Abstract
Higher plants and cyanobacteria metabolize sucrose (Suc) by a similar set of enzymes. Suc synthase (SuS, A/UDP-glucose: d-fructose 2-α-d-glucosyl transferase) catalyzes a reversible reaction. However, it is in the cleavage of Suc that this enzyme plays an important role in vivo, providing sugar nucleotides for polysaccharide biosynthesis. In cyanobacteria, SuS occurrence has been reported in heterocyst-forming strains, where it was shown to be involved also in nitrogen fixation. We investigated the presence of sequences homologous to SuS-encoding genes (sus) in recently sequenced cyanobacterial genomes. In this work, we show for the first time the presence of SuS in unicellular cyanobacterium strains (Microcystis aeruginosa PCC 7806, Gloebacter violaceus PCC 7421, and Thermosynechococcus elongatus BP-1). After functional characterization of SuS encoding genes, we demonstrated an increase in their transcript levels after a salt treatment or hypoxic stress in M. aeruginosa and G. violaceus cells. Based on phylogenetic analysis and on the presence of sus homologs in the most recently radiated cyanobacterium strains, we propose that sus genes in unicellular cyanobacteria may have been acquired through horizontal gene transfer. Taken together, our data indicate that SuS acquisition by cyanobacteria might be related to open up new ecological niches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sucrose (Suc) is biosynthesized by plants, unicellular algae, and cyanobacteria (Salerno and Curatti 2003) as part of the carbon dioxide assimilation pathway in the biosphere. In plants, it is a central molecule being essential for the allocation of carbon resources for growth and respiration and participates in a regulatory network that coordinates metabolism and development (Winter and Huber 2000; Smeekens et al. 2010). Suc metabolism and its regulation have been widely studied in plants (Winter and Huber 2000; Koch 2004; Smeekens et al. 2010; Wind et al. 2010), but less so in prokaryotic organisms. Plants and cyanobacteria metabolize Suc by a similar set of enzymes (Salerno and Curatti 2003). Suc utilization requires the breakdown of the α1-β2-glycosidic bond, which depends on the action of Suc synthase (SuS, U/ADP-glucose: d-fructose 2-α-d-glucosyl transferase, EC 2.4.1.13), a readily reversible glucosyltransferase that yields a sugar nucleotide and fructose, or of invertases that irreversibly hydrolyse Suc to glucose and fructose.
In plants SuS plays a critical function in long-distance carbon allocation, stress responses, symbiotic interactions, and in Suc to polysaccharide interconversion, being related to starch, cellulose, and callose biosynthesis (Avigad and Dey 1997; Winter and Huber 2000; Haigler et al. 2001; Koch 2004; Loreti et al. 2005; Coleman et al. 2009; Fujii et al. 2010). Whereas in plants SuS is ubiquitous, its occurrence in cyanobacteria is not widespread (Salerno and Curatti 2003). To date SuS has been only reported in the filamentous heterocyst-forming strains Anabaena (Nostoc) sp. PCC 7119 and 7120, Anabaena variabilis, Nostoc punctiforme, and N. commune (Porchia et al. 1999; Curatti et al. 2000, 2006, 2008). Although SuS catalyzes a reversible reaction, it is in the cleavage of Suc that this enzyme plays an important role in vivo. It was shown that SuS is located in the photosynthetic vegetative cells (Curatti et al. 2006), where it provides sugar nucleotides for polysaccharide biosynthesis. The participation of SuS in the synthesis of glycogen was first demonstrated by modeling the Suc metabolic network in nitrogen-fixing Anabaena filaments. Glycogen synthesis occurs through ADP-glucose (ADP-Glc) donation of glucosyl for elongation of an α-1,4-glucosidic chain. It is mainly regulated at the level of ADP-Glc synthesis catalyzed by ADP-Glc pyrophosphorylase (AGPase, EC 2.7.7.27). The flux through AGPase, as calculated by metabolic simulation, is insufficient to supply the ADP-Glc needed for glycogen and Suc production through the sucrose-phosphate synthase pathway (Cumino et al. 2002). Therefore, a concomitant production of ADP-Glc should also be ascribed to Suc cleavage by SuS in the vegetative cells (Cumino et al. 2007). Additional experimental data support that SuS is involved in the Suc to polysaccharide conversion according to nutritional and environmental signals in Anabaena strains (Curatti et al. 2008).
The present study describes the first functional characterization of SuS-encoding genes (sus) in unicellular cyanobacteria. Phylogenetic analysis suggests that modern unicellular strains might have acquired sus by horizontal gene transfer. We also investigate the effect of salt and hypoxia on sus expression in different unicellular strains. Taken together our data suggest that SuS acquisition by cyanobacteria might be related to open up new ecological niches.
Materials and methods
Biological material and cell cultures
Microcystis aeruginosa PCC 7806 and Gloeobacter violaceus PCC 7421 were routinely grown in BG11 basal medium that contains NaNO3 (Rippka et al. 1979), under white fluorescent light (30 and 10 μE m−2 s−1, respectively) at 21 ± 2 and 28 ± 2°C, respectively. Anabaena (Nostoc) sp. PCC 7119 and its derivative mutant strains (susA − and susA +, formerly named LC30 and LC60, respectively) were cultured as previously described (Curatti et al. 2002). Cells from exponential-phase cultures were harvested by centrifugation at 2,500×g for 15 min, washed with 50 mM Hepes–NaOH buffer (pH 7.5), and either immediately processed or stored at −80°C until use. The effect of NaCl was studied in exponential-phase cells grown in BG11–KNO3. Salt was added up to a final concentration of 150 or 180 mM, in G. violaceus and M. aeruginosa cultures, respectively. After the salt treatment, cells were harvested, washed, and stored at −80°C.
