Improved PCR method for the creation of saturation mutagenesis libraries in directed evolution: application to difficult-to-amplify templates. Appl Microbiol Biotechnol 81:387–397. doi:10.1007/s00253-008-1678-9
Sebková V, Unger C, Hardegger M, Sturm A (1995) Biochemical, physiological, and molecular characterization of sucrose synthase from Daucus carota. Plant Physiol 108:75–83
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi:10.1038/msb.2011.75
Son MH, Kim B-G, Kim DH, Jin M, Kim K, Ahn J-H (2009) Production of flavonoid o-glucoside using sucrose synthase and flavonoid o-glucosyltransferase fusion protein. J Microbiol Biotechnol 19:709–712
Subbaiah CC, Palaniappan A, Duncan K, Rhoads DM, Huber SC, Sachs MM (2006) Mitochondrial localization and putative signaling function of sucrose synthase in maize. J Biol Chem 281:15625–15635. doi:10.1074/jbc.M600355200
Taboada B, Ciria R, Martinez-Guerrero CE, Merino E (2012) ProOpDB: prokaryotic operon database. Nucleic Acids Res 40:D627–D631. doi:10.1093/nar/gkr1020
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tanase K, Yamaki S (2000) Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit. Plant Cell Physiol 41:408–414
Terasaka K, Mizutani Y, Nagatsu A, Mizukami H (2012) In situ UDP-glucose regeneration unravels diverse functions of plant secondary product glycosyltransferases. FEBS Lett 586:4344–4350. doi:10.1016/j.febslet.2012.10.045
Tsai C-Y (1974) Sucrose-udp glucosyltransferase of Zea mays endosperm. Phytochemistry 13:885–891. doi:10.1016/S0031-9422(00)91418-3
Vargas W, Cumino A, Salerno GL (2003) Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta 216:951–960. doi:10.1007/s00425-002-0943-x
Waffenschmidt S, Jaenicke L (1987) Assay of Reducing Sugars in the Nanomole Range with 2, 2 ’ -Bicinchoninate. Anal Biochem 165:337–340
Winter H, Huber SC (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Biochem Mol Biol 35:253–289. doi:10.1080/10409230008984165
Zervosen A, Römer U, Elling L (1998) Application of recombinant sucrose synthase-large scale synthesis of ADP-glucose. J Mol Catal B Enzym 5:25–28. doi:10.1016/S1381-1177(98)00040-X
Acknowledgments
The authors wish to thank the Special Research Fund (BOF) of Ghent University (MRP-project “Ghent Bio-Economy” and PhD-scholarship to MD), as well as the EC (FP7-project “SuSy”, grant agreement no. 613633) for financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 558 kb)
Rights and permissions
About this article
Cite this article
Diricks, M., De Bruyn, F., Van Daele, P. et al. Identification of sucrose synthase in nonphotosynthetic bacteria and characterization of the recombinant enzymes. Appl Microbiol Biotechnol 99, 8465–8474 (2015). https://doi.org/10.1007/s00253-015-6548-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6548-7