Hypoxic treatments were carried out in flasks with rubber caps and the air space was flushed with N2 for 10 min. Cells were kept under this atmosphere for 3–6 h before being collected. Growth of Anabaena sp. PCC 7119 and the susA +, and susA − derivative mutant strains was followed in cultures under standard conditions and after hypoxia. Cell turbidity was determined by OD660 nm.
Escherichia coli DH5α and BL21(DE3):pLysS (Novagen) strains were grown in Luria–Bertani medium supplemented with 30 μg ml−1 chloramphenicol and 50 μg ml−1 carbenicillin at 37°C, and used for cloning and recombinant protein production, respectively.
Isolation, manipulation, and analysis of nucleic acids
Plasmids were isolated and modified according to standard protocols (Sambrook and Russell 2001). Genomic DNA from M. aeruginosa and G. violaceus were isolated as previously described (Curatti et al. 2002). Total DNA from T. elongatus BP-1 was kindly provided by Dr Diana Kirilovsky (CEA, Institut de Biologie et Technologies de Saclay and CNRS, France). Isolation and purification of RNA was carried out using the TRIZOL reagent (Gibco–BRL/Invitrogen). RNA quality was visualized after electrophoresis in 1% agarose gels and stained with ethidium bromide.
Cloning and expression of sus genes
Homologs to Anabaena sp. PCC 7120 susA sequence (Curatti et al. 2000) were obtained from public databases (http://www.ncbi.nlm.nih.gov). Open reading frames (orfs) were retrieved from the genomes of M. aeruginosa PCC 7806 (IPF_1565), G. violaceus PCC 7421 (gvip490), and T. elongatus BP-1 (tlr1047). DNA fragments were PCR-amplified using the primer pairs described in Table 1, supplemental material. Amplification products (Ma-susA, Gv-susA, and Te-susA) were ligated into the pRSET-A vector (Invitrogen, Carlsbad, CA) between the restriction sites BamHI and KpnI (for Ma-susA), or BamHI and EcoRI (for Gv-susA), or PvuII and HindIII (for Te-susA), obtaining the recombinant plasmids pR-Ma-susA, pR-Gv-susA, and pR-Te-susA, respectively. The identity of each construct was confirmed by DNA sequencing. E. coli BL21(DE3)pLysS cells were transformed with pR-Ma-susA, or pR-Gv-susA, or pR-Te-susA, to produce the recombinant proteins His6::Ma-SuS, His6::Gv-SuS, and His6::Te-SuS, respectively. SuS activity was measured in E. coli cells transformed with pR-Ma-susA, or pR-Gv-susA, or pR-Te-susA, or with the pRSET-A vector (control). After functional characterization of the three orfs, the sequences were deposited in the GenBank (JN618991, FJ457908, and FJ457909, as susA gene from M. aeruginosa, G. violaceus, and T. elongatus, respectively).
Protein extracts and purification
Protein extracts from M. aeruginosa and G. violaceus cells were carried out from cultures at exponential phase as described (Porchia and Salerno 1996). Extracts were desalted through Sephadex G-50 columns before enzyme activity assays (Cumino et al. 2001).
His6-tagged proteins were purified by Ni-affinity chromatography (Ni-NTA Purification System). Recombinant proteins were eluted from the column with a stepwise imidazole pH 7.0 gradient (50, 100, and 150 mM). Fractions with SuS activity were pooled and concentrated in an Amicon (Newtown, PA) ultrafiltration cell. Purified enzymes were stored at −20°C (Torres and Salerno 2007). The recombinant and partially purified proteins from cyanobacterium cells were used for product identification according to Porchia et al. (1999).
Enzyme assays and western blotting
SuS activity was assayed in the Suc cleavage direction by incubating at 30°C, in 50 μl total volume, 100 mM Suc, 5 mM XDP (ADP or UDP), 100 mM Hepes–NaOH (pH 6.5), and the protein fraction to be tested. Fructose was analyzed using Somogyi–Nelson reagents or enzymatically by incubation in a mixture containing ATP-Mg2+, NADP, and the auxiliary enzymes hexokinase, phosphoglucoseisomerase, and glucose-6-phosphate dehydrogenase (Pontis et al. 1981).
For immunoblotting analyses, polypeptides were separated by SDS-PAGE on 10% polyacrylamide gels and electroblotted onto a nitrocellulose membrane (HyBond C; Amersham) as described (Cumino et al. 2007). The membranes were then probed with rabbit polyclonal antibodies (anti-7119-SuS) raised against SuS from Anabaena sp. PCC 7119 (Porchia et al. 1999).
RT-PCR and northern blot assays
For RT-PCR analysis, total RNA (1 μg) treated with DNAse (RQI Rnase-free Dnase, Promega) was reverse-transcribed using MMLV (Moloney murine leukemia virus) reverse transcriptase (Promega) and specific reverse primers (Table 1, supplemental material). PCR reactions were run on a Mastercycler® epgradient (Eppendorf) for 24 cycles of 94°C (1 min), 65°C (30 s) and 72°C (45 s), and a single step at 72°C (5 min). Standardization reactions were carried out as described (Cumino et al. 2007). As a control to monitor the relative amount of total RNA used in each RT-PCR reaction (Zhu et al. 2001), aliquots of the same RNA were reverse-transcribed in parallel and subjected to 18 cycles of PCR with 16S-RNA (Sevilla et al. 2010) or rnpB (Vioque 1992) specific primer pairs, in the case of M. aeruginosa and G. violaceus, respectively.
For northern blots, total RNA (about 30 μg) was separated in a 1.2% agarose–formaldehyde denaturing gel and immobilized in positively charged nylon membranes (0.45 μm, Nytran, Schleicher & Schuell, Keene, NH) by alkaline passive transference. A 449-bp probe for Gv-susA (from nucleotides 648 to 1,097) was generated and labeled with [α-32P]dCTP by the random primer extension system (NEN Life Science Products, Boston, MA). Prehybridization, hybridization, and exposure conditions were carried out as described by Torres and Salerno (2007).
Sequence data and phylogenetic analysis
Protein sequences with similarity to Anabaena (also named Nostoc) sp. PCC 7120 SuS-A (E value ≤10−20) were retrieved using BLAST and the nonredundant protein databases of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), the Kazusa DNA Research Institute, and the Department of Energy Joint Genome Institute (http://www.jgi.doe.gov). The sequences were aligned with Clustal W and domain organization was analyzed using InterPro database (Hunter et al. 2009) and PFAM (Finn et al. 2008). Dendrograms were constructed using the neighbor-joining method and maximum likelihood from the MEGA5 software (Tamura et al. 2011). Nonparametric bootstrapping (1,000 replicates) was used to assess tree branching support.
Results
Presence of SuS in unicellular cyanobacteria
SuS enzymes have only been identified and characterized from plants and filamentous nitrogen-fixing cyanobacteria (Salerno and Curatti 2003; Curatti et al. 2008). However, a search for homologous sequences to SuS encoding genes (sus) in cyanobacterium genomes available to date in public databases (65 in total) revealed that sus homologs are also present in unicellular strains. A BLASTp search using the deduced amino-acid sequence of Anabaena (Nostoc) sp. PCC 7120 SuS-A protein (7120-SuS) as entry allowed to retrieve homologs from 6 among 49 (12.5%) unicellular strain genomes (Table 2, supplemental material). For comparison, homologous sequences to 7120-SuS were found in 10 among 16 (62.5%) genomes of filamentous cyanobacteria, which share 71–73% identity with 7120-SuS, and 40–48% with SuS proteins from plants. Particularly, in heterocyst-forming strains, homologs to SuS encoding genes could be retrieved from the genomes of Nostoc sp. PCC 7120, N. punctiforme PCC 73102, N. azollae 0708, A. variabilis ATCC 29413, and Nodularia spumigena CCY 9414, but not from that of Cylindrospermopsis raciborskii CS-505.
The two characteristic motifs within the glucosyltransferases related to Suc metabolism are completely conserved in all the retrieved sequences (Cumino et al. 2002) (Fig. 1, supplemental material). A difference between cyanobacterium and plant SuS sequences was found in the amino-terminal region (about 100-amino acid length) that differs between plants and cyanobacteria, but it is highly conserved among each group.
Functional identification of SuS proteins in unicellular cyanobacterium strains
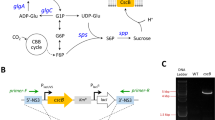
To investigate whether the putative sus genes of unicellular strains encoded SuS proteins, we characterized the orfs present in the genomes of G. violaceus PCC 7421 (Gv), M. aeruginosa PCC 7806 (Ma), and T. elongatus BP-1 (Te) by heterologous expression, resulting functional sus genes (named Ma-susA, Gv-susA, and Te-susA). The respective recombinant His6-tagged proteins were purified, and their biochemical and immunological properties were analyzed. His6::Gv-SuS, His6::Ma-SuS, and His6::Te-SuS are able to catalyze the cleavage of Suc in the presence of ADP or UDP to yield fructose and ADP-Glc or UDP-Glc (Fig. 1a) and have similar optimum pHs (6.9 ± 0.3) for this reaction, in agreement with those obtained for Anabaena SuS (Porchia et al. 1999). The three proteins were immunodetected by anti-7119-SuS polyclonal antibodies (Fig. 1b). These results confirm the presence of SuS proteins in unicellular cyanobacteria.
Biochemical and immunological characterization of His6-tagged recombinant SuS from three unicellular cyanobacterial strains. a SuS activity assayed in purified recombinant proteins: His6::Ma-SuS (from M. aeruginosa, Ma), His6::Gv-SuS (from G. violaceus, Gv) and His6::Te-SuS (from T. elongatus, Te). Activity was determined in the Suc cleavage direction using different nucleotides as substrates. The values represent the mean ± SD of two independent experiments. No SuS activity was measured in E. coli BL21(DE3):pLysS cells transformed with the empty pRSET-A vector (control). b Western blot analysis. Immunodetection of His6-SuS polypeptides in non-induced (NI) or IPTG-induced (I) E. coli cells using anti-7119-SuS polyclonal antibodies
Phylogenetic analysis
A dendrogram was constructed after multiple alignments of the full-length sequences of SuS homologs from cyanobacteria, plants, and bacteria. As shown in Fig. 2, cyanobacterium and plant SuSs cluster in two separated monophyletic groups. The plant SuS clade has been analyzed previously (Baud et al. 2004; Bieniawska et al. 2007; Jayashree et al. 2008). The cyanobacterial SuS group is subdivided into three subclades, which contain sequences of (i) filamentous strain SuSs, (ii) unicellular strain SuSs, and (iii) homologs corresponding to another glucosyltransferase (SuS-B) that is a close relative to the SuS proteins, but with different biochemical function(s) and/or properties (Curatti et al. 2002, 2006). From the two Cyanothece SuS sequences (Table 2, supplemental material), one is grouped with the unicellular strains (Cy7425-SuS) but the other is among the filamentous strains (Cy7424-SuS). As previously described, proteobacterial SuS homolgs are more closely related to the plant proteins than to the cyanobacterial SuSs (Salerno and Curatti 2003).
Phylogenetic analysis of SuS proteins. Radial representation of consensus maximum likelihood tree obtained from 1,000 replicates using the JTT + F+ Gamma model after sequence alignment of deduced amino-acid SuS sequences using a BLOSSUM 62 matrix. Cyanobacterial SuS corresponding to unicellular (white box) and filamentous (gray box) strains are indicated. An asterisk denotes a SuS protein functionally characterized (those described in this study are in bold letters). Homologous sequences to SuS-B, which do not have SuS activity, were also included. Qualitatively similar tree topologies were observed when using a neighbor-joining algorithm (not shown). Evolutionary analyses were conducted in MEGA5 using sequences from: Nodularia spumigena CCY9414 (Nspu-SuS, ZP_01631755.1, ZP_01629753.1), Microcoleus chthonoplastes PCC 7420 (Mcht-SuS, ZP_05029917.1), Cyanothece sp. PCC 7425 (Cy7425-SuS, YP_002484594.1, YP_002482482.1), Cyanothece sp. PCC 7424 (Cy7424-SuS, YP_002379025.1), Anabaena (Nostoc) sp. PCC 7120 (7120-SuS, NP_489025.1, NP_485102.1), Acaryochloris marina BIC11017 (Amar-SuS, YP_001520299.1), Gloeobacter violaceus PCC 7421 (Gv-SuS, NP_926553.1), Anabaena variabilis ATCC 29413 (Avar-SuS, YP_324253.1, YP_322796.1), Nostoc punctiforme PCC 73102, (Npun–SuS, YP_001868239.1, YP_001865476), Thermosynechococcus elongatus BP-1 (Te-SuS, NP_681838.1), Nostoc azollae 0708 (Nazo-SuS, ZP_03768709.1), Arthrospira maxima CS-328 (Amax-SuS, ZP_03271682.1), Microcystis aeruginosa PCC 7806 (Ma-SuS, CAO88728.1), Zea mays-SuS1 (Zmays-1, NP_001105194.1), Zea mays-SuS2 (Zmays-2, NP_001105194.1), Oryza sativa-SuS1 (Osat-1, NP_001050064.1), Oryza sativa-SuS2 (Osat-2, ABL74568.1), Oryza sativa-SuS3 (Osat-3, AAC41682.1), Citrus unshui-SuS1 (Cuns-1, BAA88904.1), Beta vulgaris-SuS1 (Bvul-1, ABR87939.1), Beta vulgaris-SuS2 (Bvul-2, AAK65960.1), Solanum tuberosum-SuS (Stub, AAO67719.1), Pirus pyrifolia-SuS1 (Ppir-1, BAB20799.1), Pisum sativum-SuS2 (Psat-2, O24301.1), Arabidopsis thaliana-SuS1 (Atha-1, NP_197583.1), Arabidopsis thaliana-SuS2 (Atha-2, NP_199730.1), Arabidopsis thaliana-SuS3 (Atha-3, NP_192137.1), Arabidopsis thaliana-SuS4 (Atha-4, NP_566865.2), Arabidopsis thaliana-SuS5 (Atha-5, NP_198534.2), Arabidopsis thaliana-SuS6 (Atha-6, NP_177480.1), Daucus carota-SuS1 (Dcar-1, P49035.1), Eucaliptus grandis-SuS (Egran, ABB53601.1), Medicago truncatula-SuS (Mtru, CAB40794.1), Medicago sativa-SuS (Msat, ABP88869.1), Glycine max-SuS (Gmax, P13708.2), Vicia faba-SuS (Vfab, P31926.1), Solanum tuberosum-SuS2 (Stub-2, AAO34668.1), Solanum lycopersicum-SuS (Slyc, CAA09593.1), Thioalkalivibrio sp. HL-EbGR7 (Thio, YP_002512263.1), Nitrosomonas eutropha C91 (Neut, ABI59334.1), Nitrosomonas europaea ATCC 19718 (Neur, NP_841269.1), Nitrosococcus halophilus Nc4 (Nhal, YP_003529337.1), Nitrosococcus oceani ATCC 19707 (Noce, ABA59509.1), Nitrosococcus watsonii C-113 (Nwat, YP_003762161.1), Desulfurispirillum indicum S5 (Dind, YP_004113670.1), Nitrosospira multiformis ATCC 25196 (Nmul, YP_412950.1), Acidithiobacillus ferrooxidans ATCC 23270 (Afer-23270, YP_002425980.1), Acidithiobacillus ferrooxidans ATCC 53993 (Afer-53993, YP_002219713.1), Acidithiobacillus ferrivorans SS3 (Afer-SS3, ZP_08488631.1), Denitrovibrio acetiphilus DSM 12809 (Dace, YP_003505650.1)
Effect of salt stress and hypoxia on SuS expression in G. violaceus and M. aeruginosa
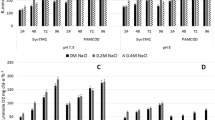
In heterocyst-forming strains (such as Anabaena sp. PCC 7119, 7120, N. punctiforme, and N. ellipsosporum) we showed that not only Suc synthesis increases but also SuS expression is higher in the presence of NaCl (Salerno, unpublished). Therefore, we decided to investigate the effect of salt on SuS gene expression in cells of unicellular strains. Exponential-phase cultures of G. violaceus and M. aeruginosa were added with NaCl up to 24 h and afterwards the effect of salt was reverted by transferring the cells to BG11–KNO3. Indeed, SuS activity increased after salt addition and the effect was reverted when cells were transferred to the basal medium (Fig. 3a, b). A similar reversion pattern was obtained when transcript levels were determined for both strains (Fig. 3c, d).
Effect of NaCl on susA expression in M. aeruginosa (Ma) and G. violaceus (Gv) cells. a, b SuS activity was assayed in protein extracts from cells cultured in BG11–KNO3 (C, control) or from cells harvested 24 h after the addition of NaCl (24 h). For reversion analysis, 24-h salt-stressed cells were transferred to BG11–KNO3 basal medium for 24 h (R). c Effect of salt on susA transcript level in G. violaceus cells analyzed by Northern blot. Total RNA was prepared from control cells grown in BG11–KNO3 basal medium (c) or from 24 h treated cells with NaCl (24 h) or from 24-h salt stressed cells transferred to BG11–KNO3 basal medium for 24 h (R). For loading control, RNA was stained with ethidium bromide (bottom panel). d Effect of salt on susA transcript level from M. aeruginosa cells analyzed by RT-PCR. Total RNA was purified from cells cultured in BG11–KNO3 (0) or from cells harvested 0.25, 0.5, 2, 10, and 24 h after the addition of NaCl. For reversion analysis, 24-h salt-stressed cells were transferred to BG11–KNO3 basal medium for 24 h (R). For loading control, amplification products corresponding to 16S rRNA were stained with ethidium bromide (bottom panel)
Following recent reports in plants on the importance of SuS expression in roots during and after exposure to hypoxia/anoxia conditions (Baud et al. 2004; Bieniawska et al. 2007, Subbaiah et al. 2007), we analyzed the effect of hypoxia on SuS gene expression in unicellular cyanobacterial strains. Transcript levels increased after 6 h of treatment either in cells of M. aeruginosa (Fig. 4a) or G. violaceus (Fig. 4b). The effect was reverted when cells were transferred to standard growth conditions for 24 h. To compare, we also investigate how hypoxia affected a heterocyst-forming strain such as Anabaena sp. PCC 7119. As shown in Fig. 4c, the increase in SuS transcript levels is not limited to unicellular cyanobacteria. We took advantage that we had a set of derivative mutants of that filamentous strain, with genetic manipulation of the SuS encoding gene (susA − and susA +, lacking or over-expressing SuS, respectively) (Curatti et al. 2002, 2006). We followed the growth of the wild-type (7119) and the two mutants after a hypoxic treatment. Under standard growth conditions the three strains showed similar growth rates and the onset of hypoxia resulted in a similar arrest of growth (Fig. 5, Fig. 2, supplemental material). However, when the oxygen supply was restored, 7119 and susA + cells immediately resume growth (Fig. 5a, c) but cells lacking SuS showed a 24-h delay in recovering growth (Fig. 5b).
Effect of hypoxia on cyanobacterial susA expression a–c Determination of susA transcript steady-state levels by RT-PCR. Aliquots of total RNA (2 μg) from M. aeruginosa (a), G. violaceus (b) and Anabaena sp. PCC 7119 (c) cells cultivated in BG11 basal medium (lane 0), or from cells harvested at different times (3 or 6 h) after a hypoxic treatment (lanes 3 and 6) were used in each RT-PCR reaction. For reversion analysis, 6-h hypoxia-treated cells were transferred to standard growth conditions for 24 h (R). For loading control, the amplification products corresponding to 16S rRNA in the cases of M. aeruginosa and G. violaceus, or to rnpB, in the case of Anabaena, are shown in the bottom panels. The PCR products were electrophoresed on 2% agarose gels and visualized after staining with ethidium bromide
Effect of hypoxia on the growth of Anabaena sp. PCC 7119 (a), susA − (b) and susA + (c) derivative mutant strains. Control cells were cultivated in BG11 under standard conditions (filled squares). Hypoxia treatment was started at time 0 for 72 h (open circles). Arrows indicate the time when hypoxic cells were transferred to standard oxygen conditions (filled circles)
Discussion
The function of SuS, a glycosyltransferase belonging to the GT4 family (Henrissat et al. 2001), as a Suc cleavage enzyme has been well established in plants and filamentous diazotrophic cyanobacteria (Winter and Huber 2000; Salerno and Curatti 2003; Curatti et al. 2008). Particularly, in these microorganisms it has been associated with the carbon flux during nitrogen fixation and with polysaccharide accumulation (Curatti et al. 2002, 2006, 2008; Cumino et al. 2007). In fact, homologous sequences to sus are found in most of the sequenced genomes of heterocyst-forming cyanobacteria, but only in a few ones of unicellular strains (Table 2, supplemental material). In this study we report the first functional identification of genes encoding SuS proteins in unicellular strains (M. aeruginosa, G. violaceus, and T. elongatus), contributing not only to the knowledge of the enzymes involved in Suc metabolism in modern cyanobacteria but also to give a better insight into the origin and evolution of this central pathway in oxygenic photosynthetic organisms.
The six unicellular cyanobacteria harboring sus homologs in their genomes (Table 2, supplemental material) are strains with unusual characteristics. T. elongatus BP-1 is a uniquely thermophilic unicellular cyanobacterium (55°C optimal growth temperature) that was isolated from a hot spring and contains a large number of heat shock proteins as well as an unusually large number of type II intron sequences (Nakamura et al. 2002; Onai et al. 2004; Kos et al. 2008). G. violaceus PCC 7421 has been isolated from calcareous rocks and tends to grow in colonies surrounded by a sticky mucous sac that plays a role in adhesion (Schneider and Jürgens 1991). It possesses a number of unique characteristics such as the absence of thylakoids. The machinery for photosynthesis is located in the cytoplasmic membrane, where the photosynthetic electron transfer system should co-exist with a respiratory system by sharing some components (Nakamura et al. 2003). The cyanobacterium Acaryochloris marina MBIC11017 has been isolated from the Prochloron-dominated colonial ascidian Lissoclinum patella off a tropical coast. It was found to be the only oxygenic photoautotroph that uses chlorophyll d as the predominant photosynthetic pigment (Swingley et al. 2008). Notably, the occurrence of sus homologous sequences seems not to be ubiquitous in strains of the genera Cyanothece and Microcystis. The homologs can only be retrieved from genomes of only two (PCC 7424 and PCC 7425) out of six Cyanothece strains and from one (PCC 7806) out of two M. aeruginosa strains. Cyanothece strains were isolated from rice fields and have the rare ability to anaerobically fix nitrogen (Porta et al. 2000). Finally, M. aeruginosa is a freshwater cyanobacterium distributed worldwide and involved in numerous proliferation events (blooms) in stratified water bodies (Visser et al. 2005). The physical and chemical microenvironments and growth conditions for M. aeruginosa strains are significantly different from that of the bulk (Ploug 2008).
Although SuS is present in G. violaceus PCC 7421, which is thought to be the most deeply rooted cyanobacterium, its occurrence in other strains (unicellular and filamentous ones) is not consistent with the phylogeny of cyanobacteria (Figs. 2, 6). This phylogenetic incongruence points to sus gene as an obvious candidate that might have been acquired by horizontal gene transfer. The fixation and long-term persistence of this acquisition suggest that SuS might confer a selective advantage on the recipient and extant cyanobacterial strains (Koonin et al. 2001).
Hypothetical evolutionary pathway from a common ancestral glucosyltransferase (GTD)-like domain to modern cyanobacterial and plant SuS. The phylogenetic relationships among species are depicted according to rRNA sequence analysis (Larsson et al. 2011). A GTD-like primordial domain (light orange bars) might have given rise to a hypothetical common-ancestral sucrose–phosphate synthase gene which might encode a SPS-like protein (red bar), involved in the Suc biosynthesis pathway (Salerno and Curatti 2003). Duplications of GTD during cyanobacterial diversification might have originated sus genes, coding for SuS proteins [GTD domain (red bar) with an amino-terminal extension (yellow bar)] in filamentous heterocyst-forming cyanobacteria. The presence of SuS homologs in a few unicellular extant cyanobacteria (Gv-SuS, Te-SuS and Ma-SuS, characterized in this study) points up to an acquisition through horizontal gene transfer. Plant SuS are depicted as a GTD domain with a different amino-terminal region (orange bar). A few proteobacteria are likely to have acquired SuS (with still unknown biochemical function) laterally from cyanobacteria and/or from plants
The accumulation of Suc in photosynthetic organisms in response to abiotic stress is likely to be the consequence of the fate of photosynthetic carbon after growth arrest. In cyanobacteria, Suc was identified as the main compatible osmolite in many fresh-water strains, and as a minor or transient part of the total compatible solute pool in more halotolerant strains that accumulate also other organic compounds (Salerno et al. 2004; Desplats et al. 2005; Hagemann 2010; Klähn and Hagemann 2011). However, Suc net accumulation, due to an increase in the Suc biosynthesis enzymes expression, is also accompanied by an enhancement in Suc degradation by SuS activity, either in filamentous (Salerno, unpublished) or in unicellular strains (this study). Therefore, a Suc cycling mechanism may be operating in cyanobacterial salt-treated cells. These sugar cycles were proposed in plants to allow a pathway’s net flux to respond to factors controlling respiration, maintaining osmotic potential, controlling sugar accumulation, and promoting sugar signaling (Rohwer and Botha 2001; Roby et al. 2002). However, although it has been widely reported in plants, its mechanisms and functions remain poorly understood (Alonso et al. 2005).
In this study, we show the first evidence of the involvement of SuS in cyanobacterial cells submitted to hypoxia. SuS transcript levels increased in M. aeruginosa and G. violaceus cells under hypoxic conditions (Fig. 4a, b). We may speculate that the presence of SuS would give any advantage to these strains, since both M. aeruginosa and G. violaceus often grow in colonies, and the accumulation of very thin layers of biomass can lead to significant gradients of oxygen within the cell aggregates (Ibelings and Maberly 1998). However, the effect of hypoxia on sus expression is not restricted to unicellular strains since similar results were obtained with the filamentous strain Anabaena sp. PCC 7119 (Fig. 4c). The possibility of submitting cultures of an Anabaena derivative mutant lacking SuS activity (susA −) to a hypoxic treatment allowed us to show that SuS is important in the recovery of growth after the stress. This indicates that SuS is involved in Suc utilization in metabolically highly active cells where ATP synthesis may be limited by low oxygen tension. In plants, a remarkable increase in Suc cleavage by SuS after the onset of hypoxia has been well documented for a variety of species and organs including cereal seeds (Guglielminetti et al. 1997), rice seedlings, and maize (Zeng et al. 1998), potato (Biemelt et al. 1999), wheat (Albrecht and Mustroph 2003), and Arabidopsis (Baud et al. 2004; Klok et al. 2002; Bieniawska et al. 2007) roots. Our results support the conclusion that SuS is involved in the response to low oxygen conditions from cyanobacteria to plants.
Finally, we propose a model on SuS origin and evolution in cyanobacteria (Fig. 6), which seems to be rather intricate. SuS might have been acquired (and lost?) several times during evolution by different linages. We hypothesize that the conservation of SuS might allow coping with different adverse environmental conditions, opening up new ecological niches.
Abbreviations
- Gv :
-
Gloebacter violaceus
- Ma :
-
Microcystis aeruginosa
- Suc:
-
Sucrose
- SuS:
-
Sucrose synthase
- sus, susA :
-
Sucrose synthase encoding gene
- Te :
-
Thermosynechoccocus elongatus
References
Albrecht G, Mustroph A (2003) Localization of sucrose synthase in wheat roots: increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta 217:252–260
Alonso AP, Vigeolas H, Raymond P, Rolin D, Dieuaide-Noubhani M (2005) A new substrate cycle in plants. Evidence for a high glucose-phosphate-to-glucose turnover from in vivo steady-state and pulse-labeling experiments with [13C]glucose and [14C]glucose. Plant Physiol 138:2220–2232
Avigad G, Dey PM (1997) Carbohydrate metabolism: storage carbohydrate. In Plant Biochemistry (Dey PM, Harborne JB eds) pp 143–204, Academic Press, London
Baud S, Vaultier MN, Rochat C (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55:397–409
Biemelt S, Hajirezaei MR, Melzer M, Albrecht G, Sonnewald U (1999) Sucrose synthase activity does not restrict glycolysis in roots of transgenic potato plants under hypoxic conditions. Planta 210:41–49
Bieniawska Z, Paul Barratt DH, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49:810–828
Coleman HD, Yan J, Mansfield SD (2009) Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci USA 106:13118–13123
Cumino A, Ekeroth C, Salerno GL (2001) Sucrose-phosphate phosphatase from Anabaena sp. strain PCC 7120: isolation of the protein and gene revealed significant structural differences from the higher-plant enzyme. Planta 214:250–256
Cumino A, Curatti L, Giarrocco L, Salerno GL (2002) Sucrose metabolism: Anabaena sucrose-phosphate synthase and sucrose-phosphate phosphatase define minimal functional domains shuffled during evolution. FEBS Lett 517:19–23
Cumino AC, Marcozzi C, Barreiro R, Salerno GL (2007) Carbon cycling in Anabaena sp. PCC 7120. Sucrose synthesis in the heterocysts and possible role in nitrogen fixation. Plant Physiol 143:1385–1397
Curatti L, Porchia AC, Herrera-Estrella L, Salerno GL (2000) A prokaryotic sucrose synthase gene (susA) isolated from a filamentous nitrogen-fixing cyanobacterium encodes a protein similar to those of plants. Planta 211:729–735
Curatti L, Flores E, Salerno G (2002) Sucrose is involved in the diazotrophic metabolism of the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett 513:175–178
Curatti L, Giarrocco L, Salerno GL (2006) Sucrose synthase and RuBisCo expression is similarly regulated by the nitrogen source in the nitrogen-fixing cyanobacterium Anabaena sp. Planta 223:891–900
Curatti L, Giarrocco LE, Cumino AC, Salerno GL (2008) Sucrose synthase is involved in the conversion of sucrose to polysaccharides in filamentous nitrogen-fixing cyanobacteria. Planta 228:617–625
Desplats P, Folco E, Salerno GL (2005) Sucrose may play an additional role to that of an osmolyte in Synechocystis sp. PCC 6803 salt-shocked cells. Plant Physiol Biochem 43:133–138
Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A (2008) The Pfam protein families database. Nucleic Ac Res 36:D281–D288
Fujii S, Hayashi T, Mizuno K (2010) Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant Cell Physiol 51:294–301
Guglielminetti L, Wu Y, Boschi E, Yamaguchi J, Favati A, Vergara M, Perata P, Alpi A (1997) Effects of anoxia on sucrose degrading enzymes in cereal seeds. J Plant Phys 150:251–258
Hagemann M (2010) Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35:87–123
Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47:29–51
Henrissat B, Coutinho PM, Davies GJ (2001) A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47:55–72
Hunter S, Apweiler R, Attwood TK et al (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37:D211–D215
Ibelings BW, Maberly SC (1998) Photoinhibition and the availability of inorganic carbon restrict photosynthesis by surface blooms of cyanobacteria. Limnol Oceanogr 43:408–419
Jayashree B, Pradeep R, Anil K, Gopal B (2008) Correlation between the sucrose synthase protein subfamilies, variations in structure and expression in stress-derived expressed sequence tag datasets. J Proteomics Bioinform 1:408–423
Klähn S, Hagemann M (2011) Compatible solute biosynthesis in cyanobacteria. Env Microbiol 13:551–562
Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES (2002) Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14:2481–2494
Koch K (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7:235–246
Koonin EV, Makarova KS, Aravind L (2001) Horizontal Gene Transfer in Prokaryotes: Quantification and Classification 1. Annu Rev Microbiol 55:709–742
Kos PB, Deak Z, Cheregi O, Vass I (2008) Differential regulation of psbA and psbD gene expression, and the role of the different D1 protein copies in the cyanobacterium Thermosynechococcus elongatus BP-1. Biochim Biophys Acta 1777:74–83
Larsson J, Nylander J, Bergman B (2011) Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evolutionary Biology 11:187
Loreti E, Poggi A, Novi G, Alpi A, Perata P (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137:1130–1138
Nakamura Y, Kaneko T, Sato S et al (2002) Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res 9:135–148
Nakamura Y, Kaneko T, Sato S et al (2003) Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res 10:137–145
Onai K, Morishita M, Itoh S, Okamoto K, Ishiura M (2004) Circadian rhythms in the thermophilic cyanobacterium Thermosynechococcus elongatus: compensation of period length over a wide temperature range. J Bacteriol 186:4972–4977
Ploug H (2008) Cyanobacterial surface blooms formed by Aphanizomenon sp. and Nodularia spumigena in the Baltic Sea: Small-scale fluxes, pH, and oxygen microenvironments. Limnol Oceanogr 53:914–921
Pontis HG, Babio JR, Salerno G (1981) Reversible unidirectional inhibition of sucrose synthase activity by disulfides. Proc Natl Acad Sci USA 78:6667–6669
Porchia AC, Salerno GL (1996) Sucrose biosynthesis in a prokaryotic organism: Presence of two sucrose-phosphate synthases in Anabaena with remarkable differences compared with the plant enzymes. Proc Natl Acad Sci USA 93:13600–13604
Porchia AC, Curatti L, Salerno GL (1999) Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably different from the plant enzymes with respect to substrate affinity and amino-terminal sequence. Planta 210:34–40
Porta D, Rippka R, Hernández-Mariné M (2000) Unusual ultrastructural features in three strains of Cyanothece (cyanobacteria). Arch Microbiol 173:154–163
Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61
Roby C, Cortes S, Gromova M, Le Bail JL, Roberts JK (2002) Sucrose cycling in heterotrophic plant cell metabolism: first step towards an experimental model. Mol Biol Rep 29:145–149
Rohwer JM, Botha FC (2001) Analysis of sucrose accumulation in the sugar cane culm on the basis of in vitro kinetic data. Biochem J 358:437–445
Salerno GL, Curatti L (2003) Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci 8:63–69
Salerno GL, Porchia AC, Vargas WA, Abdian PL (2004) Fructose-containing oligosaccharides: novel compatible solutes in Anabaena cells exposed to salt stress. Plant Sci 167:1003–1008
Sambrook J, Russell DW (2001) Molecular Cloning. A Laboratory Manual, 3rd. edition edn. Cold Spring Harbor Lab. Press, New York
Schneider S, Jürgens UJ (1991) Cell wall and sheath constituents of the cyanobacterium Gloeobacter violaceus. Arch Microbiol 156:312–318
Sevilla E, Martin-Luna B, Vela L, Bes MT, Peleato ML, Fillat MF (2010) Microcystin-LR synthesis as response to nitrogen: transcriptional analysis of the mcyD gene in Microcystis aeruginosa PCC 7806. Ecotoxicology 19:1167–1173
Smeekens S, Ma J, Hanson J, Rolland F (2010) Sugar signals and molecular networks controlling plant growth. Curr Op Plant Biol 13:273–278
Subbaiah CC, Huber SC, Sachs MM, Rhoads D (2007) Sucrose synthase: expanding protein function. Plant Signal Behav 2:28–29
Swingley WD, Chen M, Cheung PC, Conrad AL, Dejesa LC, Hao J, Honchak BM, Karbach LE, Kurdoglu A, Lahiri S (2008) Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci USA 105:2005–2010
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol doi:10.1093/molbev/msr121. MBE Advance Access published August 18, 2011
Torres LL, Salerno GL (2007) A metabolic pathway leading to mannosylfructose biosynthesis in Agrobacterium tumefaciens uncovers a family of mannosyltransferases. Proc Natl Acad Sci USA 104:14318–14323
Vioque A (1992) Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res 20:6331–6337
Visser PM, Ibelings BW, Mur LR, Walsby AE (2005) The ecophysiology of the harmful cyanobacterium Microcystis. In: Huisman J, Matthijs H, Visser P (eds) Harmful Cyanobacteria. Springer, Netherlands, pp 109–142
Wind J, Smeekens S, Hanson J (2010) Sucrose: Metabolite and signaling molecule. Phytochem 71:610–1614
Winter H, Huber SC (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Biochem Mol 35:253–289
Zeng Y, Wu Y, Avigne WT, Koch KE (1998) Differential regulation of sugar-sensitive sucrose synthases by hypoxia and anoxia indicate complementary transcriptional and posttranscriptional responses. Plant Physiol 116:1573–1583
Zhu J, Jager K, Black T, Zarka K, Koksharova O, Wolk CP (2001) HcwA, an autolysin, is required for heterocyst maturation in Anabaena sp. strain PCC 7120. J Bacteriol 183:6841–6851
Acknowledgments
We are very thankful to H.G. Pontis for insightful reading of the manuscript and for helpful discussions, and C. Fernández and M. Vidal for technical assistance. This research was funded by Grants from CONICET (PIP 134), Universidad Nacional de Mar del Plata, and FIBA.
Author information
Authors and Affiliations
Corresponding author
Additional information
M. A. Kolman, L. L. Torres contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kolman, M.A., Torres, L.L., Martin, M.L. et al. Sucrose synthase in unicellular cyanobacteria and its relationship with salt and hypoxic stress. Planta 235, 955–964 (2012). https://doi.org/10.1007/s00425-011-1542-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-011-1542-5